Abstract
Spatial and temporal subcellular localization plays critical roles in regulating protein function. Cten (C-terminal tensin like) is a member of the tensin family. Cten recruits signaling molecules, such as DLC1, to focal adhesions, modulates homeostasis of receptor tyrosine kinases, including EGFR and c-Met, and promotes cell migration. These functions are likely controlled by Cten localization at focal adhesions and/or in the cytoplasm. In addition, Cten has been detected in the nucleus by which mechanism is unknown. To this end, we have examined the distribution of Cten in various cell lines, determined primary sequence requirements for its nuclear and focal adhesion localizations, and analyzed potential roles of nuclear Cten. Our results show that a proportion of Cten translocates to nuclei in cancer cell lines and that nuclear exporting of Cten is a CRM1-dependent process. A nuclear localization sequence and a nuclear export sequence are identified within Cten. In addition, like other tensins, Cten contains two independent focal adhesion binding sites. Although further expression of recombinant Cten showed no effect on cancer cell proliferation, silencing of Cten significantly reduced cell growth. Furthermore, expression of Cten mutants either with defective nuclear export sequence or tagged with SV40 nuclear localization sequence promoted cell growth. These results suggest that nuclear Cten contributes to cancer cell proliferation. Our findings identify a molecular mechanism for regulating Cten protein trafficking in mammalian cells and provide new insights into the dynamics of focal adhesion complexes in health and disease.
Keywords: Cten, tensin, focal adhesion, nucleus
1. Introduction
Protein function and activity are highly regulated in cells. They are initially controlled by their expression levels through transcriptional/translational regulation and then by their post-translational modifications, such as phosphorylation, acetylation, glycosylation, or ubiquitination. Their binding partners also dictate their functions and activities. In many cases, accurate spatial and temporal compartmentalization of proteins in a cell adds another layer of control. All these events need to be well regulated and balanced to maintain normal cell homeostasis and function.
Cten (C-terminal tensin like) is a member of the tensin family that resides at focal adhesion sites [1, 2], which are the transmembrane junctions between the extracellular matrix and the actin cytoskeleton. Tensins interact with phosphotyrosine-containing proteins through their SH2 (Src Homologous 2) domains and bind to βintegrin tails via their PTB (PhosphoTyrosine Binding) domains [1, 2]. Tensin-1 (TNS1) binds to actin filaments in multiple ways [3] and links the actin cytoskeleton to integrin receptors at focal adhesions. Tensin-2 (TNS2) and Tensin-3 (TNS3) share conserved N- and C-terminal regions with TNS1 but their central regions are very divergent [4, 5]. On the other hand, Cten is a smaller protein and only shares its C-terminal SH2 and PTB domains with other tensins [6]. The expression pattern of Cten is also unique in that it is restricted in the normal prostate and placenta [6]. Nonetheless, Cten is overexpressed in many types of cancer, including lung, breast, colon, pancreas, and skin cancers [7]. As a member of the tensin family, Cten is mainly localized to the cytoplasmic side of focal adhesion [6]. Cytoplasmic Cten binds and recruits DLC1, a negative regulator of Rho GTPase, to the focal adhesion that is critical for DLC1’s function [8, 9]. In addition, Cten prolongs signaling cascades mediated by receptor tyrosine kinases. Up-regulated Cten directly binds to and stabilizes active c-Met [10]. It also protects active EGFR from ubiquitination and degradation by binding to c-Cbl [11], the E3 ligase that is responsible for EGFR ubiquitination and degradation. Furthermore, up-regulated Cten promotes cell migration in various cell types [11–17]. These functions of Cten are likely operated at focal adhesions and/or in the cytoplasm.
Cten is unexpectedly detected in the nucleus of various cancer cell lines [18]. It binds to β-catenin in the nucleus and contributes to the tumorigenicity of colon cancer cells [18]. It has been suggested that higher nuclear Cten levels are associated with poor survival potential for colon cancer patients [19]. However, the mechanism of nuclear localization of Cten is largely unknown. Here, we have identified primary sequences required for Cten’s nuclear targeting, nuclear exporting, as well as focal adhesion localization. By using Cten mutants that constitutively target or retain Cten proteins in the nucleus, we further demonstrate that nuclear Cten contributes to cell proliferation.
2. Materials and methods
2.1. Cell culture
All cell lines were obtained from ATCC. The cervical cancer HeLa cells, lung cancer A549 cells and colorectal cancer SW480 cells were maintained in Eagle’s Minimum Essential Medium (MEM) or Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO) containing 10 % FBS (Sigma) and 1% penicillin/streptomycin. Cells were incubated in a humidified 5% CO2 atmosphere at 37 °C.
2.2. Antibodies and reagents
Rabbit monoclonal anti-Cten (SP83) was purchased from Novus Biologicals. Mouse anti-lamin A/C and mouse anti-α-tubulin were from Santa Cruz Biotechnology and Sigma, respectively. Alexa Fluor 594 Phalloidin and secondary antibodies conjugated with Alexa Fluor 488 were from Invitrogen. Leptomycin B (LMB) was from Sigma.
2.3. Subcellular fractionation
Cells grown in 60 mm-dishes were washed with cold PBS twice, and then swollen in 200 μl of buffer A (10 mM HEPES/pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.15% NP40, and proteinase and phosphatase inhibitors) for 10 min on ice. After centrifugation at 12,000g for 30s, the supernatant was collected as cytoplasmic fraction. The pellet was washed with buffer A (without NP40) twice, and then resuspended in 60 μl of buffer B (20 mM HEPES/pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% NP40, and proteinase and phosphatase inhibitors). After 15 min incubation on ice and centrifugation at 12,000g for 10 min, the supernatant was collected as nuclear fraction. The cytoplasmic or nuclear fraction were diluted in 200 μl of buffer B or 60 μl of buffer A. Protein concentrations were determined using the Bio Rad protein assay and equal amounts of proteins from nuclear or cytoplasmic fractions were analyzed by immunoblotting.
2.4. Plasmids and DNA transfection
All fragments of Cten with the EcoRI and BamHI cloning sites were amplified by PCR and subcloned into pEGFP-C2 vector. The deleted fragments of Cten were constructed by fusion PCR and the point mutations of Cten were generated by site-directed mutagenesis. All constructs were confirmed by DNA sequencing. The plasmid DNA was transfected into cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
2.5. Immunofluorescence and confocal microscopy
For immunofluorescence microscopy, cells were grown on acid-treated coverslips, fixed with 4% paraformaldehyde/PBS for 15 min, and permeabilized with 0.1 % Triton X-100/PBS for 5 min. After blocking with 5 % goat serum, the coverslips were incubated with primary antibodies overnight, followed by appropriate secondary antibodies conjugated with Alexa Flour 488 and Alexa Fluor 594 Phalloidin for 30 min. The coverslips were mounted on slides using DAPI-containing mounting solution (Vector Laboratories) and observed using a Zeiss LSM 710 laser confocal microscopy.
2.6. Cell proliferation assay
Cells in 100 μL of growth medium were seeded in 96-well plates at a density of 2,000 cells per well and cultured at 37°C with 5% CO2. Following the culture for 24, 48, and 72h, 10 μL WST-1 solution (Roche) was added into each well and incubated for an additional 1 h period at 37°C. Subsequently, the absorbance was determined with a microplate reader at 450 nm. The experiments were performed in three separated repeats with duplicate samples in each repeat.
3. Results
3.1. Cten is a CRM1-dependent nucleocytoplasmic shuttling protein
Previously, we showed that Cten is not only a focal adhesion molecule but is also present in the nucleus [18]. To further extend our understanding of the potential function and mechanism of Cten nuclear localization, we first examined Cten distributions in various cell lines by immunoblot analysis of equal amounts of nuclear and cytoplasmic fractions (figure 1A). The proportions of nuclear Cten were between 25% and 35%. The proportions were further increased to 35% and 45%, respectively, when cells were treated with leptomycin B (LMB), an inhibitor of the nuclear export receptor CRM1 (figure 1A). By immunofluorescence staining, LMB treatment significantly enhanced the accumulation of endogenous Cten in the nuclei (figure 1B&C), indicating that the nuclear export of Cten is a CRM1 dependent process.
Figure 1. Cten is a CRM-1-dependent nucleocytoplasmic shuttling protein.
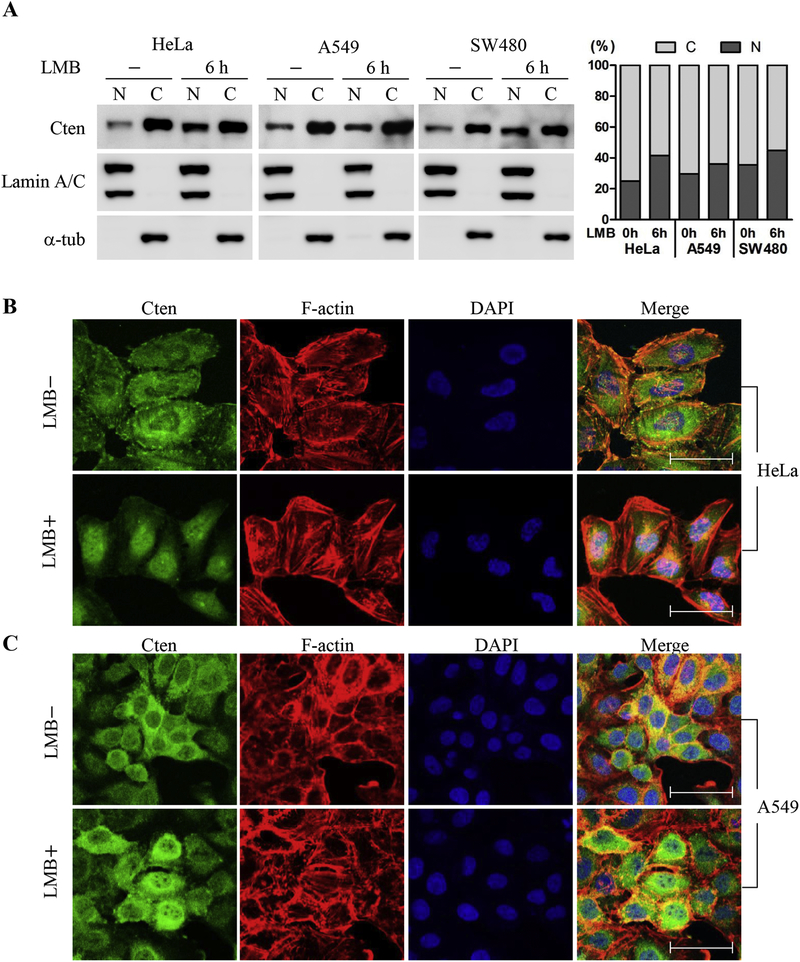
(A) Lung cancer cells A549, colorectal cancer SW480 cells, and cervical cancer HeLa cells were untreated or treated with 20 ng/mL of LMB for 6h or 24h. Nuclear and cytoplasmic fractions were prepared as description in Materials and Methods. Equal amounts of nuclear and cytoplasmic fractions (10 μg) were analyzed by immunoblotting with anti-Cten, anti-lamin A/C, or anti-α-tubulin antibodies. Right: The proportions of nuclear and cytoplasmic Cten measured by immunoblotting after subcellular fractionation in cells treated with LMB for 0h or 6h. HeLa (B) or A549 (C) cells treated with or without 20 ng/mL LMB for 6h were then fixed, permeabilized, and stained for DNA using DAPI (blue), for F-actin using Alexa Fluor 594 Phalloidin (red), and for Cten using a rabbit anti-Cten antibody followed by Alexa-488-coupled anti-rabbit IgG (green). Images were obtained by confocal laser scanning microscopy. Scale bars, 50 μm.
3.2. Identification of nuclear localization and export regions in Cten
To reveal the mechanisms regulating nucleocytoplasmic shuttling of Cten, we first searched for potential nuclear localization sequence (NLS) and nuclear export sequence (NES) in Cten using the cNLS Mapper prediction program (//nls-mapper.iab.keio.ac.jp). While Cten contains no monopartite NLS, a Cten region (aa 636–668) was predicated as a bipartite NLS (table 1). To demonstrate this NLS was functional, several Cten mutants fused with EGFP were generated for subcellular localization studies (figure 2A). The results showed that (a) EGFP-Cten (WT:1–715) proteins were mainly present in the cytoplasm and at focal adhesions and weakly in the nuclei; (b) in the presence of LMB, like endogenous Cten, EGFP-Cten proteins were markedly accumulated in the nuclei; (c) EGFP-Cten (1–570) proteins were excluded from the nuclei; (d) EGFP-Cten (438–715) proteins were predominantly in the nuclei; (e) EGFP-Cten (636–668) proteins were exclusively in the nuclei; and (f) EGFP-Cten (636–668) was able to redirect the cytoplasmic protein β-galactosidase (~120 + 30 = 150kD) to the nuclei. With these findings, we further examined the effect of 641RR to AA mutations within the NLS (636–668) on Cten subcellular localization. The nuclear localization was significantly reduced in Cten641RR/AA, especially with LMB treatment (figure 2B). The inefficiency of nuclear localization of Cten641RR/AA mutant was further confirmed by fractionation and immunoblot analyses (figure 2C). Altogether, we had identified a novel 33 aa NLS (636–668) in Cten.
Table 1. Potential NLS and NESs predicted in Cten.
NLS sequences are rich in arginine and lysine, while NESs consist of four hydrophobic amino acid residues with a defined space spanning among them. By using the cNLS Mapper protein sequence analysis program, a potential NLS and six candidate NESs (cNESs) were identified in Cten. Critical residues for NLS and cNESs are indicated in bold.
| Signal | Sequence |
|---|---|
| Cten potential NLS | 636RKVFFRRHYPLTTLRFCGMDPEQRKWQKYCKPS668 |
| Cten candidate NES | |
| cNES1 | 102FSLESLNQMILEL112 |
| cNES2 | 110LELDPTFQL118 |
| cNES3 | 146LDIKYIEV153 |
| cNES4 | 317LHSLGSVSL325 |
| cNES5 | 369MVDIPIVLI377 |
| cNES6 | 544MALALPCKLTI554 |
| NES consensus | ΦX(1–3) ΦX(2–3) ΦX(1–2)Φ (Φ: L, I, V or F and X: any amino acid) |
Figure 2. Identification of nuclear localization sequence in Cten.
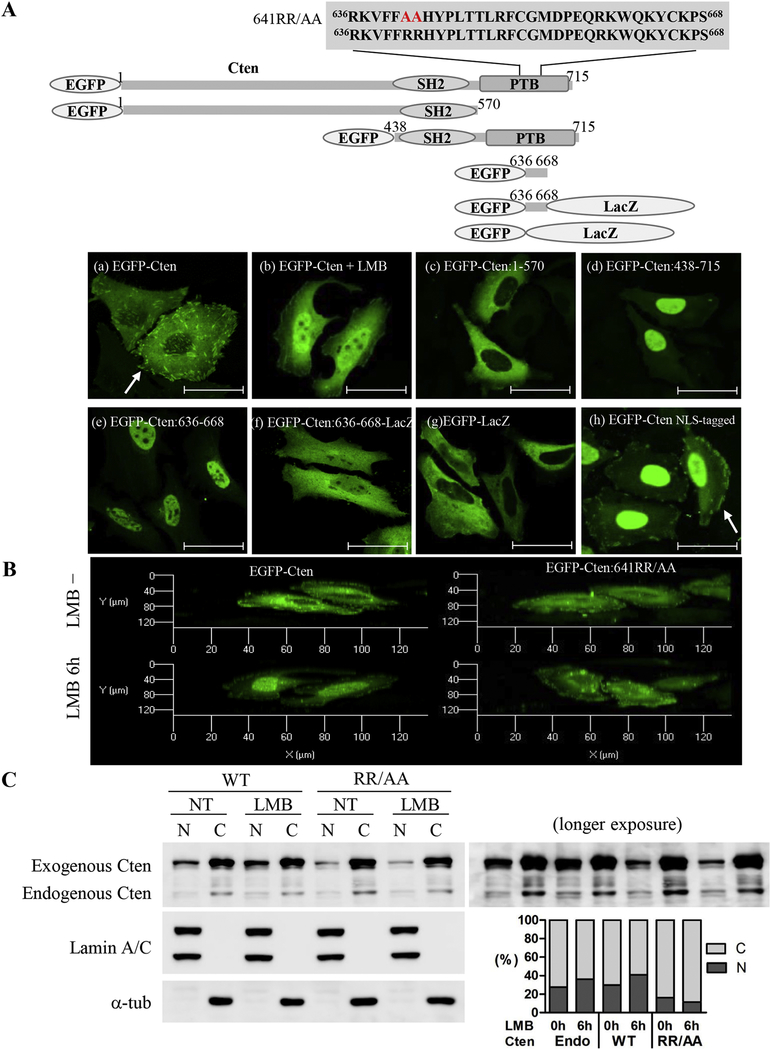
(A) Top: Schematic representation of EGFP-Cten constructs used to identify Cten’s nuclear localization sequence. The predicted NLS sequence located in the fragment 636 to 668 was shown. Bottom: HeLa cells transiently transfected with indicated constructs were fixed and visualized by confocal laser microscopy. Scale bar, 50 μm. Arrows indicate focal adhesion sites. (B) Overhead 3D reconstruction of confocal Z-stack images demonstrating the nuclear localization of Cten. HeLa cells were transiently transfected with EGFP-Cten or EGFP-Cten641RR/AA and then treated with or without 20 ng/mL LMB for 6 hours. The 3D Confocal immunofluorescence images in HeLa cells were obtained from a stack of 5–6 consecutive images (interval thickness=1 μm) centered on the middle of the nucleus. (C) HeLa cells transiently transfected with EGFP-Cten or EGFP-Cten641RR/AA were treated with or without 20 ng/mL LMB for 6 hours. Left: Equal amounts of nuclear and cytoplasmic fractions (2.5μg) were analyzed by immunoblotting using antibodies against Cten, lamin A/C, or a-tubulin. Right: Quantification of band intensities from nuclear and cytoplasmic fractions of endogenous or exogenous Cten measured by Cten immunoblotting.
Similarly, we had spotted and examined 6 potential NES within Cten (named cNES1–6) (table 1). Initial testing of a series of Cten fragments fused with EGFP indicated that Cten fragment 81–175 (containing cNES1–3) retained the nuclear export activity by showing significantly weaker nuclear localization, which was enhanced by LMB treatment (figure 3A). By testing full length Cten mutants with cNES1–3(Δ49–153), cNES1–2(Δ100–118), or cNES3(Δ139–153) deleted, we found only cNES3 deletion mutant still contained the export activity (figure 3A), indicating that the export sequence resides within cNES1 and cNES2. We further constructed and tested point mutations within cNES1–2, which were overlapped with 3 amino acids. Only cNES1(5A) mutant significantly accumulated in the nuclei (figure 3B). These results indicated that the cNES1 (102–112) is the functional NES within Cten.
Figure 3. Identification of nuclear export sequence in Cten.
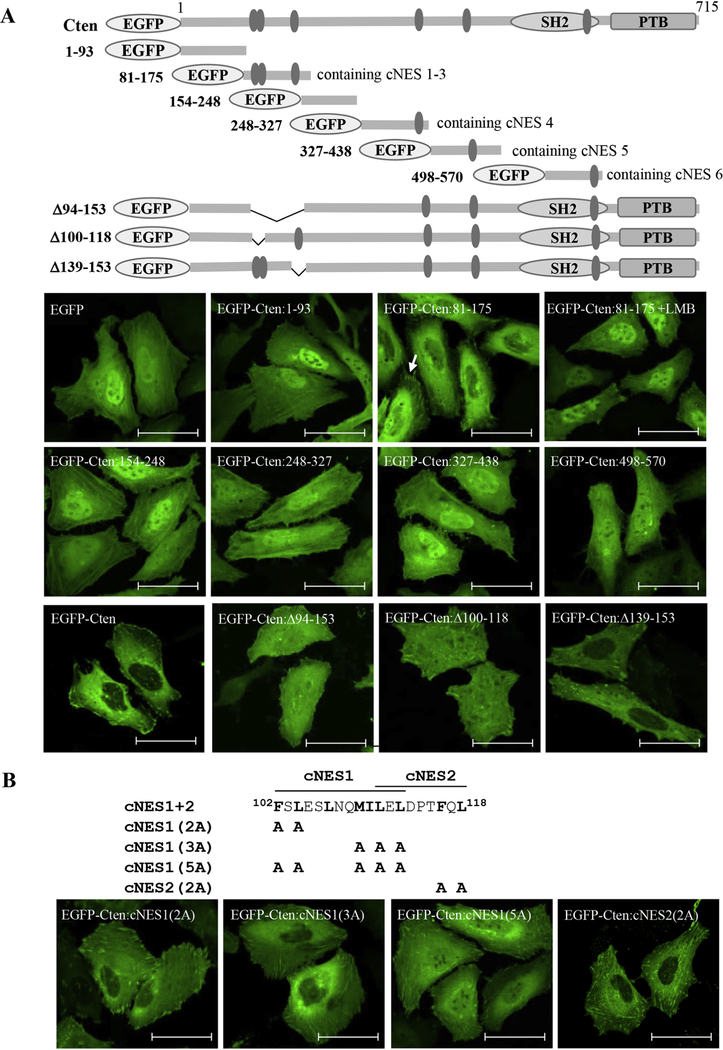
(A) Top: Schematic representation of EGFP-Cten constructs used to identify Cten nuclear export sequence. The predicted NES sequences, cNES 1–3 in fragment 81–175, cNES 4 in fragment 248–327, cNES 5 in fragment 327–438, and cNES 6 in fragment 498–570, were shown. Bottom: HeLa cells transiently transfected with indicated constructs were fixed and visualized by confocal laser microscopy. Scale bar, 50 μm. Cells treated with 20 ng/mL of LMB for 6 hours were also shown here in EGFP-Cten:81–175 expressing cells. (B) Mutations changing to alanines within cNES1 and cNES2 are indicated in the schematic diagrams. HeLa cells transfected with indicated EGFP-Cten mutants were fixed and imaged. Scale bar, 50 μm.
3.3. Cten contains two independent focal adhesion targeting sites
Since Cten only shares the C-terminal SH2 and PTB domains with other tensins and this region contains one of the two focal adhesion targeting sites in other tensins, we initially predicted Cten only has one focal adhesion binding (FAB) site. However, Cten’s SH2 and PTB (438–715) fragments, which were very efficiently targeting to the nuclei, still co-localized with vinculin at focal adhesions (figure 4A). The same fragment with a R474A mutation (SH2 dead) completely abolishes the focal adhesion localization (figure 4A), indicating that a functional SH2 domain is required for targeting to the focal adhesion, but not to the nucleus. On the other hand, the full length Cten with R474A mutation still targets to focal adhesions, suggesting the presence of a second FAB site outside the SH2 domain. In fact, Cten 81–175 fragments were also detected at focal adhesion sites (figure 3A). Finally, Cten harboring R474A mutation and aa 100–118 deletion completely lost the ability for focal adhesion localization. These results demonstrate that Cten, like other tensins [20], contains two independent focal adhesion binding sites (figure 4A). To assure all EGFP fusion constructs used in this project express correctly in transfected cells, immunoblot analysis was conducted and expected protein sizes were detected as shown in figure 4B.
Figure 4. Identification of two focal adhesion binding regions within Cten.
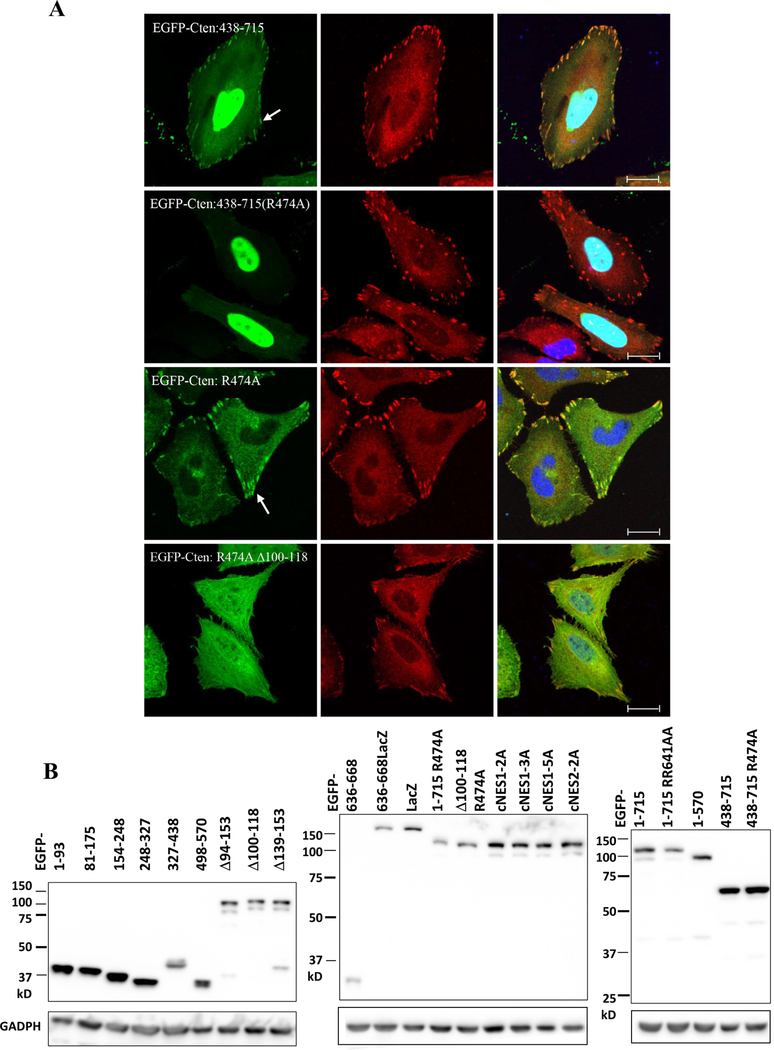
(A) HeLa cells transfected with indicated EGFP-Cten constructs were fixed and co-stained with vinculin (red) and DAPI (blue). Scale bar, 20 μm. Arrows show focal adhesion sites. (B) HeLa cells transfected with indicated EGFP-Cten constructs were immunoblotted with antibodies against EGFP or GAPDH.
3.4. Nuclear Cten promotes cell growth
To examine the role of nuclear Cten in cell growth, EGFP-Cten or EGFP-Cten cNES(5A) (NES dead) constructs were transfected into HeLa cells. While overexpression of EGFP-Cten showed no effect on cell growth (figure 5), EGFP-Cten cNES(5A) significantly promoted cell growth on Day 2 and 3. On the other hand, silencing of Cten markedly reduced cell growth (figure 5). To further examine whether nuclear localization is critical for promoting cell growth, EGFP-Cten tagged with 3 copies of the SV40 NLS was generated and tested. EGFP-Cten NLS-tagged proteins were highly concentrated within the nuclei while some remained at focal adhesion sites (figure 2A). In agreement with the result using EGFP-Cten cNES(5A), expression of EGFP-Cten NLS-tagged also enhanced HeLa cell proliferation (figure 5). These results suggest that nuclear Cten promoted cell proliferation.
Figure 5. Nuclear Cten promotes cell proliferation.
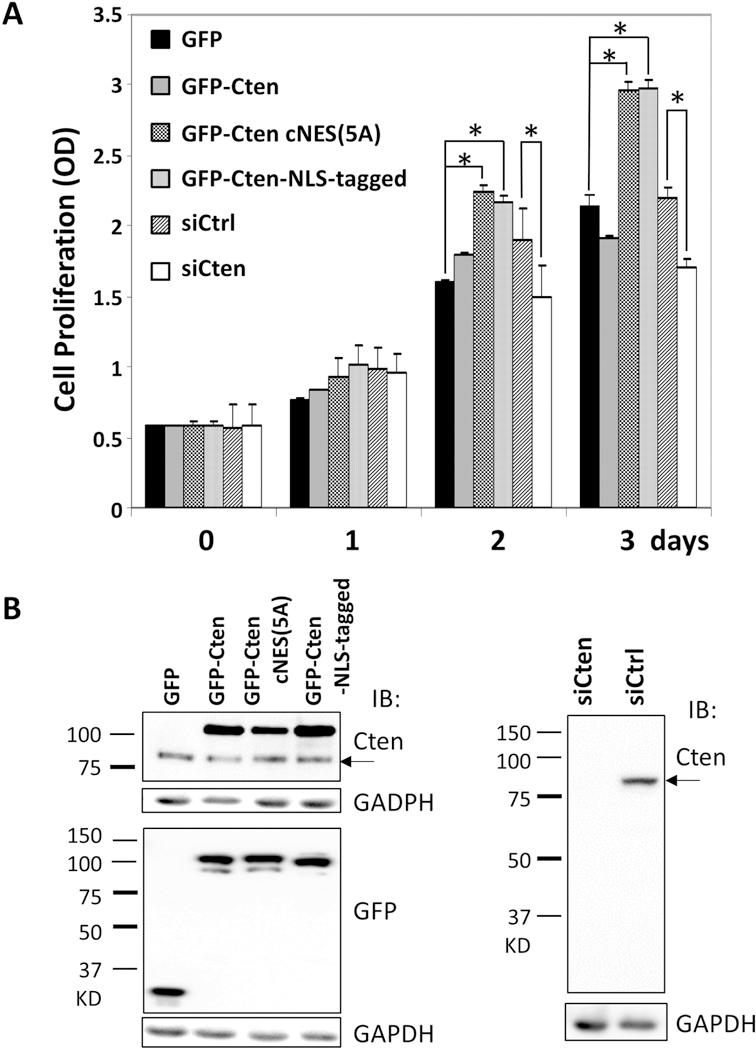
HeLa cells transfected with EGFP-Cten, EGFP-Cten:cNES1(5A) (aka NES dead) mutant, or EGFP-Cten NLS-tagged (fused with SV40 NLS sequence) or indicated siRNAs for 1 to 3 days were analyzed for cell growth by WST1 assays (A) or for protein expressions by immunoblotting with antibodies against Cten, EGFP, or GAPDH (B). * shows p<0.001. Arrows indicate endogenous Cten.
4. Discussion
In this report, we have identified a nuclear export sequence, a nuclear localization sequence, and two focal adhesion targeting regions within Cten (figure 6). We also showed that nuclear export of Cten is a CRM1 dependent process. Together with previous findings, we have a better understanding of cten’s functions related to its subcellular localizations (figure 6). Several growth factors and cytokines are known to up-regulate Cten protein levels [17] and Cten may (1) translocate to the nucleus and enhance cell proliferation, (2) localize to membrane and stabilize active EGFR and c-Met [10, 11], as well as (3) target to focal adhesions and promote cell migration/invasion. Although we have defined the primary sequences that are required for Cten’s subcellular localization, we do not know what factors determine the nuclear localization of Cten. Since nuclear ratios appear to be higher in Cten positive cancer cell lines and nuclear Cten promotes cell proliferation, whether certain cancer associated proteins, such as growth factors and receptor tyrosine kinases, could promote Cten nuclear localization is currently under investigation.
Figure 6. A schematic diagram summarizing cten’s functions related to its subcellular localizations.
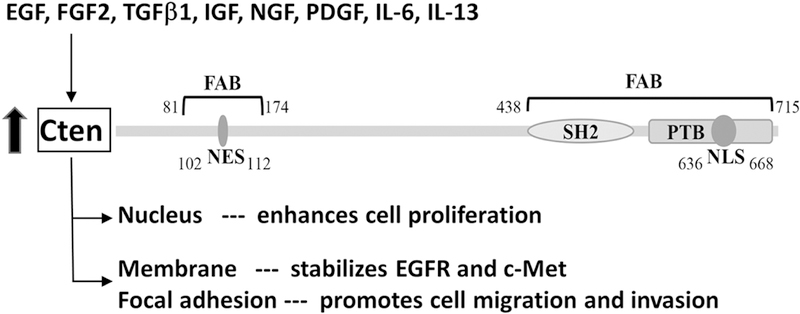
Several growth factors and cytokines are known to induce Cten expression. Two focal adhesion binding (FAB) regions, one NES and one NLS sites are identified in Cten. These sites allow Cten travel to focal adhesions as well as in-and-out of the nucleus. Cten in the nucleus appears to promote cell proliferation whereas at focal adhesions to up-regulate cell migration/invasion. Meanwhile, Cten stabilizes active EGFR and c-Met on the plasma membrane.
Regulating cell proliferation appears to be one of Cten’s functions, since silencing of Cten reduces cell growth (figure 5). However, overexpression of Cten does not further promote cell proliferation in our studies as well as other reports [11, 14, 21], except in one study showing that overexpression of Cten promotes keratinocyte proliferation via FAK and ERK [22]. It is presumably due to its low efficiency of nuclear accumulation and/or the effect is saturated by endogenous Cten expressed in cancer cell lines. Nonetheless, expression of Cten mutants either trapped or forced to be in the nuclei further enhance cell growth, strongly suggesting that it requires nuclear localization of Cten for the function.
How nuclear Cten promotes cell proliferation is currently unclear. One possibility is that Cten may associate with transcriptional complexes and either stabilize the complexes or enhance transcription activities that lead to proliferation. A known nuclear Cten binding partner is β-catenin [18], which is a co-activator of TCF transcription factor. We are currently taking a proteomics approach by identifying molecules associated with nuclear Cten to further understand the mechanism of promoting proliferation and other potential functions in the nucleus.
Earlier we showed that Cten binds to DLC1, a tumor suppressor targeting to focal adhesions, through its SH2 domain [8]. However, a functional SH2 is also required for Cten’s focal adhesion localization. How can one SH2 domain simultaneously target Cten itself to focal adhesions and bind DLC1? The identification of an additional focal adhesion binding region solves the puzzle, i.e. the SH2 binds to DLC1 while the N-terminal FAB interacts with focal adhesions.
The identification of the focal adhesion, nuclear localization and export sequences in this study has paved the way for further dissecting the physiological as well as pathological roles of subcellular Cten localizations.
Highlight.
Cten shuttles between focal adhesion and nucleus.
Cten contains one nuclear localization sequence, one nuclear export sequence, and two focal adhesion binding regions.
Nuclear Cten promotes cancer cell proliferation.
ACKNOWLEDGEMENTS
We thank Dr. Yi-Chun Liao for her initial finding on nuclear Cten. This study was supported in part by NIH grants DK097291 to YPS and CA102537 and HL139473 to SHL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors disclose no potential conflicts of interest
References
- 1.Lo SH (2004) Tensin, The international journal of biochemistry & cell biology 36, 31–4. [DOI] [PubMed] [Google Scholar]
- 2.Lo SH (2017) Tensins, Current biology : CB 27, R331–R332. [DOI] [PubMed] [Google Scholar]
- 3.Lo SH, Janmey PA, Hartwig JH & Chen LB (1994) Interactions of tensin with actin and identification of its three distinct actin-binding domains, The Journal of cell biology 125, 1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Duncan IC, Bozorgchami H & Lo SH (2002) Tensin1 and a previously undocumented family member, tensin2, positively regulate cell migration, Proceedings of the National Academy of Sciences of the United States of America 99, 733–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Y, Liao YC & Lo SH (2004) Epidermal growth factor modulates tyrosine phosphorylation of a novel tensin family member, tensin3, Molecular cancer research : MCR 2, 225–32. [PubMed] [Google Scholar]
- 6.Lo SH & Lo TB (2002) Cten, a COOH-terminal tensin-like protein with prostate restricted expression, is down-regulated in prostate cancer, Cancer research 62, 4217–21. [PubMed] [Google Scholar]
- 7.Lo SH (2014) C-terminal tensin-like (CTEN): a promising biomarker and target for cancer., Int J Biochem Cell Biol 51, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao YC, Si L, Devere White RW & Lo SH (2007) The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1, The Journal of cell biology 176, 43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian X, Li G, Asmussen HK, Asnaghi L, Vass WC, Braverman R, Yamada KM, Popescu NC, Papageorge AG & Lowy DR (2007) Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities, Proceedings of the National Academy of Sciences of the United States of America 104, 9012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muharram G, Sahgal P, Korpela T, De Franceschi N, Kaukonen R, Clark K, Tulasne D, Carpen O & Ivaska J (2014) Tensin-4-dependent MET stabilization is essential for survival and proliferation in carcinoma cells, Dev Cell 29, 421–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong SY, Shih YP, Li T, Carraway KL 3rd & Lo SH (2013) CTEN prolongs signaling by EGFR through reducing its ligand-induced degradation, Cancer research 73, 5266–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albasri A, Aleskandarany M, Benhasouna A, Powe DG, Ellis IO, Ilyas M & Green AR (2011) CTEN (C-terminal tensin-like), a novel oncogene overexpressed in invasive breast carcinoma of poor prognosis, Breast Cancer Res Treat 126, 47–54. [DOI] [PubMed] [Google Scholar]
- 13.Albasri A, Al-Ghamdi S, Fadhil W, Aleskandarany M, Liao YC, Jackson D, Lobo DN, Lo SH, Kumari R, Durrant L, Watson S, Kindle KB & Ilyas M (2011) Cten signals through integrin-linked kinase (ILK) and may promote metastasis in colorectal cancer, Oncogene 30, 2997–3002. [DOI] [PubMed] [Google Scholar]
- 14.Al-Ghamdi S, Cachat J, Albasri A, Ahmed M, Jackson D, Zaitoun A, Guppy N, Otto WR, Alison MR, Kindle KB & Ilyas M (2013) C-terminal tensin-like gene functions as an oncogene and promotes cell motility in pancreatic cancer, Pancreas 42, 135–40. [DOI] [PubMed] [Google Scholar]
- 15.Barbieri I, Pensa S, Pannellini T, Quaglino E, Maritano D, Demaria M, Voster A, Turkson J, Cavallo F, Watson CJ, Provero P, Musiani P & Poli V (2010) Constitutively active Stat3 enhances neu-mediated migration and metastasis in mammary tumors via upregulation of Cten, Cancer research 70, 2558–67. [DOI] [PubMed] [Google Scholar]
- 16.Katz M, Amit I, Citri A, Shay T, Carvalho S, Lavi S, Milanezi F, Lyass L, Amariglio N, Jacob-Hirsch J, Ben-Chetrit N, Tarcic G, Lindzen M, Avraham R, Liao YC, Trusk P, Lyass A, Rechavi G, Spector NL, Lo SH, Schmitt F, Bacus SS & Yarden Y (2007) A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration, Nat Cell Biol 9, 961–9. [DOI] [PubMed] [Google Scholar]
- 17.Hung SY, Shih YP, Chen M & Lo SH (2014) Up-regulated cten by FGF2 contributes to FGF2-mediated cell migration, Molecular carcinogenesis 53, 787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao YC, Chen NT, Shih YP, Dong Y & Lo SH (2009) Up-regulation of C-terminal tensin-like molecule promotes the tumorigenicity of colon cancer through beta-catenin, Cancer research 69, 4563–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albasri A, Fadhil W, Scholefield JH, Durrant LG & Ilyas M (2014) Nuclear expression of phosphorylated focal adhesion kinase is associated with poor prognosis in human colorectal cancer, Anticancer research 34, 3969–74. [PubMed] [Google Scholar]
- 20.Chen H & Lo SH (2003) Regulation of tensin-promoted cell migration by its focal adhesion-binding and Src Homology 2 domains, Biochem J 370, 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo SS, Lo SH & Lo SH (2005) Cleavage of cten by caspase-3 during apoptosis, Oncogene 24, 4311–4. [DOI] [PubMed] [Google Scholar]
- 22.Seo EY, Jin SP, Kim YK, Lee H, Han S, Lee DH & Chung JH (2017) Integrin-beta4-TNS4-Focal Adhesion Kinase Signaling Mediates Keratinocyte Proliferation in Human Skin, The Journal of investigative dermatology 137, 763–766. [DOI] [PubMed] [Google Scholar]


