Abstract
The use of high-throughput multiplexed screening platforms has attracted significant interest in the field of on-site disease detection and diagnostics for their capability to simultaneously interrogate single cell responses across different populations. However, many of the current approaches are limited by the spectral overlap between tracking materials (e.g., organic dyes) and commonly used fluorophores/biochemical stains, thus restraining their applications in multiplexed studies. This work demonstrates that the downconversion emission spectra offered by rare earth (RE)-doped β-hexagonal NaYF4 nanoparticles (NPs) can be exploited to address this spectral overlap issue. Compared to organic dyes and other tracking materials where the excitation and emission is separated by tens of nanometers, RE elements have a large gap between excitation and emission which results in their spectral independence from the organic dyes. As a proof of concept, two differently doped NaYF4 NPs (Europium: Eu3+ and Terbium: Tb3+) were employed on a fluorescent microscopy-based droplet microfluidic trapping array to test their feasibility as spectrally independent droplet trackers. The luminescence tracking properties of Eu3+-doped (red emission) and Tb3+-doped (green emission) NPs were successfully characterized by co-encapsulating with genetically modified cancer cell lines expressing green or red fluorescent proteins (GFP and RFP) in addition to a mixed population of live and dead cells stained with ethidium homodimer. Detailed quantification of the luminescent and fluorescent signals was performed to confirm no overlap between each of the NPs and between NPs and cells. Thus, the spectral independence of Eu3+-doped and Tb3+-doped NPs with each other and with common fluorophores highlights the potential application of this novel technique in multiplexed systems, where many such luminescent NPs (other doped and co-doped NPs) can be used to simultaneously track different input conditions on the same platform.
Keywords: Single Cell Analysis, Microfluidics, Rare Earth Elements, Nanoparticles, High Throughput Screening
Graphical Abstract
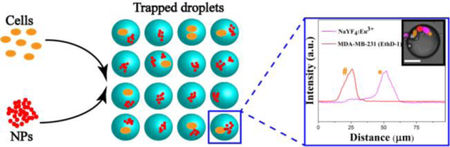
Introduction
Isolating and analyzing single cells from a heterogeneous population is essential to perform clinically relevant measurements with respect to cancer diagnostics.[1–3] Droplet microfluidics has emerged as a single cell sorting technology which offers several advantages over existing large-scale technologies like capillary electrophoresis, flow cytometry, and mass cytometry in terms of its reduced reagent costs, ease-of-use, and compatibility with fluorescent microscopy.[4, 5] Fluorescence activated cell sorting (FACS) is a commercialized flow cytometry technique, capable of simultaneous quantification up to 20 parameters in single cells based on specific light scattering and fluorescent characteristics of each cell (called fluorescence-cell barcoding).[6, 7] However, flow cytometry is often limited by its need for large sample size, the cost and size of the instrument,[8] and the use of rapid flow in the system which, when coupled with non-specific surface markers, can negatively affect cell viability. [9] There have been reports on spectral crosstalk between these fluorescent-cell barcodes which was addressed by mass cytometry, a method where each sample is labelled with a unique combination of lanthanide isotopes (called mass-tag barcoding).[6, 10] Despite having a lower throughput than flow cytometry,[4] the spectrally distinct mass-tag barcodes in mass cytometry make it an ideal system for multiplex studies. [6] However, mass cytometry is limited by its inability to sort cells during analysis coupled with a poor signal to noise ratio. [11]
The high-throughput screening (HTS) technologies discussed above offer extensive information on single cell analysis but are restrained in their ability to directly quantify the intracellular distribution of fluorescent signals. Droplet microfluidic devices enabled with spatial traps[12, 13] have recently found applications in dynamic applications like single-cell barcoding and sequencing [14, 15] (barcoding here means indexing or tracking something in a microfluidic device by giving an identity – either track a droplet/cell/molecule using some technique). One such example is to isolate and amplify single DNA molecules within droplets and barcode each of these droplets using chemically synthesized oligonucleotides [16]. Another such droplet-based technique is inDrops (indexing droplets) where cells are indexed by hydrogel beads bearing DNA primer barcodes for single cell RNA sequencing.[17] While these microfluidic devices have emerged as powerful tools in profiling cellular heterogeneity, they make use of expensive barcoding antibodies and involve time-consuming reactions. Other droplet barcoding techniques utilizing organic dyes[18, 19] are also limited in their ability to simultaneously quantify different cellular outputs due to the spectral overlap of these dyes with common biochemical stains.[20–22] These issues have resulted in some limitations in the use of droplet microfluidics in multiplexed applications where simultaneous tracking of cellular outputs to different input conditions on a single platform is required.
Advances in nanomaterials have produced a new class of fluorescent labels by conjugating semiconductor quantum dots (QDs) with biorecognition molecules.[23–25] Because of their size-tuneable absorptions/emissions, and high fluorescence quantum yield, QDs are considered a better alternative to organic dyes.[21, 22] Although being widely used as fluorescent barcodes in platforms such as FACS[26, 27] and droplet microfluidics[28], there have been concerns regarding QDs’ cytotoxicity, with heavy metals being used in their crystal structure.[29] Rare earth (RE)-doped materials are a class of phosphors that have received attention for their potential applications in bio-imaging, sensing, and therapeutics.[30] Unlike organic dyes and common fluorophores, RE dopants offer a wide range of characteristic emission lines in the visible spectrum stemming from the f-f intraband transitions, including red (Eu3+, Sm3+), yellow (Dy3+, Er3+), green (Tb3+, Er3+, Ho3+), and blue (Dy3+, Tm3+) making them suitable as spectrally independent labels.[30] Additionally, the large gap between excitations and emissions of the RE elements serves as an added advantage for spectral independence when compared to the closer excitations and emiss ions of organic dyes.[30, 31] Also, RE phosphors have been shown to exhibit greater biocompatibility over QDs.[32] Two RE luminescence mechanisms are possible, downconversion (DC) and upconversion (UC), which are analogous to Stokes and anti-Stokes shift, respectively. In UC materials, luminescence occurs by sequential absorption of lower energy photons followed by a higher energy emission. UC phosphors have been extensively exploited for in vivo bio-imaging, photodynamic therapy, and drug delivery, primarily due to their minimal photodamage to living organisms and excitation within the biologically transparent window.[33] On the other hand, the unexplored, diverse energy levels available with DC make these elements suitable for multiplex tracking purposes for in vitro studies. The DC process stems from the unique optoelectronic properties of the RE ions in which higher energy photons such as UV radiation is converted to lower energy visible light, resulting in narrow and spectrally independent emission peaks that is suitable for tracking purposes in biological studies.[34]
This work describes the use of spectrally independent RE-doped nanoparticles (NPs) as droplet trackers using DC luminescence imaged by fluorescent microscopy. The synthesized RE-doped β-NaYF4 NPs were successfully characterized for structure, morphology, spectral properties, and biocompatibility. Europium (Eu3+) and Terbium (Tb3+) were chosen as RE dopants to track droplets due to their narrow, but spectrally independent, emission peaks. A series of single cell and NP co-encapsulation studies were performed in a microfluidic droplet trapping array to confirm the spectral independence of Eu 3+-doped and Tb3+-doped NPs with green fluorescent protein (GFP), red fluorescent protein (RFP), and ethidium homodimer-1 (EthD-1). In terms of broad comparison, GFP and RFP are the two main fluorescent labels used in biological purposes. While many variants (e.g., FAM, Alexa Fluor, Texas Red, Rhodamine) are used due to sensitivity and ease of conjugation, all these fluorescent probes have similar excitation/emission range as that of GFP and RFP.[35] Upon UV excitation, the RE dopants in NPs exhibited unique emission peaks (Eu3+-doped NP in red and Tb3+-doped NP in green) that did not overlap with commonly used fluorophores. While this work demonstrates the feasibility of two RE-doped NPs, this tracking technique can be expanded with several different doped and co-doped NPs[30], thus implementing the diversity of downconversion luminescence in multiplexed screening. The results from this study confirm the potential for luminescent NPs as droplet trackers and provides a justification for their use in a multiplexed device, thus highlighting the successful first ever use of these NPs as spectrally independent droplet trackers on a fluorescent microscopy stage.
Experimental
Synthesis and Characterization of RE-doped Luminescent NPs
A simple one-step hydrothermal process was implemented to synthesize NaYF4:RE3+ nanostructures. All the chemical reagents were used as received without further purification. In typical synthesis, 2 mmol of RE(NO3)3 6H2O (RE =Y3+, Eu3+, Tb3+, 99.9% Alfa Aesar) was dissolved in 1 mL HCl (36.5–38.0%, VWR) for 1 h to maintain the acidic environment required for forming hexagonal crystals. 5% mole concentration of dopants (Eu3+ and Tb3+) was used for the doped samples. The solution was then transferred to a 20 mL Teflon liner containing 15 mL of DI H2O. Next, 8 mmol of Na3 citrate (ACS grade, VWR) (RE precursors: Na3 citrate = 1:4) was added to the above mixture as a chelating agent to control the size and morphology of the NPs. After 30 min of vigorous stirring, 5 mL of a 5M NaF (USP grade, VWR) aqueous solution was added to the precursor solution as a fluoride source. The contents were thoroughly mixed for 15 min. The Teflon liner was transferred into an autoclave for subsequent heating at 180 °C for 1 h. The products were then collected and washed with DI H2O and EtOH (ACS grade, VWR) several times to remove the organic impurities and neutralize the solution. Finally, the samples were dried at 100 °C, and the NPs were annealed at 400 °C for 2 h to ensure successful incorporation of dopants to their respective crystal sites.
The synthesized NPs were characterized for their structure, size, morphology, and composition using X - ray Diffraction spectroscopy (XRD), Scanning Electron Microscopy (SEM), and Energy Dispersive X-ray spectroscopy (EDX or EDS). XRD measurements were performed on an Empyrean PANalytical X-ray diffractometer using Cu Kα1 (λ=1.54 Å) as radiation source, with a step size of 0.05° in the scanning range of 5°−70°. SEM imaging was done using an FEI Quanta 3D FIB microscope operated at 5 kV accelerating voltage. This instrument was equipped with an EDAX detector which was used to identify the elements present in the sample. The sample was prepared by casting a drop of the aqueous product on the double - sided carbon tape attached to the sample holder. All the dried samples were sputtered with Pt for 4 min to make the sample conductive for measurements. The NP luminescence was characterized using Photoluminescence (PL) spectrometer. An APTI QM-40 spectrofluorometer with a PMT detector and a 75W xenon arc lamp as light source was employed for PL measurements. The scans were performed with a band pass of 2 nm at a scanning rate of 4 nm/s in the range of 400 −700 nm.
Microfluidic Device Design and Fabrication
The droplet microfluidic trapping device consisted of two layers (Fig. 1): the bottom main flow channel and the top trapping array. The device had two inlet channels: one for the oil phase (230 μL/h) and one for the aqueous phase (90 μL/h) which converged at a flow-focusing junction to encapsulate cells and NPs in ~180 pL discrete aqueous droplets in a continuous oil phase. The oil phase was Novec 7500 oil (3M) with 0.2% Neat 008 fluoro-surfactant (Ran Biotechnologies). The fluoro-surfactant was used to stabilize droplet formation and prevent droplet aggregation. Vertical fins were incorporated into the bottom layer of the trapping array to increase the residence time of the droplets and aid in trapping. The trapping array consisted of a 787-member grid with each trap having a 70 μm diameter. The fluidic channels were 40 μm in height and the traps were imprinted 40 μm into the PDMS above the fluidic channel (total of 80 μm in height). The methods for PDMS replication and device preparation are included in the Supporting Information. The resultant devices were used for on-chip encapsulation experiments. To initiate droplet generation, Tygon tubing (Cole Palmer) was directly connected to oil and aqueous syringes fixed on two dual infusion syringe pumps (Harvard apparatus) and were inserted into the device inlet ports. The device was mounted on the stage of a fluorescent DMi8 inverted microscope (Leica Microsystems) to visualize droplet formation and trapping.
Fig. 1.
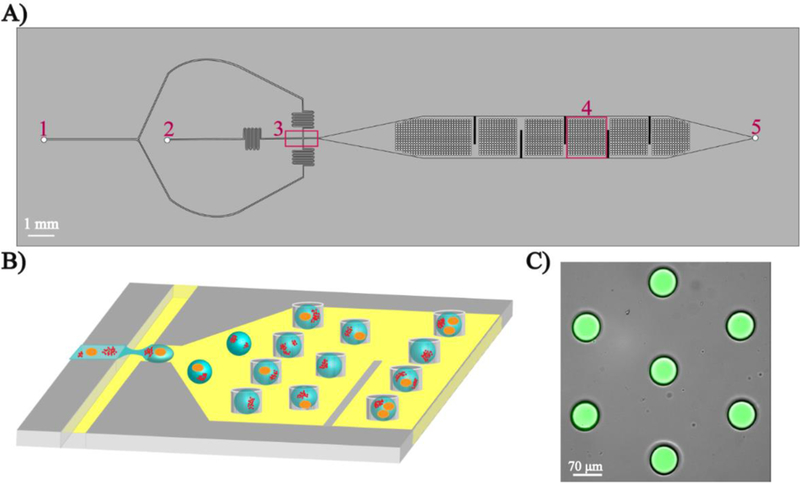
Microfluidic droplet trapping array. A) Top view of the device showing two inlets for carrier oil (1), cells and NPs in aqueous buffer (2), a flow-focusing junction (3), the droplet trapping array (4), and the single outlet (5). B) Schematic of the generation and trapping of individual droplet containing single cells and NPs. C) Overlay fluorescent microscopy image showing aqueous droplets containing the fluorescent dye 5(6)-carboxyfluorescein trapped in the device.
Cell Culture
The MDA-MB-231 cells and the red fluorescent protein (RFP)-expressing MDA-MB-231 cells were cultured in DMEM (Corning) supplemented with 10% v/v HyClone Cosmic Calf Serum (VWR Life Sciences Seradigm), 1% MEM Essential Amino Acids (Quality Biological Inc.), 1% MEM Non-Essential Amino Acids (Quality Biological Inc.), 1 mM Sodium Pyruvate (Thermo Fisher Scientific) and 6 µL insulin/500 mL media (Insulin, Human Recombinant dry powder - Sigma Aldrich). The green fluorescent protein (GFP)-expressing HeLa cells were cultured in DMEM Media with 10% foetal bovine serum (FBS - VWR Life Sciences Seradigm).
Off-chip Viability Assay
MDA-MB-231 cells were seeded at a density of 1 × 104 cells/mL in 12 well plates and incubated for 3 days. On the day of experiment, each well (in duplicate) were subjected to the viability assay at timepoints of 0 min, 30 min, 1 h, 2 h, 4 h, 6 h, 8 h, 10 h and 12 h. Each pair of wells was washed with 1 mL of 1X PBS (PBS: 137 mM NaCl, 10 mM Na2HPO4, 27 mM KCl, and 1.75 mM KH2PO4 at pH 7.4) followed by the addition of 1 mL of a 10 mg/mL NP slurry in extracellular buffer (ECB: 20 mM HEPES, 140 mM NaCl, 5 mM KCl, 1 mM MgCl2.6H2O, 1 mM CaCl2.2H2O, 5 mM D-Glucose at pH 7.4) with 1% FBS at specified timepoints over 12 h. After 12 h, the NP slurry was aspirated. After aspiration of NP slurry, the cells were not washed to avoid removal of any dead cells. 500 µL of a reagent stain mixture (2.5 µM Calcein AM and 4 µM EthD-1 in 1X PBS) was added to each well and incubated for 15–20 min followed by imaging for viability. This assay was performed only with 10 mg/mL NP slurry since this was the maximum concentration used for all on-chip encapsulation experiments and the results obtained for any lower NP concentrations are inclusive of those obtained from the 10 mg/mL study.
Microfluidic Droplet Tracking using RE-doped NPs
A 10 mg/mL slurry of NPs in ECB was used for on-chip encapsulation experiments. In order to prevent particle aggregation and settling, the slurry was sonicated for 1 h before each experiment. The RE-doped NPs were co-encapsulated with cells (GFP-HeLa cells, RFP-MDA-MB-231 cells, and MDA-MB-231 cells stained with EthD-1 at a cell density of 4.5 ×106 cells/mL) in ECB droplets which were isolated in the microfluidic droplet trapping array. For EthD-1 biochemical staining, MDA-MB-231 cells were binned into two groups. The first group (live) was resuspended in ECB and incubated at 37 °C prior to encapsulation. The second group (dead) was incubated at 42 °C for 1 h[36] in a heat block to kill all the cells. The live and dead cells were mixed into a heterogeneous population and incubated with 2 µM EthD −1 at 37 °C for 15–20 min off-chip prior to injection into the device along with the NP slurry. A fluorescent DMi8 inverted microscope (Leica microsystems) with a digital CMOS camera C11440 (Hamamatsu Photonics K.K.) and LAS X software 3.3.0 were used to image and analyse fluorescent and luminescent signals of cells and NPs. The following excitation/emission filters (Chroma Tech. Corp) were used: fluorescein isothiocyanate - FITC (λex: 440–520 nm and λem: 497–557 nm); rhodamine (λex: 536–556 nm and λem: 545–625 nm); filter set (λex: 370–420 nm and λem: 605–645 nm) for Eu3+ doped NPs; and filter set 2 (λex: 325–355 nm and λem: 505–565 nm) for Tb3+ doped NPs. The qualitative imaging was followed by a series of robust quantitative analysis. A manual line scan region of interest (ROI) was drawn across droplets containing both cells and NPs to quantify their luminescent and fluorescent signals. The acquired fluorescent intensities of cells and NPs from all filter sets were then normalized and compared in terms of Signal: Noise (S:N) ratio values.
| (1) |
Equation (1) was used to calculate normalized fluorescent signal (NFS) where X is the fluorescent signal, µ is the mean fluorescent signal, and σ is the standard deviation. The NFS values were compared with noise to give S:N ratio values. These S:N ratio values were further analysed for statistical significance through single/two-tailed hypotheses tests using SAS software.
On-chip Dose Response Studies
MDA-MB-231 cells were initially seeded at a density of 4.5 × 106 cells/mL in sterile 100 mm x 20 mm cell culture dishes. The cells were allowed to adhere and spread in the first 24 h. After 24 h, the media in each dish was swapped with 3 mL of drugged media. Three different concentrations of Paclitaxel (PTX) were tested with MDA-MB-231 cells: 10 µM, 50 µM and 100 µM. An off-chip drug treatment protocol was followed where each dish was treated with one of the three drug concentrations and the cellular response was later observed on-chip after 24 h, 48 h, or 72 h incubation. On the day of on-chip analysis, the cells were scraped from the surface of the culture dish using a sterile cell lifter (Corning) and transferred to a 15 mL tube. It is to be noted that no aspiration and centrifugation steps were included at this point, to avoid loss of floating dead cells after respective drug treatments. The cells were incubated with 2 µM EthD −1 at 37 °C for 20 min prior to on-chip encapsulation. Before injecting the sample into the device, 10 mg/mL of NP slurry in DMEM was added to the existing 3 mL of stained cell sample. This cell and NP slurry was injected into the microfluidic device and imaged for the single cell response across the trapping array. Three different NPs were used for the three different drug concentrations: Eu3+-doped NPs for 10 µM PTX, Tb3+-doped NPs for 50 µM PTX and undoped NPs for 100 µM PTX. No-drug control experiments were conducted in tandem for data validation. After each experiment, cellular viability and droplet tracking data was collected by imaging the trapping array using the fluorescent DMi8 inverted microscope and fluorescent filters rhodamine, filter set 1, and filter set 2.
Results and Discussion
Synthesis and Characterization of Hexagonal β-NaYF4 NPs for DC Applications
As previously reported,[37, 38] β-NaYF4 crystal is frequently employed for optoelectronic applications due to its low phonon energy, and high local crystal asymmetry that results in stark splitting of energ y levels, leading to efficient luminescence. The formation of hexagonal β-NaYF4 crystal structures is dependent on several parameters such as citrate concentration, pH of the solution, annealing temperature and reaction time.33,[39] The SEM image of the synthesized particles are shown in Fig. 2A which verifies the monodisperse formation of sub-micron sized hexagonal NaYF4 particles. The average diameter and length of the NPs were 180 nm and 310 nm, respectively. The crystal phase of NaYF 4 was identified from XRD patterns (Fig. 2B) and indexed to standard β-NaYF4 (JCPS: 17–6069), indicating that no other impurity phase was formed. Particle composition was verified with elemental analysis (EDX – Fig. 2C). Characteristic lines for Na, Y, and F along with characteristic and for Eu3+ and Tb3+ were identified. Due to low doping concentrations, EDX measurements for Eu 3+ and Tb 3+ were scaled by factor of 10, illustrating the presence of each dopant in the crystal lattice. In order to be consistent with the characteristic properties of the NPs synthesised from different batches, the above-mentioned steps were repeated to ensure batch to batch reproducibility of the synthesized NPs. This verified the chemical stability of NaYF4 NPs which has been previously discussed elsewhere.[40] Room-temperature DC luminescence excitation and emission spectra of β-NaYF4:RE3+ (RE3+: Eu3+, Tb3+) are shown in Fig. 2D. As it can be seen from this figure, the two different NPs can be excited in near UV region (350 −400 nm) and emit primarily in green (Tb3+) and red (Eu3+) regions of visible light. It is noteworthy that the absorption and excitation profile of these RE elements are similar, as previously reported.[41] For fluorescent microscopy, a 605–645 nm window filter-set was chosen to detect the 5D0 → 7F2 transition peak centred at 617 nm for Eu3+. Similarly, for Tb3+, a 505–565 nm window filter-set was chosen to detect the 5D4 → 7F5 transition peak centred at 545 nm.
Fig. 2.
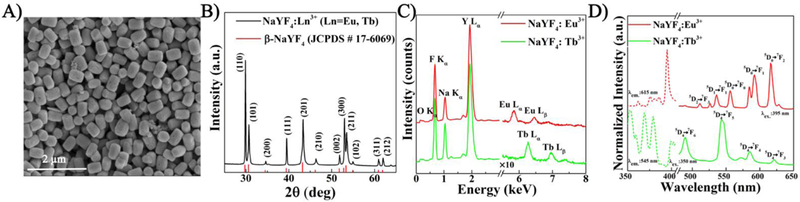
Characterization of RE-doped NPs. (A) The SEM image shows hexagonal NPs roughly 150 nm in diameter and 400 nm in length. (B) The XRD scan is indexed to the β-NaYF4 crystal structure (ICDD:04-016-7458) without the presence of any contamination based on (C) EDX fluorescence. (D) Photoluminescence excitation (dotted lines) and emission (solid lines) of Eu3+ and Tb3+ doped NaYF4 NPs demonstrates the characteristic Eu3+ and Tb3+ emission peaks when excited with 395 nm and 350 nm light.
Once the nanoparticles were successfully synthesized and characterized, their biocompatibility was verified to quantify the effect of droplet tracking on cell viability, since experiments involved UV excitation of the NPs and peripheral cell-NP interactions. UV initiated cellular/genetic damage was ruled in this application since a very low-dose, near UV excitation was used for both the NPs (800 ms of 395 nm exposure for Eu3+-NP and 800 ms of 330 nm exposure for Tb3+-NP). This has been reported previously where the effect of UV exposure times, up to 5 min, did not statistically affect the cell survival rate or did not alter their genetic expression.[42, 43] Secondly, the bio-inertness of NPs was verified in order to validate their use with cells for on-chip studies via an off-chip viability assay using a model breast cancer cell line, MDA-MB-231 (Fig. 3). The number of live and dead cells were counted as a measure to quantify the NP biocompatibility, resulting in a ~99% MDA-MB-231 cells viability (547 of 553 cells) after 12 h of incubation with Eu3+-doped NPs. Similar results were obtained for Tb3+-doped NPs (see Electronic Supplementary Material (ESM) Fig. S1A) with a ~99% viability (586 of 595 cells). These results were compared to a no-NP control experiments (see ESM Fig. S1B) which confirmed that the RE-doped NPs were biologically inert to the cells. This is in agreement with previous studies on biocompatibility of rare-earth based NPs[44], demonstrating a clear advantage of these RE-doped NPs over quantum dots (QDs).[29] It is to be noted here that, in this work, the luminescent NPs are only used for cellular tracking and that the large average diameter and length of the NPs (180 nm and 310 nm) restrict them from traversing across the cell membrane.[45]
Fig. 3.
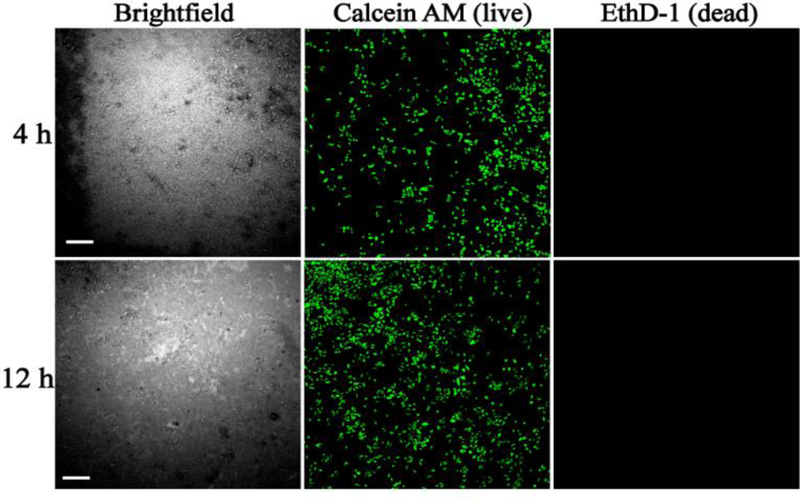
Determination of Eu3+-doped luminescent NPs on cellular viability. MDA-MB-231 cells were incubated with Eu3+-doped NaYF4 NPs at 37°C for 4 h (top row) and 12 h (bottom row) followed by live/dead staining using Calcein AM (green, middle column) and EthD-1 (red, right column). Scale bar is 300 µm.
Co-encapsulation of RE-doped NPs and Single Cells in Microfluidic Trapping Array
Once it was confirmed that the RE-doped NPs were biocompatible, the next step was to assess their ability to be used as droplet trackers in the microfluidic droplet trapping array. Stable droplets containing Eu3+-doped (Fig. 4A) and Tb3+-doped (Fig. 4B) NPs were generated and trapped in the microfluidic device. A prominent signal was observed from the NPs using their respective filters by fluorescent microscopy which demonstrated the feasibility of utilizing these phosphors in a droplet microfluidic platform. The luminescent nanoparticles were then tested for spectral independence with commonly used fluorophores.
Fig. 4.
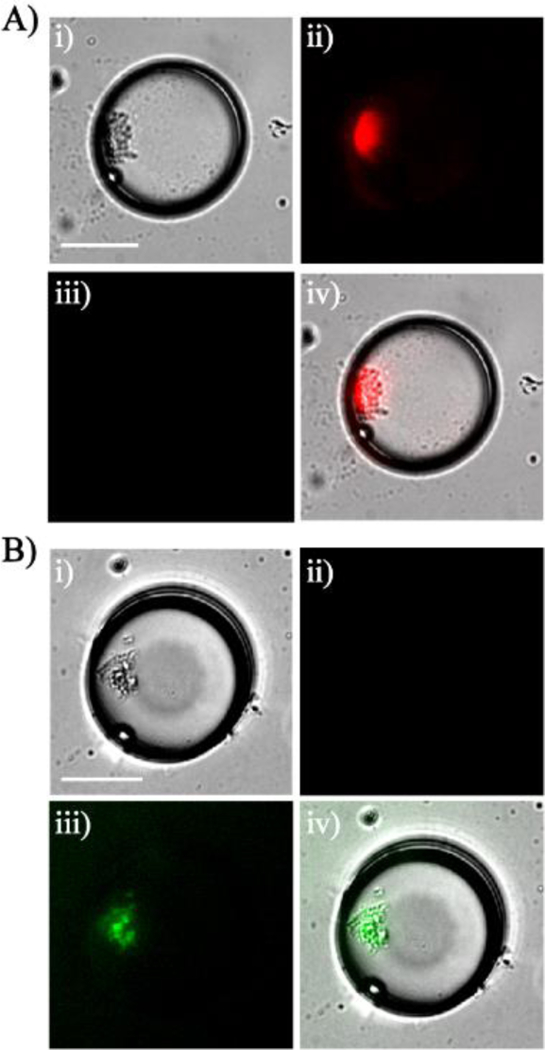
RE-doped NPs encapsulated in aqueous droplets in a microfluidic device. Encapsulation of Eu3+-doped NPs (A) and Tb3+-doped NPs (B). Images are: (i) brightfield, (ii) fluorescence with filter set 1, (iii) fluorescence with filter set 2 and, (iv) overlay. Scale bar is 35 µm.
The majority of fluorescent probes derived from fluorescein, rhodamine, cyanine, boron-dipyrromethene (BODIPY), acridine, xanthene, and coumarin have green and red emission chromophores.[35] Fluorophores that emit in the green and red have several advantages in terms of high excitation coefficients and high fluorescent quantum yield,[35, 46] thus making the two colors preferable. Hence, in this work, green and red fluorophores [green fluorescent protein (GFP) and red fluorescent protein (RFP)] and red emitting viability stain (EthD-1) were tested as an example to highlight the utility of this tracking platform. Eu 3+-doped NPs were co-encapsulated with GFP-expressing HeLa cells to observe their distinct fluorescent signals. The distant positions of selected emission lines for Eu3+ (centred at 617 nm, Fig. 2D) and GFP (centred at 509 nm) resulted in no spectral overlap between the Eu3+-doped NPs and the GFP-expressing HeLa cells (Fig. 5A). A similar experiment was performed with RFP-expressing MDA-MB-231 (emission peak centred at 588 nm). Although the selected Eu3+ emission peaks fell within the range of rhodamine filter collecting the RFP signal (λem = 545–625 nm), no Eu3+ signal was detected in this filter (see ESM Fig. S2). This is due to the fact that the excitation wavelength of rhodamine filter (λex: 536–556 nm) being outside of the Eu3+ excitation window, unlike with quantum dots where the broad absorption windows of QDs overlap with that of many biochemical stains.[47, 48]
Fig. 5.
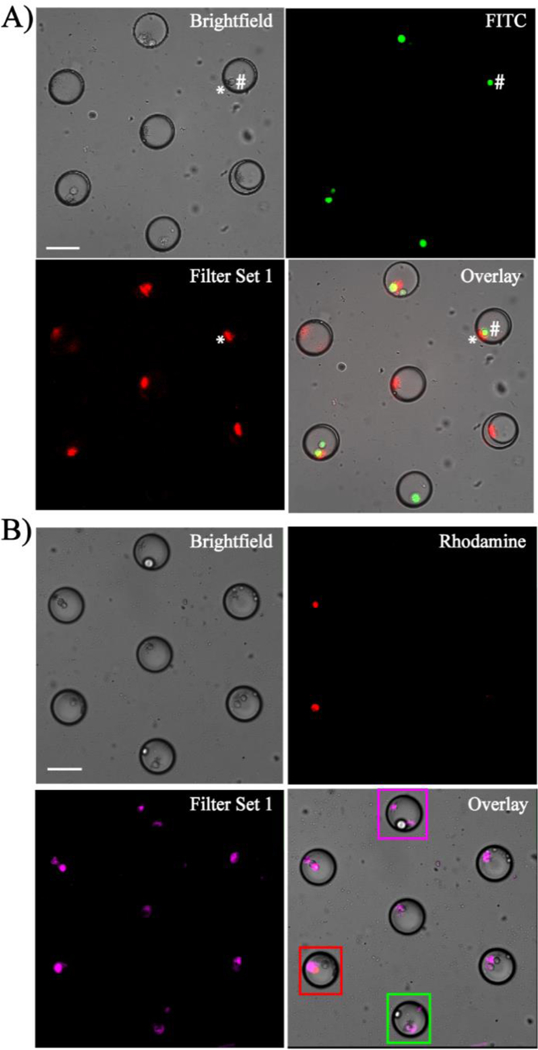
Spectral independence of Eu3+-doped NPs with fluorescently-tagged cells observed in a microfluidic device. (A) Eu3+-doped NPs co-encapsulated with GFP-HeLa cells. * denotes NPs and # denotes GFP-HeLa cell across the droplet. (B) Co-encapsulation of EthD-1 (dead stain)-stained MDA-MB-231 cells with Eu3+-doped NPs. Dead cells are shown in red and live cells remained colourless. Magenta box identifies a droplet with only NPs, red box identifies a dead cell with NPs, and green box shows a live cell with NPs. In (B), Eu3+-doped NPs are depicted in magenta to distinguish from the dead cells (red). Scale bar is 70 µm.
To validate the feasibility of luminescent NPs as droplet trackers, a mixed population of live and dead MDA-MB-231 cells were stained with EthD-1 and co-encapsulated with Eu3+-doped NPs in the microfluidic device (Fig. 5B). The microfluidic trapping array was easily able to compartmentalize single MDA-MB-231 cells and NPs within droplets, thus enabling clear tracking of single cells based on their viability. The concentration of EthD-1 played a key role in obtaining a spectrally independent signal from the dead cells, with 2 µM identified as the optimal concentration due to a slight spectral bleed-through observed due to broad excitation window of the EthD-1 at higher concentrations. The results observed with Tb3+-doped NPs are shown in Fig. S3 (see ESM). Although the selected Tb3+ peak (centered at 543 nm) fell within the emission range of FITC filter collecting the GFP signal (λem = 497–557 nm), no NP signal was detected from the FITC filter due to its higher excitation window. Additionally, no overlap was observed between Tb3+-doped NPs and RFP or EthD-1-stained cells due to their non-interfering ranges of emission filters (ESM Fig. S3). Further, it was observed that the Eu3+- and Tb3+-doped NPs were only detected in their respective filter sets. This highlights their potential to simultaneously track cells with two input conditions on a multiplexed microfluidic trapping array platform which is outside the scope of this paper.
Quantification of Spectral Independence between Fluorescent stains and RE-doped NPs
All droplet generation experiments yielded a ~99% droplet trapping efficiency with each NP - encapsulation experiment (Fig. 4), resulting in ~98% of the droplets containing NPs and ~2% empty droplets. The fluorescent microscopy images from all cell/NP co-encapsulation experiments were processed and analysed manually to categorize four different droplet subpopulations: (1) droplets with NP aggregates, droplets with cells, (3) droplets with NP and cells, and (4) empty droplets. Single cell encapsulation efficiencies from cases 2 and 3 followed a Poisson distribution as previously described. [49] An example analysis from three GFP-expressing HeLa and Eu3+ doped NP co-encapsulation experiments (n=787 droplets from each experiment), yielded an average of 28±1 (4%) empty droplets, 310±36 (43%) droplets with NPs, 356±37 (50%) droplets with NPs and cells and 21±7 (3%) droplets with cells (Figure. 6). A similar distribution was observed for all cell/NPs co-encapsulation experiments, subjected to no deviations from the specified cell density and NP concentration values. Until now, all the above reported results have been qualitatively evaluated by visual inspection of the cells and NPs within the aqueous droplets. In effort to confirm that the signals observed were distinct and reproducible, the fluorescent and luminescent signals of the cells and NPs from their respective filter sets were measured across each of the droplet diameter. Results from GFP-HeLa/Eu3+-doped NP co-encapsulation and RFP-MDA-MB-231/Eu3+-doped NP co-encapsulation experiments are shown in Fig. 7A-B. Fig. 7A-i represents a droplet with only a GFP-HeLa cell, where the cell gave a distinct signal in the FITC filter. Here, no signal was detected from the filter set 1 due to the absence of Eu3+-doped NPs. In 7A-ii, a droplet with single GFP-HeLa cell co-encapsulated with Eu3+-doped NPs is shown. The cells and NPs exhibited distinct emission spectra contributing to clearly defined and distinct signals from their respective filter sets. Similarly, for an RFP-MDA-MB-231/Eu3+ co-encapsulation experiment, the cells and NPs gave distinct signals from their respective filter sets as shown in Fig. 7B (RFP from rhodamine filter and Eu3+ NPs from filter set 1). Next, a population of live and dead MDA-MB-231 cells were stained with EthD-1 and co-encapsulated with Eu3+-doped NP inside droplets. The outcome of this experiment was categorized into six droplet subpopulations: droplets with 1) only NPs, 2) only live cell(s), 3) only dead cell(s), 4) live cell(s) with NPs, 5) dead cell(s) with NPs, and 6) a combination of live and dead cells with NPs co-encapsulated inside the same droplet. Example cases from subpopulations (4) and (5) are shown in Fig. 7C. In the case of co-encapsulating a live cell and NPs (Fig. 7C-i), the Eu3+ NP gave a prominent signal from filter set 1 while the live cells did not contribute to any fluorescent signal in rhodamine filter due to the lack of EthD-1 uptake. Conversely for a droplet co-encapsulating a dead cell and NP, the dead cell (presence of EthD-1) and Eu3+-doped NP gave distinct signals in their respective filter sets (Fig. 7C-ii). Similar analyses for spectral independence of Tb3+-doped NPs with GFP, RFP, EthD-1 are represented in Fig. S4 (see ESM).
Fig. 6.
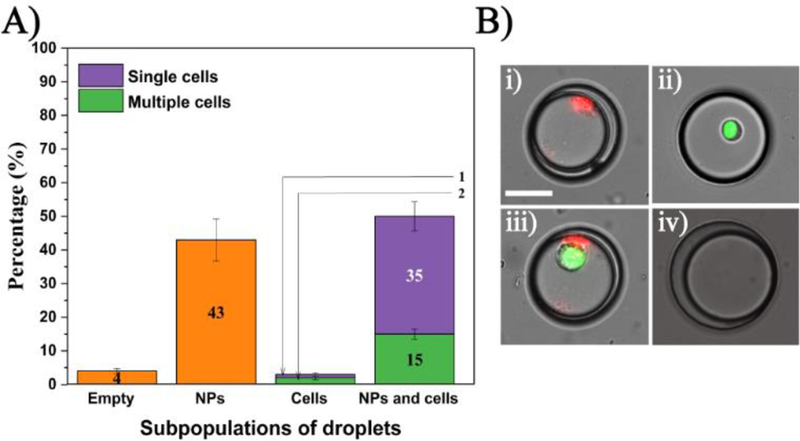
Distribution of co-encapsulated subpopulations of cells and NPs in trapped droplets. (A) Percentage of each subpopulation (n = 2325 droplets total). B) Images of each subpopulation including droplets with: (i) NP, (ii) cell, (iii) NP and cells, and (iv) empty droplet. Scale bar is 35 µm.
Fig. 7.
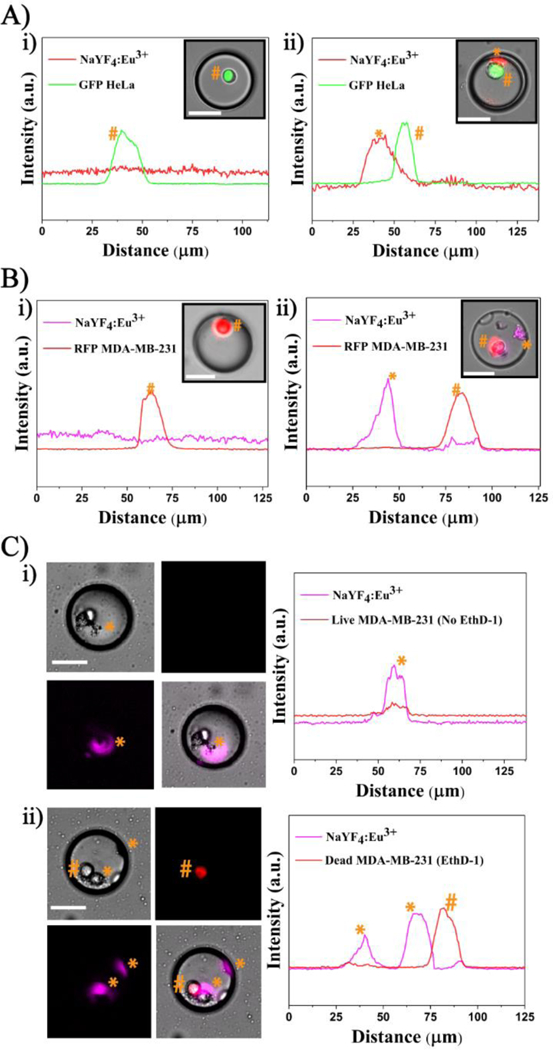
Spectral independence of Eu3+-doped NP with common fluorescent markers: (A) GFP and (B) RFP. Line scan across droplets showing a significant signal from the GFP-HeLa cell in (A)-i; spectral independence of Eu3+ NP with GFP- HeLa cell in (A)-ii. RFP MDA-MB-231 cell in (B)-i; RFP MDA-MB-231 cell with Eu3+ NP in (B)-ii. (C) A population of live and dead MDA-MB-231 cells co-encapsulated with Eu3+-doped NP. Line scan across a droplet having a live cell with NP in (C)-i and dead cell with NP in (C)-ii. * denotes NP and # denotes cell and their corresponding signal intensities across the droplet. Note: Eu3+ NPs are depicted in magenta to distinguish from RFP and dead stain (red). Scale bar is 35
Numerical validation of spectral independence between luminescent NPs and fluorophores was performed. A sample size of n=50 droplets was analysed for each of the subpopulations shown in Fig. 7 and ESM Fig. S4. The acquired fluorescence intensities of cells and NPs, in terms of NFS, from all filter sets were recorded and compared with each other in terms of S:N ratio values. Tables S1 and S2 (see ESM) show the spectral independence validation of Eu3+-doped NPs and Tb3+-doped NPs with GFP, RFP and EthD-1. For different droplet subpopulations co-encapsulating fluorescent cells and Eu3+-doped NPs, the NPs had a prominent average S:N ratio value of ~5:1 only observable with filter set 1 (ESM Table S1). Similarly, the fluorescent cells recorded distinct average S:N ratio values from only their respective filter sets: GFP in FITC filter, RFP and EthD-1 in rhodamine filter, respectively. Similar numerical validation was done for the spectral independence of Tb3+-doped NPs (ESM Table S2). The Tb3+-doped NP had a distinct average S:N ratio value of ~3:1 from only its filter set 2, while the fluorescent cells recorded distinct signals from only their filter sets. Moreover, the Eu3+-doped NPs could not be detected above the noise using filter set 2 and likewise for Tb3+-doped NPs with filter set 1. Thus, these measurements demonstrate the potential of using RE doped NPs as a probe to track multiple input conditions on a single platform. Moreover, statistical assessment of the average S:N values for the two RE3+-doped NPs and fluorescent cells were performed using single/two-tailed hypotheses tests to determine the average threshold S:N range of luminescent NPs and fluorescent cells collected from all filter sets. The resultant p - values of the hypotheses tests from Tables S3–S5 and S6–S8 (see ESM) highlight the distinct emission spectra from RE3+-doped NPs, GFP, RFP and EthD-1. Specifically, the p-values highlight the clear and consistent identification of NPs and cells based on luminescent spectra and their ability to act as secondary identifiers for potential applications in droplet trackers for multiplexed platforms and other biological systems.
Dose Response Analysis of MDA-MB-231 cells to Paclitaxel using Luminescent NPs
Single cell analysis techniques have been instrumental in identifying low-occurrence, drug resistant subpopulation of cells. Microfluidic droplet trapping arrays can quantify the intracellular distribution of fluorescence across single cells and directly visualize cells to minimize the number of false positives which occur during flow cytometry studies. However, tracking the inputs into these devices has been limited by overlapping fluorophores used for droplet tracking and biological interrogation. Thus, upon numerical validation and statistical characterization of spectral properties of the two luminescent NPs, the capabilities of the microfluidic droplet tracking platform were evaluated for a real time application quantifying the single cell responses to different therapeutic doses. MDA-MB-231 cells were treated off-chip with three different doses of Paclitaxel (PTX), a microtubule inhibitor, for three different incubation times (24 h, 48 h, or 72 h). The single cell dose response was tracked in the device using three corresponding NPs (Eu3+-doped NP for 10 µM, Tb3+-doped NP for 50 µM, and undoped NP for 100 µM PTX) in the microfluidic droplet array. The experiments at each of the three incubations times were performed simultaneously on the fluorescent microscopy stage to demonstrate the ability of the luminescent materials to discretely track cell viability after drug treatment with three different doses over a period of 72 h. A representative image of droplet tracking from the 48 h time point is shown in Fig. S5 (see ESM), where a heterogeneous response was observed in terms of live cells (colourless due to the absence of EthD-1 uptake) and dead cells (red due to EthD-1 uptake) as tracked by Eu3+-doped NP for 10 µM PTX (ESM Fig. S5A), Tb3+-doped NP for 50 µM PTX (ESM Fig. S5B), and undoped NP for 100 µM (ESM Fig. S5C). To determine if the results from these tracking experiments conformed with previously studied drug effects on MDA -MB-231 cells, the resultant heterogeneous dose-dependent cellular responses from duplicate droplet tracking-drug experiments were compared with no drug positive control experiments. Fig. 8 shows the mean MDA-MB-231 cell viability data (in percentage) with standard deviations for the three different concentrations of PTX tracked by three corresponding NPs and no-drug positive control at different time points. For the tracked data, the cell viability gradually decreased based on dose and time dependent treatment for the three drug concentrations: 103 ± 40 viable cells out of 142 ± 48 cells (71.34%) to 128 ± 95 viable cells out of 212 ± 140 cells (54.59%) for the Eu3+-doped NP tracked-10 µM PTX, 147 ± 69 viable cells out of 260 ± 105 cells (54.69%) to 136 ± 58 viable cells out of 287 ± 66 cells (40.46%) for the Tb 3+-doped NP-tracked 50 µM PTX and 50 ± 1 viable cells out of 116 ± 27 cells (45.01%) to 32 ± 6 viable cells out of 124 ± 8 cells (21.63%) for the undoped NP tracked-100 µM PTX during 24h to 72 h. Alternatively, healthy cell viability average of around ~85% was observed throughout for the no-drug control population. The three different dose responses were statistically compared with the positive control with a null hypothesis being the average difference of cell viability between dose-dependent responses and control is zero. After a series of one-tailed t-test (with α=0.05), the null hypothesis was rejected in all cases based on the significant p-values as shown in Fig. 8. The findings from these droplet tracking studies were similar to previously reported drug studies.[50] These findings demonstrate the potential of luminescent nanomaterials coupled with a microfluidic droplet trapping array as an approach to provide insight on therapeutic effectiveness and dose-dependent heterogeneous single cell response at the same time. While the studies presented here were limited by a single-input microfluidic droplet trapping, the luminescent nanomaterials characterized here can be incorporated into multiple input droplet microfluidic devices[51–53] to exploit their full potential to track droplets, the results of which will be reported in the future. Nevertheless, the successful results from the single-input droplet tracking experiments emphasizes the capability of expanding this in multiplicity by combining multi-inputs and similarly doped NPs to simultaneously track different input conditions at the same time.
Fig. 8.
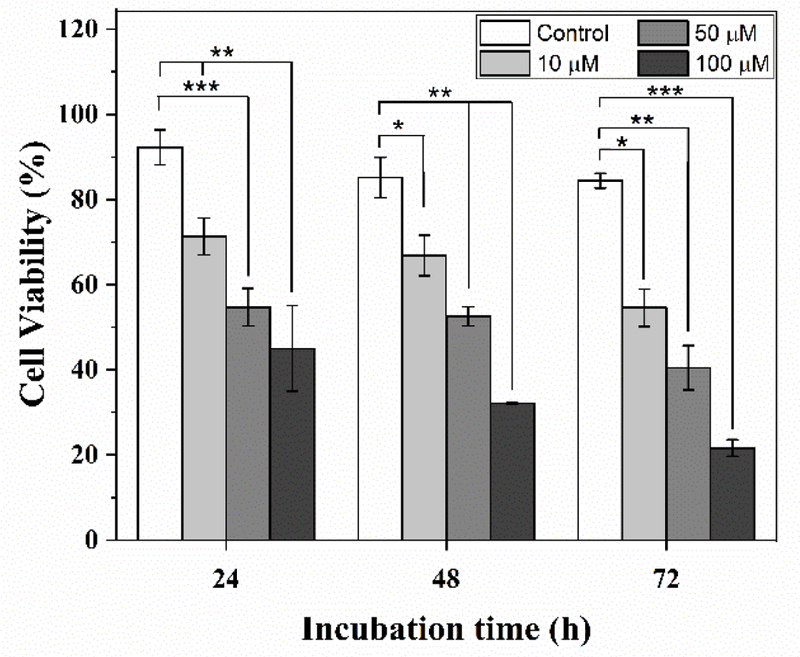
Dose and time dependent viability response of MDA-MB-231 cells to PTX. Cells were treated with 10 µM, 50 µM and 100 µM PTX off-chip and observed on-chip at 24 h, 48 h and 72 h using droplet microfluidic tracking system. Significant differences compared to no-drug positive control treatment. *p<0.1, **p<0.05, ***p<0.01 are indicated.
Conclusions
A series of droplet tracking experiments co-encapsulating RE (Eu3+ and Tb3+)-doped NPs and fluorescently labelled cells was successfully performed using a single-input microfluidic device. This platform utilized low-cost luminescent RE-doped NPs in a droplet microfluidic device instead of expensive, overlapping barcodes (antibodies and dyes) to demonstrate, for the first time, their ability to track droplets in a spectrally independent manner using fluorescent microscopy. This work found that the DC emission spectra of Eu3+-doped NP (in red) and Tb3+-doped NP (in green) did not overlap with common fluorophores (GFP, RFP) and fluorescent biochemical stain (EthD-1), thus overcoming the limitations of commonly used tracking materials. The fluorescence and luminescence signals of cells and NPs were quantified for spectral independence through a series of signal measurements and statistical single/two-tailed hypotheses tests. As a result of this robust quantification, the spectral independence of these RE - doped NPs was well ascertained. Furthermore, these biocompatible and spectrally independent RE-doped luminescent NPs were used to quantify single cell dose response of MDA-MB-231 cells to PTX, where each of the NPs was able to visually isolate and distinctly track single cancer cells challenged with specific doses of drug. The strength of this tracking system to perform both population-based and single-cell analysis of cancer cells to identify distinct subpopulations of cells, including drug resistant ones was highlighted. While the data presented in this paper utilized only two RE dopants in a single-input microfluidic droplet trapping array, this novel nanoparticle-microfluidic hybrid platform has significant potential for multiplexing to increase the number of parameters which are simultaneously screened and trapped for an improved statistical response. Additionally, this hybrid platform poses a clear advantage to perform multiplexed, dynamic cellular measurements including the intracellular distribution of biochemical stains.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institute of Biomedical Imaging and Bioengineering, R03EB02935 (ATM), the National Science Foundation, CBET1509713 (ATM), and the Louisiana Board of Regents, LEQSF (2016–19)-RD-A-03, (JAD/PD). The authors would like to thank Dr. Nancy Albritton (University of North Carolina) for providing the GFP-expressing HeLa cells and Dr. Elizabeth Martin (LSU) for providing the MDA-MB-231 cells and RFP-expressing MDA-MB-231 cells. The authors would like to thank Riad Elkhanoufi (LSU) for assistance with device fabrication.
Biographies

Manibarathi Vaithiyanathan is a 4th year Ph.D. student working in Dr. Melvin’s Lab at Louisiana State University. Her research interest focuses on developing experimental and computational tools for single cell analysis to address healthcare (cancer diagnosis) and environmental issues. Currently, she is developing a microfluidic platform to understand phosphorus fixation in single algae cells.

Khashayar R. Bajgiran is a 3rd year PhD student at Louisiana State University Cain Department of Chemical Engineering, where he works at Melvin’s and Dorman’s lab. His area of focus is on luminescent nanoparticles and droplet microfluidics for cancer drug screening and sensing applications. Currently he is working on developing a platform capable of multiplex screening of cancer single cells response to chemotherapeutics.

Pragathi Darapaneni is a graduate student working under Dr. James Dorman in Louisiana State University. Her research is focussed on tuning the optoelectronic properties of transition metal doped solids using weak fields so as to deploy them in the rare earth based applications such as LEDs, bio-probes, and anti-counterfeit technologies.

Nora Safa is a 5th year PhD student at LSU Cain Department of Chemical Engineering, set to graduate in December 2018. Her work involves developing a library of short sequence cell penetrating peptides, a droplet microfluidic platform and a unique cell-permeable reporter to facilitate screening of DUB activity in single cancer cells. Her work has direct application in drug discovery and point of care diagnostics.

Adam Melvin is an Assistant Professor in the Cain Department of Chemical Engineering at Louisiana State University. Dr. Melvin received his Ph.D. in Chemical Engineering from North Carolina State University and was an NIH postdoctoral fellow at the University of North Carolina at Chapel Hill in the Departments of Chemistry and Biomedical Engineering. His current research interests include biochemical engineering, microfluidics, single cell analysis, chemical biology, and cancer metastasis.

James A Dorman is an Assistant Professor in Chemical Engineering at Louisiana State University. His research interests are on the luminescent and electronic properties of rare earth and transition metal doped oxide nanomaterials. His work with these materials has focused on engineering energy transfer mechanisms and recombination pathways for tailored emission spectra and enhancing luminescent efficiencies.
Footnotes
Compliance with Ethical Standards
Conflicts of Interest
There are no conflicts to declare.
References
- 1.Saadatpour A, Lai S, Guo G, Yuan G-C. Single-cell analysis in cancer genomics. Trends Genet 2015;31(10):576–86. 10.1016/j.tig.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tellez-Gabriel M, Ory B, Lamoureux F, Heymann M-F, Heymann D. Tumour Heterogeneity: The Key Advantages of Single-Cell Analysis. Int J Mol Sci 2016;17(12):2142 10.3390/ijms17122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsoucas D, Yuan G-C. Recent progress in single-cell cancer genomics. Curr. Opin. Genet. Dev 2017;42:22–32. 10.1016/j.gde.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider T, Kreutz J, Chiu DT. The Potential Impact of Droplet Microfluidics in Biology. Anal. Chem 2013;85(7):3476–82. 10.1021/ac400257c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang L, Cheng Y, Zhao Y. Emerging Droplet Microfluidics. Chem. Rev 2017;117(12):7964–8040. 10.1021/acs.chemrev.6b00848. [DOI] [PubMed] [Google Scholar]
- 6.Newell EW, Cheng Y. Mass cytometry: blessed with the curse of dimensionality. Nat. Immunol 2016;17:890 10.1038/ni.3485. [DOI] [PubMed] [Google Scholar]
- 7.O’Donnell EA, Ernst DN, Hingorani R. Multiparameter Flow Cytometry: Advances in High Resolution Analysis. Immune Netw 2013;13(2):43–54. 10.4110/in.2013.13.2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyatt Shields Iv C, Reyes CD, Lopez GP. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015;15(5):1230–49. 10.1039/C4LC01246A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu P, Zhang W, Xin H, Deng G. Single Cell Isolation and Analysis. Front Cell Dev Biol 2016;4:116 10.3389/fcell.2016.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell 2016;165(4):780–91. 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single-cell technologies for monitoring immune systems. Nat. Immunol 2014;15(2):128–35. 10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Basu S, Wohlfahrt KJ, Lee SF, Klenerman D, Laue ED et al. A microfluidic platform for trapping, releasing and super-resolution imaging of single cells. Sens. Actuator B-Chem 2016;232:680–91. 10.1016/j.snb.2016.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holton AB, Sinatra FL, Kreahling J, Conway AJ, Landis DA, Altiok S. Microfluidic Biopsy Trapping Device for the Real-Time Monitoring of Tumor Microenvironment. PLOS ONE 2017;12(1):e0169797 10.1371/journal.pone.0169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 2017;12(1):44–73. 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- 15.Ma J, Zhan L, Li RS, Gao PF, Huang CZ. Color-Encoded Assays for the Simultaneous Quantification of Dual Cancer Biomarkers. Anal Chem 2017;89(16):8484–9. 10.1021/acs.analchem.7b02033. [DOI] [PubMed] [Google Scholar]
- 16.Lan F, Haliburton JR, Yuan A, Abate AR. Droplet barcoding for massively parallel single-molecule deep sequencing. Nat. Commun 2016;7:11784 doi: 10.1038/ncomms11784 . 10.1038/ncomms11784https://www.nature.com/articles/ncomms11784#supplementary-information. https://www.nature.com/articles/ncomms11784#supplementary-information . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015;161(5):1187–201. 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C-H, Miller MA, Sarkar A, Beste MT, Isaacson KB, Lauffenburger DA et al. Multiplexed Protease Activity Assay for Low Volume Clinical Samples Using Droplet Based Microfluidics and Its Application to Endometriosis. J. Am. Chem. Soc 2013;135(5):1645–8. 10.1021/ja307866z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rane TD, Zec HC, Wang TH. A barcode-free combinatorial screening platform for matrix metalloproteinase screening. Anal Chem 2015;87(3):1950–6. 10.1021/ac504330x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J, Hu M, Zhan P, Peng X. Energy transfer cassettes based on organic fluorophores: construction and applications in ratiometric sensing. Chem. Soc. Rev 2013;42(1):29–43. 10.1039/C2CS35273G. [DOI] [PubMed] [Google Scholar]
- 21.Abdel-Mottaleb MMA, Beduneau A, Pellequer Y, Lamprecht A. Stability of fluorescent labels in PLGA polymeric nanoparticles: Quantum dots versus organic dyes. Int. J. Pharm 2015;494(1):471–8. 10.1016/j.ijpharm.2015.08.050. [DOI] [PubMed] [Google Scholar]
- 22.Montón H, Nogués C, Rossinyol E, Castell O, Roldán M. QDs versus Alexa: reality of promising tools for immunocytochemistry. J Nanobiotechnology 2009;7(1):4 10.1186/1477-3155-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen C-Y, Xie H-Y, Zhang Z-L, Wu L-L, Hu J, Tang M et al. Fluorescent/magnetic micro/nano-spheres based on quantum dots and/or magnetic nanoparticles: preparation, properties, and their applications in cancer studies. Nanoscale 2016;8(25):12406- 10.1039/C5NR08534A. [DOI] [PubMed] [Google Scholar]
- 24.Leng Y, Wu W, Li L, Lin K, Sun K, Chen X et al. Magnetic/Fluorescent Barcodes Based on Cadmium-Free Near-Infrared-Emitting Quantum Dots for Multiplexed Detection. Adv. Funct. Mater 2016;26(42):7581–9. 10.1002/adfm.201602900. [DOI] [Google Scholar]
- 25.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat. Biotechnol 2004;22:969 10.1038/nbt994. https://www.nature.com/articles/nbt994#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 26.Duan N, Wu S, Yu Y, Ma X, Xia Y, Chen X et al. A dual-color flow cytometry protocol for the simultaneous detection of Vibrio parahaemolyticus and Salmonella typhimurium using aptamer conjugated quantum dots as labels. Anal. Chim. Acta 2013;804:151–8. 10.1016/j.aca.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 27.Buranda T, Wu Y, Sklar LA. Quantum dots for quantitative flow cytometry. Methods Mol Biol 2011;699:67–84. 10.1007/978-1-61737-950-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghrera AS, Pandey CM, Ali MA, Malhotra BD. Quantum dot-based microfluidic biosensor for cancer detection. Appl. Phys. Lett 2015;106(19):193703 10.1063/1.4921203. [DOI] [Google Scholar]
- 29.Wegner KD, Hildebrandt N. Quantum dots: bright and versatile in vitro and in vivo fluorescence imaging biosensors. Chem. Soc. Rev 2015;44(14):4792–834. 10.1039/C4CS00532E. [DOI] [PubMed] [Google Scholar]
- 30.Gai SL, Li CX, Yang PP, Lin J. Recent Progress in Rare Earth Micro/Nanocrystals: Soft Chemical Synthesis, Luminescent Properties, and Biomedical Applications. Chemical Reviews 2014;114(4):2343–89. 10.1021/cr4001594. [DOI] [PubMed] [Google Scholar]
- 31.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008;5:763 doi: 10.1038/nmeth.1248 . 10.1038/nmeth.1248https://www.nature.com/articles/nmeth.1248#supplementary-information. https://www.nature.com/articles/nmeth.1248#supplementary-information . [DOI] [PubMed] [Google Scholar]
- 32.Dong H, Du SR, Zheng XY, Lyu GM, Sun LD, Li LD et al. Lanthanide Nanoparticles: From Design toward Bioimaging and Therapy. Chem. Rev 2015;115(19):10725–815. 10.1021/acs.chemrev.5b00091. [DOI] [PubMed] [Google Scholar]
- 33.Haase M, Schafer H. Upconverting Nanoparticles. Angewandte Chemie-International Edition 2011;50(26):5808–29. 10.1002/anie.201005159. [DOI] [PubMed] [Google Scholar]
- 34.Eliseeva SV, Bunzli JCG. Lanthanide luminescence for functional materials and bio-sciences. Chem. Soc. Rev 2010;39(1):189–227 10.1039/b905604c. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira E, Bértolo E, Núñez C, Pilla V, Santos HM, Fernández‐Lodeiro J et al. Green and Red Fluorescent Dyes for Translational Applications in Imaging and Sensing Analytes: A Dual‐Color Flag. ChemistryOpen 2018;7(1):9–52. 10.1002/open.201700135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Park HJ, Park C-S, Oh E-T, Choi B-H, Williams B et al. Response of Breast Cancer Cells and Cancer Stem Cells to Metformin and Hyperthermia Alone or Combined. PLoS ONE 2014;9(2):e87979 10.1371/journal.pone.0087979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Liu Q, Feng W, Sun Y, Li F. Upconversion Luminescent Materials: Advances and Applications. Chem. Rev 2015;115(1):395–465. 10.1021/cr400478f. [DOI] [PubMed] [Google Scholar]
- 38.Chen G, Qiu H, Prasad PN, Chen X. Upconversion Nanoparticles: Design, Nanochemistry, and Applications in Theranostics. Chem. Rev 2014;114(10):5161–214. 10.1021/cr400425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li CX, Yang J, Quan ZW, Yang PP, Kong DY, Lin J. Different microstructures of ss-NaYF4 fabricated by hydrothermal process: Effects of pH values and fluoride sources. Chem. Mater 2007;19(20):4933–42. 10.1021/cm071668g. [DOI] [Google Scholar]
- 40.Wang F, Liu XG. Recent advances in the chemistry of lanthanide-doped upconversion nanocrystals. Chem. Soc. Rev 2009;38(4):976–89. 10.1039/b809132n. [DOI] [PubMed] [Google Scholar]
- 41.Dorman JA, Choi JH, Kuzmanich G, Bargar JR, Chang JP. Optimizing the crystal environment through extended x-ray absorption fine structure to increase the luminescent lifetimes of Er3+ doped Y2O3 nanoparticles. J. Appl. Phys 2012;111(8). 10.1063/1.3702789. [DOI] [Google Scholar]
- 42.Sabnis A, Rahimi M, Chapman C, Nguyen KT. Cytocompatibility studies of an in situ photopolymerized thermoresponsive hydrogel nanoparticle system using human aortic smooth muscle cells. J. Biomed. Mater. Res. Part B Appl. Biomater 2009;91(1):52–9. 10.1002/jbm.a.32194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong DY, Ranganath T, Kasko AM. Low-dose, long-wave UV light does not affect gene expression of human mesenchymal stem cells. PloS one 2015;10(9):e0139307 10.1371/journal.pone.0139307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naczynski DJ, Tan MC, Zevon M, Wall B, Kohl J, Kulesa A et al. Rare-earth-doped biological composites as in vivo shortwave infrared reporters. Nat. Commun 2013;4:2199 10.1038/ncomms3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang NJ, Hinner MJ. Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. Methods Mol. Bio 2015;1266:29–53. 10.1007/978-1-4939-2272-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu P, Yan X-P. Doped quantum dots for chemo/biosensing and bioimaging. Chem. Soc. Rev 2013;42(12):5489–521. 10.1039/C3CS60017C. [DOI] [PubMed] [Google Scholar]
- 47.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu. Rev. Biomed. Eng 2005;7:55–76. 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 48.Chan WCW, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr. Opin. Biotechnol 2002;13(1):40–6. 10.1016/S0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 49.Collins DJ, Neild A, deMello A, Liu A-Q, Ai Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip 2015;15(17):3439–59. 10.1039/C5LC00614G. [DOI] [PubMed] [Google Scholar]
- 50.Kern KM, Schroeder JR. Comparison of Cantharidin Toxicity in Breast Cancer Cells to Two Common Chemotherapeutics. Int. J. Breast Cancer 2014;2014:423059 10.1155/2014/423059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muluneh M, Kim B, Buchsbaum G, Issadore D. Miniaturized, multiplexed readout of droplet-based microfluidic assays using time-domain modulation. Lab Chip 2014;14(24):4638–46. 10.1039/C4LC00819G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu S-Q, Zhang Y-X, Zhu Y, Du W-B, Yao B, Fang Q. Multifunctional picoliter droplet manipulation platform and its application in single cell analysis. Anal. Chem 2011;83(19):7570–6. 10.1021/ac201678g. [DOI] [PubMed] [Google Scholar]
- 53.Bui M-PN, Li CA, Han KN, Choo J, Lee EK, Seong GH. Enzyme kinetic measurements using a droplet-based microfluidic system with a concentration gradient. Anal. Chem 2011;83(5):1603–8. 10.1021/ac102472a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


