Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant and lethal disease with few treatment options. Steroid receptor coactivator-3 (SRC-3, also known as NCOA3, AIB1, pCIP, ACTR, RAC3, TRAM1) sits at the nexus of many growth signaling pathways and has been pursued as a therapeutic target for breast, prostate and lung cancers. In this study, we find that SRC-3 is overexpressed in PDAC and inversely correlates with patient overall survival. Knockdown of SRC-3 reduces pancreatic cancer cell proliferation, migration and invasion in vitro. Additionally, inhibition of SRC-3 using either shRNA or a small molecule inhibitor can significantly inhibit tumor growth in orthotopic pancreatic cancer mouse models. Collectively, this study establishes SRC-3 as a promising therapeutic target for pancreatic cancer treatment.
1. INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant and lethal disease with poor prognosis. It is the twelfth most common type of cancer and the fourth leading cause of cancer-related death in the United States [1]. The 5-year survival rate is less than 8% and the median survival is around 6 months from the time of diagnosis [2]. It is now recognized that highly malignant and metastatic PDAC develops from local precursor lesions called pancreatic intraepithelial neoplasia (PanIN) [3]. Molecular and pathological analyses have revealed an ordered series of genetic alterations during the progression of PanIN to PDAC. In addition to the well-characterized signature somatic mutations in KRAS, TP53, CDKN2A and SMAD4 [4, 5], accumulating studies have revealed a large number of gene alterations, either at the genetic or expression levels, in PDAC [6]. A comprehensive genetic analysis of 24 PDAC cases found an average of 63 exonic alterations in this cohort and overexpression of 541 genes, which converge on 12 key cellular signaling pathways [7]. However, the contribution of most alterations other than KRAS, TP53, CDKN2A and SMAD4 to PDAC development is largely unknown.
Steroid receptor coactivators (SRCs), also known as nuclear receptor coactivators (NCOAs), are a family of transcription coactivators, including SRC-1 (also known as NCOA1), SRC-2 (also known as NCOA2, GRIP1, TIF2) and SRC-3 (also known as NCOA3, AIB1, pCIP, ACTR, RAC3, TRAM1) [8]. They interact with nuclear receptors (NRs) to recruit additional transcription co-regulators, such as histone modifiers and chromatin remodeling modules, acting as a scaffold for the assembly of multi-subunit coactivator holocomplexes [8]. In addition to NRs, SRCs are also co-activators for multiple transcription factors, such as NF-κB, Smad3, p53 and β-catenin/TCF and have been demonstrated to regulate various signaling pathways in both physiological and pathological contexts [8]. Each member of the SRC family has been reported to be overexpressed or amplified in multiple cancer types, particularly in steroid hormone-related cancers such as breast cancer [9, 10], ovarian cancer [10] and prostate cancer [11, 12]. Among the SRCs, SRC-3 is more frequently reported and more extensively studied in cancer than the other two members. Overexpression of SRC-3 mRNA has been reported in a range from 13% to 64% of breast cancers by different research groups [10, 13, 14]. SRC-3 also has been found to be amplified in a range between 4.8 to 9.5% in different breast cancer cohorts [10, 15]. Higher SRC-3 expression is associated with larger tumor size [10, 15], higher tumor grade [16] and poorer disease-free survival (DFS) in estrogen receptor-positive breast cancer patients treated with adjuvant tamoxifen [13]. Transgenic mouse models and cell line studies demonstrated that SRC-3 promotes cancer cell proliferation and tumor metastasis [17–21]. High SRC-3 expression or gene amplification also was documented in endometrial carcinoma [14, 22], ovarian cancer [10, 15], prostate cancer [23] and hepatocellular carcinoma [24], which was pathologically associated with high tumor grade, poor prognosis or DFS.
Overexpression and amplification of SRC-3 in pancreatic cancer was first reported by Henke et al. in 2004 [25]. Immunohistochemistry and in-situ hybridization showed a correlation between SRC-3 expression and PanIN/PDAC progression. SRC-3 expression was detected rarely in normal pancreas tissues (4.5%), but with an increased frequency in pancreatitis (14.3%) and low-grade PanIN (23.1%). Even higher frequencies and levels were found in high-grade PanIN (81.1%) and PDAC (64.5%) samples. Amplification of the SRC-3 gene was found in 37% of PDAC specimens. The association of SRC-3 expression with tumor progression in pancreatic lesions implicates SRC-3 in PDAC development. However, no further evidence was reported since then. Given the oncogenic role of SRC-3 and its association with prognosis in a variety of cancers, we propose that SRC-3 contributes to PDAC oncogenesis and progression and that it should be considered as a therapeutic target for pancreatic cancer treatment.
2. MATERIALS AND METHODS
2.1. Patient data analysis
We obtained mRNA expression data and patient survival data from Firehose (https://gdac.broadinstitute.org/). We evaluated the correlations between mRNA expression of SRC3 gene and Pancreatic Adenocarcinoma (PADD) patients’ overall survival time. The 1/3 samples with the highest gene expression were defined as “SRC-3 high” high group (N = 59), and the 1/3 samples with the lowest gene expression were defined as “SRC-3 low” group (N = 60), and these two groups were used in survival analysis. The R package “survival” was used to perform the overall survival analysis and produce Kaplan-Meier survival plots. A log-rank test was used to assess the significance.
2.2. Cell lines
PANC-1, Capan-1, Capan-2, and AsPc-1 cell lines were purchased from American Type Culture Collection (ATCC). HPDE, CFPAC, HPAC, HS766T, Panc-28, BxPC3, L3.6PL, MDA-PCAT, Miapaca-2 and MPanc-96 cell lines were courtesy of Dr. Craig Logsdon lab. A549, H1975, H1299 and MCF-7 cell were kindly provided by Dr. David Lonard’s laboratory. All cell lines in the experiments were validated by short tandem repeat (STR) DNA analysis and were negative for mycoplasma.
HPDE cells were grown in keratinocyte serum-free medium (ThermoFisher Scientific Inc. Cat#17005–042, Ref.). PANC-1, Capan-1, Capan-2, CFPAC, HPAC, HS766T, Panc-28, L3.6PL, MDA-PCAT, Miapaca-2, MPanc-96, A549 and MCF-7 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM). The left AsPc-1, BxPC3, H1975 and H1299 cells were maintained in RPMI-1640 medium. Both DMEM and RPMI-1640 medium were supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 mg/mL streptomycin. All cell lines were cultured in a humidified incubator containing 5% CO2 at 37°C.
Stable cell lines PANC-1-shCtrl, PANC-1-shSRC-3, and PANC-1-Ind-shSRC-3 were made by two steps of lentiviral infection to fulfil the experimental purposes for both in vitro tests and in vivo mouse models. In the first step, PANC-1 cells were infected by lentivirus made from a luciferase expression plasmid, pLenti-CMV-V5-LUC-Blast (from Addgene Co.), and then the stable cells were selected under culture medium containing 10 μg/mL of blasticidin. In the second step, PANC-1 cells with stable luciferase expression were infected with lentivirus made from plasmids, pLKO-shScramble, pLKO-shSRC-3.1, pLKO-shSRC-3.2, and pLKO-Tet-OnshSRC-3 to obtain PANC-1-shCtrl, PANC-1-shSRC-3.1, PANC-1-shSRC-3.2, and PANC-1-Ind-shSRC-3 stable cells, respectively. Condition medium containing 2 μg/mL of puromycin was used to select cells that has pLKO plasmid integration. Capan-2-shCtrl, Capan-2-shSRC-3.1 and Capan-2-shSRC-3.2 stable cells were established by single infection of the corresponding pLKO lentiviruses described above.
2.3. Plasmids and antibodies
SRC-3 specific shRNA expression plasmids pLKO-shSRC-3.1 with targeting sequence 5’-TTCCACCTCCTAGGGATATAA-3’, and pLKO-shSRC-3.2 with targeting sequence 5’-GGATCAGAAGGCAGGATTATA-3’, as well as pLKO-shScramble were purchased from Sigma-Aldrich. For the purpose to induce the shSRC-3 expression by doxycycline, pLKO-Tet-On-shSRC-3 plasmid was constructed by inserting an oligo fragment containing the same targeting sequence as pLKO-shSRC-3.2 into the pLKO-Tet-On vector through AgeI and EcoRI restrict sites. Antibodies, anti-SRC–3 (Cat# 2126), anti-Ki-67 (Cat# 9027), anti-E-Catherin (Cat# 3195), anti-N-Cadherin (Cat# 13116), anti-MMP-2 (Cat# 13132), anti-MMP-7 (Cat# 71031), anti-CDK6 (Cat# 3136), anti-Cyclin D1(Cat# 2978), anti-Cyclin D3 (Cat# 2936) and anti-β-Actin (Cat# 8457) were all obtained from Cell Signaling Technology Inc.. Antibodies, anti-β-Catenin (Cat# sc-7199), anti-c-Myc (Cat# sc-42), anti-E2F1 (Cat# sc-251), anti-Cyclin A (Cat# sc-53231), and anti-Bcl-2 (Cat# sc-783) were all obtained from Santa Cruz Biotechnology Inc.
2.4. Quantitative RT-PCR
RNA was isolated from cultured cells with TRIzol reagent. 1μg RNA was reverse transcribed into cDNA with iScript reverse transcription supermix (Bio-Rad). Relative gene expression was analyzed using quantitative PCR. PCR was performed with iTaq Universal SYBR® Green Supermix (Bio-Rad) using a Bio-Rad iCycler. Genes were amplified with specific primers by an initial denaturation at 95°C for 5 min, followed by 4 0 cycles at 95°C for 30 sec and 60°C for 1 min. Relative gene e xpression levels was analyzed using the 2−ΔΔCT method. RPS6 gene served as an internal reference.
2.5. Western blotting
Cultured cells were lysed with a radioimmunoprecipitation assay (RIPA) buffer containing 1x complete protease inhibitor cocktail (Roche). Protein concentration was determined by Braford assay. Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes. Membranes were blocked with 5% non-fat dry milk for 1hr at room temperature and then incubated with primary antibody diluted in 5% non-fat dry milk (rabbit anti-SRC-3 1:1000, rabbit anti-β-actin 1:5000) at 4°C overnight. After washing with PBST, blots were incubated in anti-rabbit IgG-HRP (1:10000) in 5% non-fat dry milk for 1hr at room temperature. Blotting signals were detected with enhanced chemiluminescent (ECL) detection reagent (GE Healthcare).
2.6. Cell proliferation and MTT assays
For each tested cell line, 2,000 cells were seeded in each well of a 96-well plate and cell viability was determined by MTT assay. For MTT assays, cells were covered with 100 μL of DMEM containing 500 μg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). After incubation at 37 °C in the i ncubator for 1 to 3 hours, 100 μL of DMSO were used to dissolve converted formazan and its concentration was determined by absorbance spectrophotometry at a wavelength of 570 nm.
2.7. Cell migration and invasion assays
2.7.1. In vitro migration assay
Costar 24-transwell assay plates (Becton Dickinson Labware Inc. Bedford, MA) were used for migration assays. The lower compartment was loaded with 0.6 mL of 2% FBS DMEM to serve as a chemoattractant. 2.5×104 cells from each cell line were then seeded with 0.6 mL of serum-free medium in the upper chambers. After 8 hours of incubation, the filter was washed with PBS, cleansed with a cotton swab, and then fixed with methanol. Hematoxylin staining was performed to visualize the cells on the filter. After this, the filter was cut out of the chambers and mounted on a slide and cells were counted under a microscope. The experiments were repeated 3 times.
2.7.2. In vitro invasion assay
24-well Biocat matrigel invasion chambers (Becton Dickinson Labware Inc. Bedford, MA) were used for chemoinvasion assays. 0.75 mL of DMEM with 10% FBS located in the lower compartment served as a chemoattractant. 2.5×104 cells from each cell line were seeded in the upper compartment, which contained 0.5 mL of serum-free medium. All tested samples were assayed in triplicate. After 16 hours of incubation in a humidified incubator with 5% CO2 at 37°C, the cells that passed through the membrane were fixed with methanol and stained with hematoxylin. Cell numbers in five predetermined fields on each membrane were counted under a microscope. The statistical significance of the differences in invasion rate between cell lines were measured by students’ t-test.
2.8. Immunohistochemistry
Tumors were cut into 2mm × 2mm cubes and fixed in 4% paraformaldehyde. The fixed samples were dehydrated, embedded into paraffin blocks and cut into 5μm thick sections. For immunohistochemistry, the paraffin sections were de-paraffinized and rehydrated in xylene and ethanol, then boiled in sodium citrate pH 6.0 antigen retrieval buffer at 95°C for 20min. Sta ining was performed using an IMMPRESS HRP polymer reagent kit (Vector Lab) with sections blocked and incubated with primary antibodies (anti-SRC-3 1:200, rabbit anti-Ki-67 1:4000,) at 4°C overnight. After three w ashes with PBS, the sections were incubated with secondary antibodies at room temperature for 30min. After three washes, the sections were developed with the DAB Peroxidase (HRP) Substrate Kit (Vector Lab).
2.9. In vivo orthotopic tumor progression model
To establish orthotopic pancreatic tumors, 0.5 × 106 PDAC cells were injected into the pancreas of SCID or nude mice (Charles River Laboratories, female, 6–7 weeks of age) through a standard live surgery process [26]. All the animal studies were conducted with the approval of the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine (BCM). All the mice were sacrificed in a CO2 chamber. In addition, any animals that showed loss of mobility, weight loss or other symptoms related to tumor growth were euthanized. Tumors were allowed to reach >20% of body weight. All animals were sacrificed and tumors were harvested with a volume of no more than 1500 mm3. In addition, any animal that displayed immobility, huddled posture, inability to eat, ruffled fur, self-mutilation, vocalization, wound dehiscence, hypothermia, or greater than 20% weight loss were euthanized according to BCM guidelines.
To test whether SRC-3 is required to maintain PDAC tumor growth, an orthotopic pancreatic cancer mouse model was developed by injecting a total of 0.5 × 106 PANC-1-Ind-shSRC-3 cells into the pancreas head through mouse live surgery. After 14 days of tumor growth, mice were randomly split into two groups (N=7). One group were fed with water containing 200 ppm of doxycycline. The doxycycline water was replenished every 3 days. The tumor size was monitored weekly by IVIS live imaging.
2.10. Cell cytotoxicity assay
PDAC cells (5000 to 7000 cells per well) were seeded in 96-well plates in medium supplemented with 10% FBS. The next day, when cells reached 70% to 80% confluence, bufalin (Santa Cruz Co.) was added to achieve an array of final concentrations (0.2, 0.5, 1, 2, 5, 10, 20, 50, 100, 500 and 1000 nM). After 72 hours of treatment, cell viability was measured by MTT assay. Cell viabilities relative to untreated control cells were plotted using Graphpad Prism. The IC50 values for bufalin were calculated based on the Hill-Slope model.
2.11. Statistical analysis
Data are presented in mean ± SEM. The Student’s t-test was used for comparison between two groups and one-way ANOVA and Tukey post-hoc tests were used for comparison among three or more groups with P<0.05 considered statistically significant.
3. RESULTS
3.1. SRC-3 is overexpressed in PDAC and inversely correlates with survival
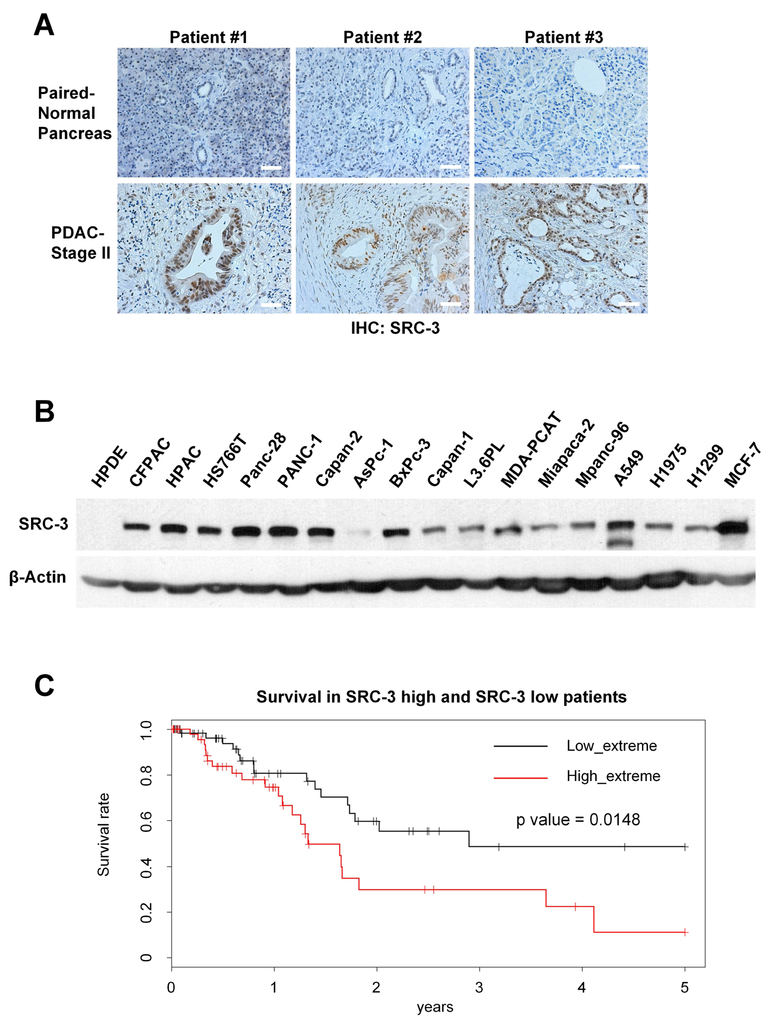
To measure SRC-3 protein levels in pancreatic cancer tissues, we performed immunohistochemistry against SRC-3 on paired PDAC and adjacent normal pancreatic tissues from 11 patients with stage II PDAC. SRC-3 protein levels were found to be significantly higher in 9 PDAC tissues, compared to the corresponding adjacent normal pancreatic tissues (Figure 1A). We also examined SRC-3 expression in a normal pancreatic ductal cell line (HPDE) and a series of PDAC cell lines by Western blot. SRC-3 was highly expressed in most of the PDAC cell lines but was hardly detected in HPDE cells (Figure 1B). Then we analyzed mRNA-seq data from PDAC tumors in The Cancer Genome Atlas (TCGA). Based on the expression value for SRC-3, we divided patients into “SRC-3 high” (top 1/3) and “SRC-3 low” groups (bottom 1/3). The overall survival time is significantly shorter (P=0.0148) in patients with high SRC-3 levels compared to those with low SRC-3 levels (Figure 1C).
Fig. 1. SRC-3 overexpression correlates with the poor prognosis in PDAC patients.
(A) SRC-3 protein expression levels in tissues from PDAC patients were detected by immunohistochemistry (IHC) assays. SRC-3 protein is highly expressed in 9 of 11 tested PDAC tissues but not detectable in all PDAC paired adjacent normal pancreas tissues. SRC-3 IHC staining of tissues from 3 PDAC patients is presented here (scale bar: 50 μm). (B) SRC-3 protein levels, as measured by Western-blotting, are significantly high in most pancreatic cancer cell lines, and are comparable with lung cancer cell lines A549, H1975 and H1299, and breast cancer cell line MCF-7, but not detectable in normal pancreas duct cell line HPDE. (C) TCGA patients with a higher SRC-3 mRNA expression level have obviously shorter overall survival time than those with a lower SRC-3 level (P < 0.0148, log-rank test).
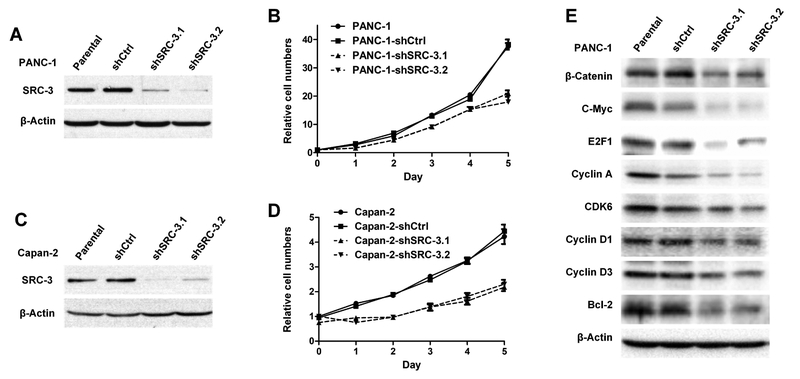
3.2. Knockdown of SRC-3 reduces pancreatic cancer cell proliferation
To study the role of SRC-3 in PDAC, SRC-3 was stably knocked down with lentivirus-mediated shRNAs in the PANC-1 and Capan-2 PDAC cell lines that normally overexpress SRC-3 at high levels. Western analysis showed that SRC-3 was knocked down effectively with either shRNA (shSRC-3.1 and shSRC-3.2) in PANC-1 (Figure 2 A) and Capan-2 (Figure 2C) cells. MTT cell viability assays showed that knock down of SRC-3 slowed down cell proliferation in both PANC-1 (Figure 2B) and Capan-2 (Figure 2D) cells, suggesting that SRC-3 played a pro-proliferative role in pancreatic cancer. Accordingly, several signaling molecules, β-Catenin, c-Myc, E2F1, Cyclin A, CDK6, Cyclin D1, Cyclin D3 and Bcl-2, which regulates cell proliferation or survival were found to be downregulated in SRC-3 knock-down PANC-1 cells (Figure 2E).
Fig. 2. SRC-3 silencing by shRNA causes decreasing cell proliferation rate in pancreatic cancer cell lines.
(A) SRC-3 expression in PANC-1 cells, as detected by Western -blotting, is effectively knocked down by shRNAs labeled as SRC-3.1 and SRC-3.2, which have different SRC-3 targeting sequences. (B) SRC-3 silenced cell lines PANC-1-shSRC-3.1 and Panc-shSRC-3.2 show a slower growth rate than PANC-1-shCtrl and parental PANC-1 cell lines. The cell viability was measured by MTT assay. Each time point has 6 repeats. (C) SRC-3 expression in Capan-2 cell line is also significantly knocked down by shSRC-3.1 and shSRC-3.2 shRNA. (D) SRC-3 silenced Capan-2 cell lines, Capan-2-shSRC-3.1 and Capan-2-shSRC-3.2, also display a significantly decreased proliferation rate, when comparing with Capan-2-shCtrl and parental Capan-2 cell lines. (E) A set of signaling molecules in PANC-1, PANC-shCtrl, PANC-1-shSRC-3.1 and PANC-1-shSRC-3.2 were detected by Western blotting assay, and found to be down-regulated in SRC-3 knockdown PANC-1 cells.
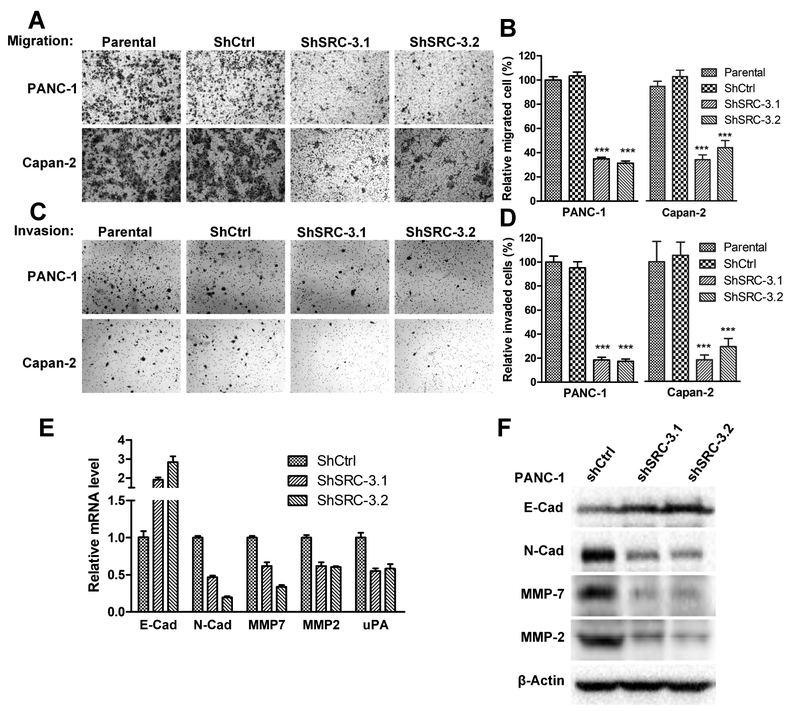
3.3. Knockdown of SRC-3 inhibits pancreatic cancer cell migration and invasion
SRC-3 has been shown to promote metastasis by enhancing cell migration and invasion [27–29]. PANC-1-shSRC-3.1 and -shSRC-3.2 cells migrated significantly less than PANC-1 and PANC-1-shCtrl control cells in a migration assay (Figure 3A). Consistently, Capan-2-shSRC-3.1 and -shSRC-3.2 cells also migrated less than Capan-2 and Capan-2-shCtrl cells (Figure 3A), demonstrating that knockdown of SRC-3 inhibited the migratory ability of pancreatic cancer cells (Figure 3B). Knockdown of SRC-3 also inhibited cell invasion in PANC-1 and Capan-2 cells (Figures 3C and 3D). E-cadherin mRNA expression was increased while N-cadherin was downregulated (Figure 3E) in SRC-3 knockdown PANC-1 cells. Matrix Metalloproteinase 7 (MMP-7), MMP-2 and uPA mRNA levels also were downregulated in SRC-3 knockdown PANC-1 cells (Figure 3E). The changes of protein levels for E-cadherin, N-cadherin, MMP-7 and MMP-2 is consistent with the changes of their mRNA levels (Figure 3F). It should be noted that we were unable to detect uPA protein in either PANC-1-shCtrl or PANC-1-shSRC-3 cell lines, possibility due to the low protein level of uPA or the low-affinity for the antibody used. The expression changes in E-cadherin, N-cadherin, MMPs and uPA suggest that SRC-3 is involved in pancreatic cancer cell migration and invasion, at least in part, through the regulation of cell junctions and metalloprotease activity.
Fig. 3. SRC-3 silencing reduces cell migration and invasion potential in PDAC cell lines.
(A) A trans-well migration assay was performed to test the migration ability of PDAC cells, including SRC-3 silenced cell lines PANC-1-shSRC-3.1 and PANC-1-shSRC-3.2, control cell lines PANC-1 and PANC-1-shCtrl. The cells migrated through trans-well membrane were stained with hematoxylin. Photographs were taken at a magnification 200X. (B) The number of migrated cells in 5 different fields of trans-well membrane photographed under microscope was counted. Comparing to parental cells (100%), the number of migrated PANC-1-shSRC-3.1, PANC-1-shSRC-3.2, Capan-2-shSRC-3.1 and Capan-2-shSRC-3.2 cells reduced to 34.9%, 31.2%, 36.0% and 46.5% correspondingly (One-way ANOVA test, ***, P<0.001). (C) All the cell lines in migration assay were also tested by Matrigel invasion assay. The invaded cells were stained with hematoxylin and then photographed under microscope (200X of magnification). (D) For each cell line, the number of invaded cells from 5 different microscopic fields were counted. Comparing to parental cells (100%), the number of invaded PANC-1-shSRC-3.1, PANC-1-shSRC-3.2, Capan-2-shSRC-3.1 and Capan-2-shSRC-3.2 cells reduced to 18.5%, 17.4%, 18.2% and 29.1% correspondingly (One-way ANOVA test, *** P<0.001). (E) The mRNA levels of a set of genes involved in migration and invasion were measured by quantitative RT-PCR. Genes N-Cadherin, MMP7, MMP2 and uPA, have significantly down-regulated mRNA expression in PANC-1-shSRC-3.1 and PANC-1-shSRC-3.2 cells. However, gene E-Cadherin exhibits a reverse trend. (F) The protein levels of E-Cadherin, N-Cadherin, MMP-7 and MMP-2 were measured by Western blotting. The trends of protein level alteration in PANC-1-shSRC-3.1 and PANC-1-shSRC-3.2 cells are consistent with the changes in the corresponding mRNA levels.
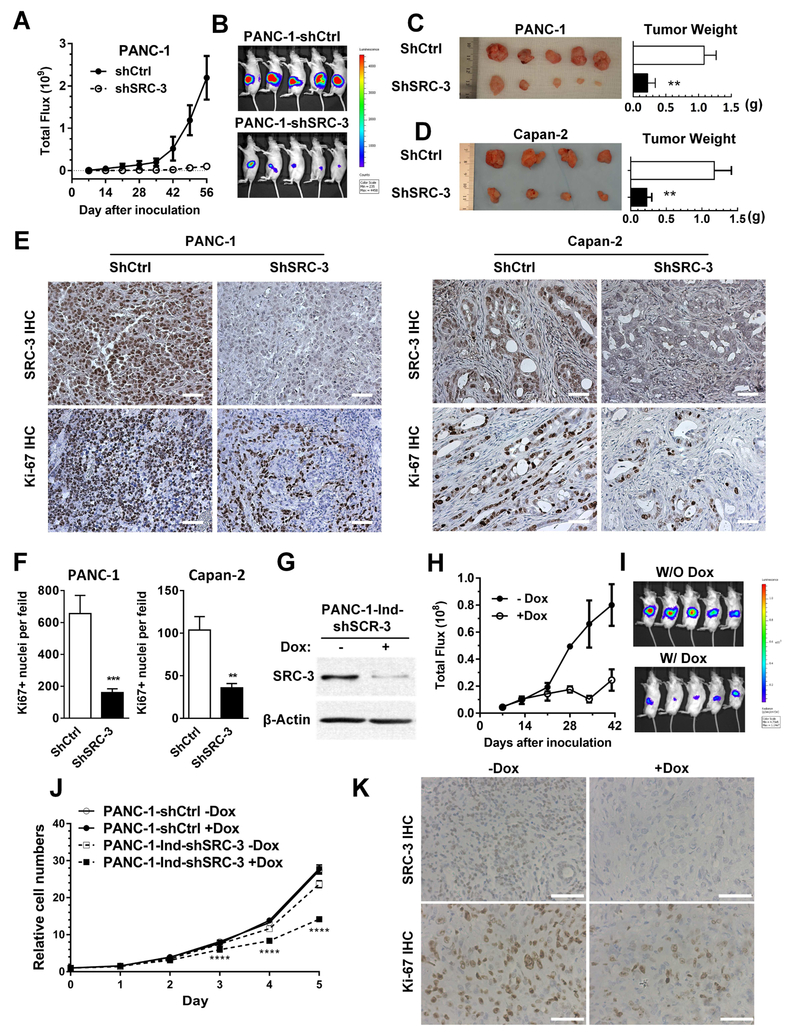
3.4. Knockdown of SRC-3 inhibits orthotopic tumor growth
In vitro experiments revealed that SRC-3 plays a key role in cell proliferation, migration and invasion in pancreatic cancer cells, strongly suggesting that it has an oncogenic role in pancreatic cancer. To examine whether SRC-3 plays an oncogenic role in vivo, control and SRC-3 knockdown PANC-1 (stably expressing firefly luciferase) and Capan-2 cells were inoculated into the pancreas of nude mice (n = 6 per group) to form orthotopic tumors. For the PANC-1 model, tumor growth was monitored through luminescence imaging of the luciferase activities in vivo from day 7 to day 56 after tumor inoculation. Luminescence imaging showed that the SRC-3 knockdown tumors (PANC-1-shSRC-3) grew much more slowly than the control tumors (PANC-1-shCtrl) (Figure 4A and 4B). The average weight of SRC-3 knockdown (PANC-1-shSRC-3) tumors was significantly lower than that of control (PANC-1-shCtrl) tumors on day 56 (Figure 4C). Lower SRC-3 expression and less Ki-67 positive cells were observed in the SRC-3 knockdown tumors than the corresponding controls (Figure 4E and 4F), indicating less proliferation in SRC-3 knockdown tumors. In the Capan-2 model, SRC-3 knockdown (Capan-2-shSRC-3) tumors similarly had a lower tumor weight than control tumors (Capan-2-shCtrl) (Figure 4 D). Ki-67 positive cells were less frequently observed in SRC-3 knockdown tumors than controls (Figure 4 E and 4F). The results from both orthotopic models suggest that SRC-3 promotes tumor formation and growth in vivo, which was consistent with its pro-proliferation role in vitro.
Fig. 4. SRC-3 silencing suppresses tumor growth in orthotopic PDAC mouse models.
(A) PANC-1-shCtrl and PANC-1-shSRC-3 cells with stable expression of luciferase were orthotopically injected into the pancreas of SCID mice. The tumor growth was monitored under IVIS live imaging system. Total flux (photons/second) was recorded. PANC-1-shSRC-3 group shows a dramatically suppressed tumor growth compared to the PANC-1-shCtrl group. (B) A live image of mouse tumors was taken right before sacrifice. (C) Tumors were harvested, photographed, and weighted. The statistic results of tumor weights are presented as mean±SEM, PANC-1-shCtrl group has a value of 1.09±0.18 g, whereas PANC-1-shSRC-3 group has a value of 0.22±0.11 g. Those values show a statistically significant difference by t-test, **, p<0.01. (D) A set of orthotopic PDAC mouse models were established by using Capan-2-shCtrl and Capan-2-shSRC-3 cells. After 8 weeks of tumor growth, tumors were harvested. The statistic values of tumor weights from Capan-2-shCtrl and Capan-2-shSRC-3 groups are 1.17±0.24 g and 0.23±0.7 g, respectively. A significant difference is revealed by t-test, **, p<0.01. (E) The SRC-3 and Ki-67 protein levels were measured by IHC. Mouse tumors established from PANC-1-shSRC-3 and Capan-2-shSRC-3 exhibits hardly detectable SRC-3 expression and less Ki-67 positive cells compared to the corresponding controls (scale bar: 50 μm). (F) The Ki-67-positive cells from multiple tumors (N>3) of each group were counted based on the photos taken under microscope. The numbers are 655.9±113.7 of PANC-1-shCtrl group, 161.0±3.4 of PANC-1-shSRC-3, 103.8±15.8 of Capan-2-shCtrl, and 36.0±4.9 of Capan-2-shSRC-3 (**, p<0.01, ***, p<0.001, by t-test). (G) Cell line PANC-1-Ind-shSRC-3 was constructed by introducing a doxycycline inducible anti-SRC-3 shRNA into PANC-1-Luciferase cells. The Western blotting results showed that SRC-3 expression levels in PANC-1-Ind-shSRC-3 cells were significantly knocked down when treated with doxycycline. (H) A set of orthotopic PDAC mouse models were made by inoculating PANC-1-Ind-shSRC-3 cells into SCID mouse pancreases. After 14 days of tumor growth, mice were randomly split into 2 groups (n =7). One group was feed with water containing doxycycline (200 ppm). The tumor size was monitored and recorded once a week by IVIS live imaging system. The tumor growth is remarkably delayed in the mouse group feed with doxycycline. (I) A live image of mouse tumors was pictured right before sacrifice to show the visible differences of tumor size between the groups feed with and without doxycycline. (J) The growth curves of PANC-1-Ind-shSRC-3 in the presence or absence of 1 μg/mL of doxycycline were measured by MTT assays. The original values of day 0 was set as 1. Doxycycline does not affect the growth of control cell line PANC-1-shCtrl, but significantly inhibits PANC-1-Ind-shSRC-3 cell growth. (K) An IHC staining was performed to demonstrate the SRC-3 protein levels and the amount of Ki-67 positive cells on above mouse tumors established with PANC-1-Ind-shSRC-3 cells. The tumor tissues from mice feed with doxycycline shows undetectable SRC-3 expression and less amount of Ki-67 positive cells. One representative photo from each group is presented (scale bar: 50 μm).
We further tested whether SRC-3 is required to maintain pancreatic cancer growth by using a doxycycline inducible SRC-3 shRNA system. When PANC-1-Ind-shSRC-3 cell lines were treated with doxycycline of 1μg/mL, SRC-3 protein levels were significantly reduced (Figure 4G). We tested the PANC-1-Ind-shSRC-3 cell growth in the presence or absence of doxycycline (1μg/mL), compared to PANC-1-shCtrl cells. As shown in Figure 4J, doxycycline has no effect on PANC-1-shCtrl cell growth, but significantly inhibits PANC-1-IndshSRC-3 cell growth. PANC-1-Ind-shSRC-3 cells were orthotopically injected into the pancreas of SCID mice. Fourteen days after injection, when tumors were formed, the experimental mouse group was given doxycycline containing water. Compared to the control group, the tumor in the doxycycline (200 ppm) treated group had significantly smaller sizes (Figures 4H and 4I) and displayed obviously lower SRC-3 expression levels and less Ki-67 positive cells (Figure 4K). It should be noted that doxycycline at 200 ppm does not inhibit PANC-1 cell growth in vivo [30]. Collectively, our data suggests that SRC-3 is required to maintain pancreatic cancer growth.
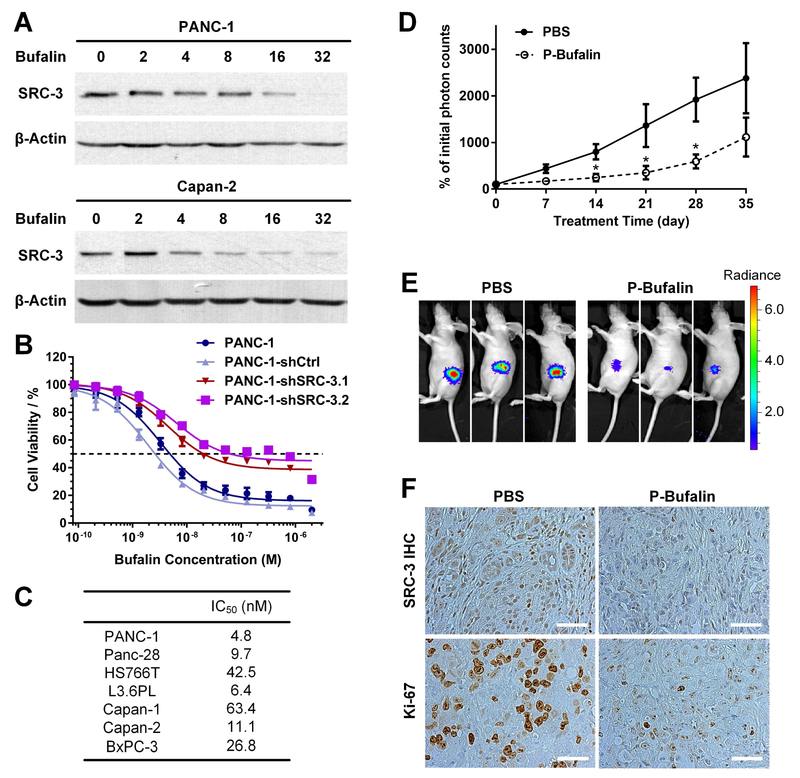
3.5. Specific inhibition of SRC-3 by the SMI, bufalin, potently reduces PDAC cell viability
Our previous work showed that bufalin can specifically bind to and inhibit SRC-3 activity by promoting SRC-3 protein degradation [31, 32]. Both bufalin nanoparticles and phospho-bufalin, a prodrug form of bufalin, can induce a significant inhibition of breast cancer cell viability and breast tumor growth [31, 32]. Based on these findings, we anticipated that bufalin also will have a therapeutic effect on PDAC. First, we analyzed SRC-3 protein levels and PDAC cell viability after bufalin treatment. We found that bufalin caused SRC-3 protein down-regulation in a dose-dependent manner. When bufalin was applied at a concentration of 32 nM, after 24 hours of treatment, the SRC-3 protein in PANC-1 cells became hardly detectable by Western analysis (Figure 5A). Similar SRC-3 inhibitory effects of bufalin also were observed in all tested Capan-2 cell lines (Figure 5A). In cell viability assays, bufalin demonstrated a high cytotoxicity toward PDAC cells with IC50 values in a low nM range, between 4.8 to 63.4 nM for all the tested PDAC cell lines (Figure 5B and 5C). However, SRC-3 knockdown cells became more resistant to bufalin-induced cytotoxicity comparing to the parental cells (Figure 5B). Even with high concentrations (2μM) of bufalin treatment, there are still a significant portion of PANC-1-shSRC-3 cells alive. The lost sensitivity of SRC-3 knock-down cells to bufalin further suggests that bufalin induced cytotoxicity is through inhibition of SRC-3 (Figure 5B).
Fig. 5.
Inhibition of SRC-3 by bufalin blocks PDAC tumor growth. (A) Bufalin down-regulates SRC-3 expression in pancreatic cancer cell lines. PANC-1 and Capan-2 cells were treated with different doses of bufalin (0, 2, 4, 8, 16 and 32 nM) for 24 hours. The SRC-3 protein levels were measured by Western blotting assay. (B) Bufalin induces toxicity in PANC-1 cells through the SRC-3 pathway. To achieve 50% of cell death (the dotted line), the bufalin concentrations required are 4.8, 2.6, 20.3 and 63.3 nM in PANC-1, PANC-1-shCtrl, PANC-1-shSRC-3.1 and PANC-1-shSRC-3.2 cells, respectively, which shows that SRC-3 silenced cells are more resistant to bufalin treatment. The cells were treated with different doses of bufalin for 72 hours. The cell viability was measured with MTT assay. Each dose of bufalin treatment has six repeats. (C) All tested pancreatic cancer cell lines are highly susceptible to bufalin treatment with IC50 in the low nM range. (D) Bufalin blocks PDAC tumor growth in an orthotopic mouse model. The mouse model was created through injecting PANC-1-Luciferase cells into pancreas of Nude mice. One group (n=8) were treated with phospho-bufalin (0.75 mg/kg, I.P. injection) three times per week from the beginning of second week to the end of fifth week. The control group (n=8) was injected with PBS. The tumor growth rate was determined by luciferase live imaging assay. (E) One luciferase live image of mouse tumors was presented to exhibit that p-bufalin administered group has significantly smaller tumor burden at the end of therapy. (F) The p-bufalin treated mouse tumor tissues has reduced SRC-3 expression and less Ki-67 positive cancer cells, as demonstrated by immunohistochemistry assay (scale bar: 50 μm).
3.6. 3-phospho-bufalin inhibits tumor growth in an orthotopic PDAC model
So far, all FDA-approved drugs for pancreatic cancer treatment can only produce marginal responses for patients and the developing new drugs for pancreatic cancer treatment is an urgent unmet need. The success of bufalin-NP and phospho-bufalin in blocking breast tumor growth [31, 32] encouraged us to test their therapeutic effects in a mouse PDAC orthotopic xenograft model. PANC-1-Luc cells (0.5×106 cells per mouse) were injected into the pancreas of nude mice (4–5 weeks). The mice were randomized into two groups based on luciferase imaging (n = 8 per group). One week after tumor inoculation, the treatment groups began receiving phospho-bufalin (0.75 mg/kg) three times per week while the control group received PBS. Tumor volumes were measured once per week by luciferase live imaging. As shown in Figure 5D and 5E, phosphobufalin can significantly inhibit PDAC tumor growth. Also, in phospho-bufalin treated PDAC tumors, lower SRC-3 expression and less Ki-67 positive cells were observed (Figure 5F).
4. DISCUSSION
SRC-3 has been recognized as an oncogene in a variety of cancers, promoting cancer development by enhancing proliferation, migration and invasion. Although SRC-3 was found to be amplified and overexpressed in neoplastic pancreas and PDAC lesions, its role in PDAC development was not characterized. In this study, we found that SRC-3 was overexpressed in human PDAC samples and negatively correlated with patient survival. We also demonstrated that SRC-3 contributed to the oncogenic phenotypes of pancreatic cancer cells, including proliferation, migration and invasion in vitro and tumor formation in vivo, which was concordant with its role in breast, prostate and lung cancer.
SRC-3 is a transcriptional co-activator, which cooperates with a wide range of transcription factors and transcriptional co-regulators. The oncogenic programs driven by SRC-3 in different cancers can vary depending on the transcription factors with which it co-operates. SRC-3 is known or suggested to interact with a variety of signaling pathways that are crucial in PDAC. For instance, the KRAS pathway is the most commonly upregulated pathway in pancreatic cancer and KRAS is frequently mutated, while the upstream receptor HER2 also is often overexpressed in PDAC. Oncogenic KRAS signaling is essential for initiation, progression and maintenance of PDAC [33, 34]. The role of SRC-3 in the Her2 pathway (which is upstream of Ras) has been substantiated in mouse genetic models. SRC-3 deficiency suppresses v-H-ras-induced and Neu/Her2-induced breast cancer in mice [18, 20]. Moreover, SRC-3 is required for downstream signaling through Her2, including Akt and JNK activation [20]. It would be most interesting to see whether SRC-3 is required for KRAS-induced PanIN and PDAC development as well. KRAS mutations have long been considered as undruggable, and most of the therapies against KRAS signaling have been based on the targeting of its downstream effector pathways, such as the Raf/MEK/ERK and PI3K/Akt pathways [35]. If SRC-3 is required for KRAS signaling in PDAC, it will be a promising therapeutic target since small-molecule inhibition of SRC-3 has been shown to inhibit breast cancer development in mouse models [31, 32].
SRC-3 also regulates other key signaling pathways crucial for PDAC development. For instance, SRC-3 interacts with p53 and represses p53-mediated transactivation in vitro [36]. Also, SRC-3 abrogates p53 function by regulating its deubiquination and stability via TRAF4 [37]. SRC-3 also interacts with Notch and enhances Notch signaling in colorectal cancer [38]. SRC-3 also was found to be recruited to the Iκ-B promoter upon TNF-α-induced NF-κB activation [39]. Comprehensive genome-wide analyses of PDACs by next generation sequencing has revealed marked genetic heterogeneity in individual tumors. The lack of success in conventional and single-target therapy in PDAC cancers likely can be attributed to inter-tumoral and possibly intra-tumoral genetic or molecular heterogeneity [6]. Because SRC-3 is a pleiotropic regulator of multiple cancer growth pathways, targeting SRC-3 may represent an effective strategy to overcome the impediment in cancer therapy brought about by tumor heterogeneity-related resistance to drugs that target specific molecular pathways.
Additionally, it should be noted that bufalin was chosen as a ‘tool compound’ in this study. Due to the cardiotoxicity of bufalin, its translational potential is limited. Recently, we have developed a new class of SRC-3 SMI, SI-2, which is highly potent with minimal cardiotoxicity [40–42]. We are in the process of testing SI-2 and its analogs in PDAC models and will report our findings in due course.
Table 1.
The list of primers used in quantitative RT-PCR
| Gene | Primers | Primer sequences |
|---|---|---|
| E-Cadherin | E-Cadherin-F | 5’-GCCCTTTCTGATCCCAGGTC-3’ |
| E-Cadherin-R | 5’-TAGCCTGGAGTTGCTAGGGT-3’ | |
| N-Cadherin | N-Cadherin-F | 5’-GACTGCTGCTCTTTGTGGGT-3’ |
| N-Cadherin-R | 5’-GCTCGTGGTTTTGCTTCCTC | |
| MMP7 | MMP7-F | 5’-ACATTGTGTGCTTCCTGCCA-3’ |
| MMP7-R | 5’-AGCTTCTCAGCCTCGAATGT-3’ | |
| MMP2 | MMP2-F | 5’-CTACGATGGAGGCGCTAATGG-3’ |
| MMP2-R | 5’-CTTGGGGCAGCCATAGAAGG-3’ | |
| uPA | uPA-F | 5’-TCCACCTGTCCCCGCAG-3’ |
| uPA-R | 5’-GCAGTTGCACCAGTGAATGT-3’ | |
| RPS6 | RPS6-F | 5’-AAGGAGAGAAGGATATTCCTGGAC-3’ |
| RPS6-R | 5’-AGAGAGATTGAAAAGTTTGCGGAT-3’ |
Highlights.
SRC-3 high expression is correlated with the poor prognosis in PDAC patients.
SRC-3 silencing reduces PDAC cell proliferation, migration and invasion.
SRC-3 silencing blocks the PDAC tumor growth in an orthotopic mouse model.
SRC-3 specific inhibitor bufalin significantly delays PDAC tumor growth in mice subjected to treatment.
Acknowledgments
This work is supported in part by National Institutes of Health Grants R01GM115622, R01CA207701, and R21CA213535 (to J.W.), HD076596 (to D.M.L.), DK059820 (to B.W.O.), R01CA175486 and U24CA209851 (to H.L.), RP100348, RP101251, and RP170500 (to B.W.O.), Susan G. Komen Foundation Grant PG12221410 (to B.W.O.); the Integrative Molecular and Biomedical Sciences (IMBS) Program (NIH 5 T32 GM008231) (to M.Z.); the Center for Comparative Medicine, the Cytometry and Cell Sorting Core, the Center for Drug Discovery, and the Dan L. Duncan Cancer Center at Baylor College of Medicine; and the Texas Medical Center Digestive Diseases Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
J.C., D.M.L., B.W.O., and J.W. are coinventors of a patent application related to this work. D.M.L., B.W.O., and J.W. are cofounders and hold stock in CoActigon, Inc., which is developing steroid receptor coactivator inhibitors for clinical use.
References
- [1].Howlader N, Noone A, Krapcho M, Miller D, Bishop K, Kosary C, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis D, Chen H, Feuer E, Cronin K.e., SEER Cancer Statistics Review, National Cancer Institute, 2017. [Google Scholar]
- [2].Jemal A, Siegel R, Xu J, Ward E, Cancer statistics, 2010, CA Cancer J Clin, 60 (2010) 277–300. [DOI] [PubMed] [Google Scholar]
- [3].Koorstra JB, Feldmann G, Habbe N, Maitra A, Morphogenesis of pancreatic cancer: role of pancreatic intraepithelial neoplasia (PanINs), Langenbecks Arch Surg, 393 (2008) 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bardeesy N, DePinho RA, Pancreatic cancer biology and genetics, Nat Rev Cancer, 2 (2002) 897–909. [DOI] [PubMed] [Google Scholar]
- [5].Ryan DP, Hong TS, Bardeesy N, Pancreatic adenocarcinoma, N Engl J Med, 371 (2014) 1039–1049. [DOI] [PubMed] [Google Scholar]
- [6].Samuel N, Hudson TJ, The molecular and cellular heterogeneity of pancreatic ductal adenocarcinoma, Nat Rev Gastroenterol Hepatol, 9 (2012) 77–87. [DOI] [PubMed] [Google Scholar]
- [7].Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW, Core signaling pathways in human pancreatic cancers revealed by global genomic analyses, Science, 321 (2008) 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu J, Wu RC, O’Malley BW, Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family, Nature reviews. Cancer, 9 (2009) 615–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fleming FJ, Myers E, Kelly G, Crotty TB, McDermott EW, O’Higgins NJ, Hill AD, Young LS, Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1, J Clin Pathol, 57 (2004) 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS, AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer, Science, 277 (1997) 965–968. [DOI] [PubMed] [Google Scholar]
- [11].Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM, A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy, Cancer Res, 61 (2001) 4315–4319. [PubMed] [Google Scholar]
- [12].Agoulnik IU, Vaid A, Bingman WE 3rd, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL, Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression, Cancer Res, 65 (2005) 7959–7967. [DOI] [PubMed] [Google Scholar]
- [13].Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, Wong J, Allred DC, Clark GM, Schiff R, Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer, Journal of the National Cancer Institute, 95 (2003) 353–361. [DOI] [PubMed] [Google Scholar]
- [14].Glaeser M, Floetotto T, Hanstein B, Beckmann MW, Niederacher D, Gene amplification and expression of the steroid receptor coactivator SRC3 (AIB1) in sporadic breast and endometrial carcinomas, Horm Metab Res, 33 (2001) 121–126. [DOI] [PubMed] [Google Scholar]
- [15].Bautista S, Valles H, Walker RL, Anzick S, Zeillinger R, Meltzer P, Theillet C, In breast cancer, amplification of the steroid receptor coactivator gene AIB1 is correlated with estrogen and progesterone receptor positivity, Clin Cancer Res, 4 (1998) 2925–2929. [PubMed] [Google Scholar]
- [16].Hudelist G, Czerwenka K, Kubista E, Marton E, Pischinger K, Singer CF, Expression of sex steroid receptors and their co-factors in normal and malignant breast tissue: AIB1 is a carcinoma-specific co-activator, Breast Cancer Res Treat, 78 (2003) 193–204. [DOI] [PubMed] [Google Scholar]
- [17].List HJ, Lauritsen KJ, Reiter R, Powers C, Wellstein A, Riegel AT, Ribozyme targeting demonstrates that the nuclear receptor coactivator AIB1 is a rate-limiting factor for estrogen-dependent growth of human MCF-7 breast cancer cells, J Biol Chem, 276 (2001) 23763–23768. [DOI] [PubMed] [Google Scholar]
- [18].Kuang SQ, Liao L, Zhang H, Lee AV, O’Malley BW, Xu J, AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice, Cancer Res, 64 (2004) 1875–1885. [DOI] [PubMed] [Google Scholar]
- [19].Kuang SQ, Liao L, Wang S, Medina D, O’Malley BW, Xu J, Mice lacking the amplified in breast cancer 1/steroid receptor coactivator-3 are resistant to chemical carcinogen-induced mammary tumorigenesis, Cancer Res, 65 (2005) 7993–8002. [DOI] [PubMed] [Google Scholar]
- [20].Fereshteh MP, Tilli MT, Kim SE, Xu J, O’Malley BW, Wellstein A, Furth PA, Riegel AT, The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice, Cancer Res, 68 (2008) 3697–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, O’Malley BW, Xu J, The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression, Mol Cell Biol, 28 (2008) 5937–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Balmer NN, Richer JK, Spoelstra NS, Torkko KC, Lyle PL, Singh M, Steroid receptor coactivator AIB1 in endometrial carcinoma, hyperplasia and normal endometrium: Correlation with clinicopathologic parameters and biomarkers, Mod Pathol, 19 (2006) 1593–1605. [DOI] [PubMed] [Google Scholar]
- [23].Gnanapragasam VJ, Leung HY, Pulimood AS, Neal DE, Robson CN, Expression of RAC 3, a steroid hormone receptor co-activator in prostate cancer, Br J Cancer, 85 (2001) 1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang Y, Wu MC, Sham JS, Zhang W, Wu WQ, Guan XY, Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray, Cancer, 95 (2002) 2346–2352. [DOI] [PubMed] [Google Scholar]
- [25].Henke RT, Haddad BR, Kim SE, Rone JD, Mani A, Jessup JM, Wellstein A, Maitra A, Riegel AT, Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma, Clinical cancer research : an official journal of the American Association for Cancer Research, 10 (2004) 6134–6142. [DOI] [PubMed] [Google Scholar]
- [26].Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE, Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice, Nature protocols, 4 (2009) 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Long W, Yi P, Amazit L, LaMarca HL, Ashcroft F, Kumar R, Mancini MA, Tsai SY, Tsai MJ, O’Malley BW, SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration, Molecular cell, 37 (2010) 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yan J, Erdem H, Li R, Cai Y, Ayala G, Ittmann M, Yu-Lee LY, Tsai SY, Tsai MJ, Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression, Cancer Res, 68 (2008) 5460–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, Solis LM, Wistuba II, Tsai SY, Tsai MJ, O’Malley BW, ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion, J Clin Invest, 122 (2012) 1869–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Higashihara T, Yoshitomi H, Nakata Y, Kagawa S, Takano S, Shimizu H, Kato A, Furukawa K, Ohtsuka M, Miyazaki M, Sex Determining Region Y Box 9 Induces Chemoresistance in Pancreatic Cancer Cells by Induction of Putative Cancer Stem Cell Characteristics and Its High Expression Predicts Poor Prognosis, Pancreas, 46 (2017) 1296–1304. [DOI] [PubMed] [Google Scholar]
- [31].Song X, Zhang C, Zhao M, Chen H, Liu X, Chen J, Lonard DM, Qin L, Xu J, Wang X, Li F, O’Malley BW, Wang J, Steroid Receptor Coactivator-3 (SRC-3/AIB1) as a Novel Therapeutic Target in Triple Negative Breast Cancer and Its Inhibition with a Phospho-Bufalin Prodrug, PLoS One, 10 (2015) e0140011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang Y, Lonard DM, Yu Y, Chow DC, Palzkill TG, Wang J, Qi R, Matzuk AJ, Song X, Madoux F, Hodder P, Chase P, Griffin PR, Zhou S, Liao L, Xu J, O’Malley BW, Bufalin is a potent small-molecule inhibitor of the steroid receptor coactivators SRC-3 and SRC-1, Cancer Res, 74 (2014) 1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA, Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism, Cell, 149 (2012) 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M, Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice, J Clin Invest, 122 (2012) 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Eser S, Schnieke A, Schneider G, Saur D, Oncogenic KRAS signalling in pancreatic cancer, Br J Cancer, 111 (2014) 817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lee SK, Kim HJ, Kim JW, Lee JW, Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53, Mol Endocrinol, 13 (1999) 1924–1933. [DOI] [PubMed] [Google Scholar]
- [37].Yi P, Xia W, Wu RC, Lonard DM, Hung MC, O’Malley BW, SRC-3 coactivator regulates cell resistance to cytotoxic stress via TRAF4-mediated p53 destabilization, Genes Dev, 27 (2013) 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mo P, Zhou Q, Guan L, Wang Y, Wang W, Miao M, Tong Z, Li M, Majaz S, Liu Y, Su G, Xu J, Yu C, Amplified in breast cancer 1 promotes colorectal cancer progression through enhancing notch signaling, Oncogene, 34 (2015) 3935–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gao Z, Chiao P, Zhang X, Lazar MA, Seto E, Young HA, Ye J, Coactivators and corepressors of NF-kappaB in IkappaB alpha gene promoter, J Biol Chem, 280 (2005) 21091–21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Song X, Chen J, Zhao M, Zhang C, Yu Y, Lonard DM, Chow DC, Palzkill T, Xu J, O’Malley BW, Wang J, Development of potent small-molecule inhibitors to drug the undruggable steroid receptor coactivator-3, Proceedings of the National Academy of Sciences of the United States of America, 113 (2016) 4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rohira AD, Yan F, Wang L, Wang J, Zhou S, Lu A, Yu Y, Xu J, Lonard DM, O’Malley BW, Targeting SRC Coactivators Blocks the Tumor-Initiating Capacity of Cancer Stem-like Cells, Cancer Res, 77 (2017) 4293–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gates LA, Gu G, Chen Y, Rohira AD, Lei JT, Hamilton RA, Yu Y, Lonard DM, Wang J, Wang SP, Edwards DG, Lavere PF, Shao J, Yi P, Jain A, Jung SY, Malovannaya A, Li S, Shao J, Roeder RG, Ellis MJ, Qin J, Fuqua SAW, O’Malley BW, Foulds CE, Proteomic profiling identifies key coactivators utilized by mutant ERalpha proteins as potential new therapeutic targets, Oncogene, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]