Abstract
Breast cancer metastasis is a multi-step process and requires cells to overcome anoikis. Anoikis is defined as cell-death that occurs due to loss of cell adhesion. During the course of cancer progression, tumor cells acquire resistance to anoikis. However, mechanisms of anoikis resistance are not clear. Human epidermal growth receptor 2 (HER2) overexpressing breast tumors are known to be highly aggressive and metastatic. The mechanisms correlating HER2 with metastasis are poorly understood. We observed increased anoikis resistance in HER2 overexpressing breast cancer cells. In addition, we identified that HER2 overexpression was also associated with increased sonic hedgehog (SHH) signaling especially GLI2, and that inhibition of SHH pathway suppressed anoikis resistance. GSK3β is known to facilitate proteasome-mediated degradation of GLI2. Moreover, we observed that silencing of GLI2 resulted in reduced migration and invasion of HER2 overexpressing cells. Anoikis resistant HER2 overexpressing cells also showed increased rate and extent of metastasis in vivo, as compared to wild type anoikis resistant cells. Taken together, this study indicates a novel role of HER2/GSK3β/GLI2 axis in anoikis resistance and metastasis, and that GLI2 could be a potential target for anti-cancer therapies.
Keywords: Sonic hedgehog (SHH), GSK3β, GLI2, HER2, Anoikis resistance, Breast cancer metastasis
1. Introduction
Anoikis is a form of cell death, initiated due to absence of extracellular matrix surrounding epithelial cells [1]. Cancer cells acquire anoikis resistance to survive under anchorage-independent conditions and this is also one of the hallmarks of cancer essential for metastasis [2]. Cancer cells adopt different survival strategies for anchorage-independent growth, such as, undergoing epithelial-to-mesenchymal transition, manipulating integrin signaling, hyper-activation of receptor tyrosine kinases and reactive oxygen species mediated pro-survival signaling [3]. These phenomena facilitate the dissemination of tumor cells to distant organs by promoting cell survival during transit.
HER2 is an oncogene belonging to the class of receptor tyrosine kinases. HER2 has gained significant importance in breast cancer therapeutics due to its association with poor prognosis in patients [4]. Anti-HER2 therapy mainly, Herceptin has been shown to be responsive in 26% HER2 positive breast cancer patients. However, complete remission is seen in only 7–8% of patients, mainly because patients exhibit resistance to therapy at some point of treatment [5]. Underlying mechanisms of resistance in HER2 positive tumors are poorly understood, hindering the rational drug design for these tumors. Two studies suggest the requirement of HIF1α and integrin α5 for anoikis resistance in HER2 positive tumors [6, 7]. Another study shows EGFR-mediated anoikis resistance, which in turn was regulated by HER2 [8, 9]. In addition, role of HER2 was demonstrated in mammospheres forming ability of breast cancer cells [10]. However, significance of HER2 in anoikis resistance has not been clearly defined yet, possibly due to lack of complete information on the downstream targets of HER2.
Sonic hedgehog (SHH) signaling has recently gained significant importance as a target in several cancer types. SHH signaling plays an important role in breast development and it is also known to induce mammary tumorigenesis [11, 12]. GLI2 is a transcription factor belonging to Gli-Kruppel family [13]. It is known to be one of the mediators in Hedgehog (HH) pathway. Patched and smoothened are the cell surface membrane receptors involved in hedgehog signaling. In the absence of hedgehog ligands, SMO is inactivated by PTCH1 that causes inactivation of GLI transcription factor by proteolytic processing. Hence hedgehog binding to PTCH1 releases SMO, leading to nuclear translocation and activation of GLI1, GLI2 and GLI3 [14] [15]. A recent study showed that GLI2 expression was associated with overall poor survival in breast cancer patients [16]. However, role of SHH pathway is least understood and more studies are needed to investigate its significance in cancer.
Previous studies from our laboratory provided the first evidence for the role of SHH signaling in anoikis resistance in ovarian cancer cells through GLI1 [17]. Recently, we have shown a unique role of STAT3 in anoikis resistance in metastasis of melanoma and pancreatic cancer [18, 19]. A prior study correlated SHH mRNA and other players of sonic hedgehog signaling pathway in breast carcinoma with HER2 expression [20]. In the present study, we evaluated the cross-talk of HER2 with SHH pathway and its role in anoikis resistance in breast cancer cells. We observed that anoikis resistance in breast cancer cells was mediated by HER2. Our results showed activated SHH in HER2 overexpressing cells, suggesting a link between HER2 and SHH signaling. Furthermore, we found that HER2 expression protected GSK3β-mediated proteasomal degradation of GLI2. Hence the cross-talk of HER2 with SHH through increased GLI2 activity promoted cell survival and anoikis resistance. To the best of our knowledge, this is the first report on the crosstalk of HER2 with SHH signaling and its role in anoikis resistance in breast cancer.
2. Material and methods
2.1. Ethics Statement:
All the mice experiments were conducted in accordance with the ethical standards and according to approved protocol by Institutional Animal Care and Use Committee (IACUC).
2.2. Cell culture:
Human breast carcinoma cell lines MDA-MB-231 and MDA-MB-231 cells with HER2 overexpression (HH) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin, Streptomycin, and Neomycin (PSN). The HH cells were maintained in the medium described above in the presence of 300μg/ml zeocin. SKBR3 cells were maintained in RPMI supplemented with 10% FBS and 1% PSN. We transfected HH cells with luciferase using lentiviral transfection technique (Genecopoeia, Rockville, MD). MDA-MB-231 cells were purchased from Calipers (PerkinElmer, Waltham, MA) and these cells were transfected with luciferase. HH cells were kindly provided by Dr. Patricia Steeg (NIH, Bethesda, MD, USA) and Dr. Quentin Smith (Texas Tech University Health Sciences Center, Amarillo, TX, USA). SKBR3 cells were kindly provided by Dr. Marc Antonyak (Cornell University, Ithaca, NY, USA). The cells were maintained and passaged in culture as described by us previously [21–24]. Periodically, all the cell lines were authenticated by short tandem repeats (STR) analysis in our core facility. All the cell lines were authenticated from 2016–2017.
2.3. Reagents and chemicals:
GLI2 shRNA was obtained from Genecopoeia, Rockville, MD. GLI2 overexpression plasmid was obtained from Addgene, Cambridge, MA [25]. The chemicals, poly(2-hydroxyethyl methacrylate) (poly-HEMA), sulforhodamine B, and antibody against actin were obtained from Sigma-Aldrich (St. Louis, MO). SHH protein was obtained from R&D Systems (Minneapolis, MN). Cyclopamine, a SHH pathway inhibitor was purchased from Enzo Life Sciences (Farmingdale, NY). MG-132, a proteasomal inhibitor was purchased from Selleckchem (Houston, TX). XIX, IM-12, a GSK3β inhibitor was purchased from EMD Millipore (Billerica, MA). Nucleofector transfection system and reagents were obtained from Lonza (Allendale, NJ).
2.4. Anoikis Assay:
Anoikis assay was performed as described by us previously [17]. Briefly, sterile petridishes were coated with poly-HEMA (20mg/ml in absolute ethanol) to prevent cells from adhering to the base of culture dishes. This was done to mimic the anchorage independent growth conditions for cells. 0.2×106 cells were plated in DMEM media in poly-HEMA-coated plates. After 48h, cells were collected by centrifugation and counted using trypan blue. Further, equal number of live cells were re-plated uniformly in 24 (50,000 cells) or 96 (5,000 cells) well plates without poly-HEMA coating and incubated at 37°C. For determination of cell survival under anchorage-independent conditions, plates were processed for Sulforhodamine B (SRB) assay after 4h incubation, as described by us previously [17, 18]. The average values from each group were used to calculate anoikis resistance.
2.5. Cell proliferation assay:
To analyze proliferation capacity under anchorage-independent conditions, cells cultured under anchorage-independent conditions were collected after 48h. Live cells were counted using trypan blue and equal numbers of cells were plated in 96 well plates. The plates were incubated for 24h to allow cell proliferation. The plates were analyzed for cell density using SRB assay.
2.6. Agar drop mammosphere assay:
In addition to above densitometric method for cell proliferation, we performed mammosphere formation assay to assess growth potential of cells cultured under anchorage-independent conditions. The cells were cultured under anchorage-independent conditions for 48h and then collected by centrifugation and suspended in agar solution. Following this, cells re-suspended in agar medium were cultured in normal culture medium for 25–30 days. Mammosphere images were taken using digital single-lens reflex (DSLR) camera (Nikon) to determine mammosphere formation capacity of cells. Immunostaining for HER2 was performed in mammospheres formed by HH cells as described below.
2.7. GLI2 knockdown and overexpression:
MDA-MB-231, HH and SKBR3 cells were transfected with GLI2 shRNA or GLI2 overexpressing plasmid and their respective empty vectors were used for controls using nucleofector system, as per manufacturer’s protocol. After transfection, cells were grown under anchorage independent conditions for 48h and processed for anoikis assay or western blotting. In all the experiments, scrambled shRNA, scrambled siRNA or an empty vector was used as control.
2.8. Western Blot Analysis:
After specified treatment of cells, whole cell lysates were prepared using 4% (w/v) CHAPS buffer. The protein concentration was quantified using Bradford assay. Equal amount of cell lysates were subjected to SDS-PAGE and the segregated proteins were transferred onto PVDF membrane. The membranes were then probed with primary antibodies overnight against HER2 (Cell Signaling #2165, rabbit mAb), GLI2 (Cell Signaling #2585, rabbit mAb), PTCH2 (Cell Signaling #2470, rabbit mAb), PTCH1 (Cell Signaling #2468, rabbit mAb), SHH (Cell Signaling #2207, rabbit mAb), SUFU (Cell Signaling #2520, rabbit mAb), C-caspase 3 (Cell Signaling #9661S, rabbit mAb), Ubiquitin (Cell Signaling #3936, mouse mAb), PCNA (Cell Signaling #13110, rabbit mAb), LaminB (Santa Cruz #sc6216, goat polyAb) and Actin (Sigma #A5441, mouse mAb) followed by incubation with anti-mouse, anti-rabbit or anti-goat secondary antibody HRP conjugate for 2 hours and detected by chemiluminescence as described by us previously [21, 22, 26]. All the antibodies were purchased from Cell Signaling Technologies (Danvers, MA) and were diluted in 1:1000 except LaminB (1:500) and actin (Sigma Aldrich, 1:2000). Blots were quantified using UN-SCAN-IT gel 7.1 software.
2.9. PCR analysis:
Total RNA was extracted from cells using TRIzol reagent (Life Technologies, Inc., Carlsbad, CA) and cDNA was synthesized, as described by us previously [27, 28]. For polymerase chain reaction (PCR), 2 μl of single-stranded cDNA, 1XPCR buffer, 2.5 mM dNTP mix, 1.5mM MgCl2, 20pmol each of forward and reverse primer and 1U Taq DNA polymerase (Thermo Fisher, Pittsburgh, PA) in a final reaction volume of 25μl was subjected to reaction in a thermal cycler (Thermo Fisher, Pittsburgh, PA) (Supplementary Table 1). The PCR products were separated on a 1.5% agarose gel, stained with 0.5 mg/mL ethidium bromide and visualized under UV light.
2.10. Cytosolic and nuclear fractionation:
The fractionation of cells was achieved using NE-PER kit (Thermo, Rockford, IL), as per the manufacturer’s instructions. The cell extracts were then subjected to western blot analysis.
2.11. Immunofluorescence:
Cells were plated in a 24-well plate on a coverslip at a density of 0.1*106cells/well and allowed to attach overnight. Cells on coverslips or in agar gel (mammosphere assay) were rinsed twice with PBS for 5 min each, fixed with 4% formaldehyde solution for 25–30 minutes, rinsed permeabilized with triton X100 solution (0.02%) for 15min and blocked with 5% goat serum for 60 min. Eventually, cells were probed with antibodies specific to GLI2 or HER2 overnight at 4°C. Next day cells were washed with PBS and incubated with Alexa Flour 594 secondary antibody specific for rabbit IgG (Invitrogen, Carlsbad, CA) for 1h at room temperature. The cells plated on coverslips were washed with PBS three times and the nucleus was counterstained with DAPI. After mounting coverslips on slides, cells were imaged under microscope (Olympus Inc.). Mammospheres in agar gel were imaged using confocal multiphoton microscope (Nikon, Melville, NY).
2.12. HER2 silencing:
The shRNA for HER2 and scrambled shRNA (Genecopoeia, Rockville, MD) were transfected in HH and SKBR3 cells using nucleofection, as per the manufacturer’s instructions. The cells were selected using puromycin selection marker and sub-cultured for further experiments. The cells were used within initial three passages after transfection.
2.13. Immunoprecipitation:
This was performed as described by us previously [27, 29]. Briefly, protein was collected by lysing MDA-MB-231 and HH cells using RIPA buffer. Immunoprecipitation was performed using primary antibody for ubiquitin. The samples were prepared and western blot analysis was performed and the membranes were probed for GLI2.
2.14. Wound healing and cell invasion:
Cell migration and invasion was performed as described by us earlier [30]. HH and SKBR3 cells were transfected with GLI2 or scrambled shRNA using nucleofection as per manufacturer’s instructions. The cells were plated for wound healing or invasion assay as described earlier by us. Cell migration was estimated by wound width and invasion was assessed by number of cells on the other side of the membrane.
2.15. In vivo anoikis model:
Female athymic nude mice (4–6 weeks old) obtained from Charles River (Wilmington, MA, USA) were maintained under specific pathogen-free conditions. The use of athymic nude mice and their treatment was approved by IACUC, Texas Tech University Health Sciences Center, and the experiments were conducted in strict compliance with the regulations. MDA-MB-231 or HH cells transfected with luciferase were cultured under anchorage independent conditions for 48h. Another set of anoikis resistant HH cells were also transfected with shRNA for GLI2 using nucleofection. These cells were cultured for additional 24h under anchorage-independent condition. The cells from each set were washed three times with PBS. Viable cells were counted by trypan blue dye exclusion assay. Approximately 5 × 106 viable cells from each group were re-suspended in 1ml PBS and 100μl of this suspension was injected intravenously in athymic nude mice through tail vein. Each group had 6 mice. Mice were imaged periodically using non-invasive live animal imaging system (Calipers, PerkinElmer, Waltham, MA) [23]. Mice were euthanized at the end of the experiment, and lungs and livers were removed carefully, weighed and imaged for luminescence signal. The organs were fixed in 4% paraformaldehyde overnight at room temperature and processed for immunohistochemistry or H& E staining.
2.16. Immunohistochemistry:
The immunohistochemistry (IHC) was performed as previously described by us [24]. Briefly, paraffin-embedded tissues were sectioned into 5μm thick sections using microtome (Leica Microsystems Inc., Buffalo Grove, IL). After deparaffinization and rehydration, antigens were retrieved by boiling the sections in 10 mM sodium citrate buffer (pH 6.0). The slides were washed with distilled water and incubated in 3% hydrogen peroxide methanol solution. The sections were then washed, blocked in 200 μl of blocking solution (5% goat serum diluted) and incubated with anti–HER2 (1:150) (Abcam, Cambridge, MA) or anti-GLI2 (1:50) (Cell signaling, Danvers, MA) overnight at 4°C. Next day primary antibody was removed and the sections were washed with wash buffer followed by 30 minute incubation with Ultravision ONE HRP polymer (Thermofisher scientific, Rockland, IL) as per the manufacturer’s instructions. Subsequently, sections were washed with wash buffer and incubated with DAB Plus chromogen for 15–20 minutes. The sections were counterstained with hematoxylin and dehydrated. The slides were mounted using Permount (Thermofisher scientific, Rockland, IL) and analyzed under a bright field Olympus microscope (Olympus America Inc).
2.17. Statistical Analysis:
Statistical analysis was performed using Prism 6.0 (GraphPad software Inc., San Diego, CA, USA). Results were represented as means ± SD (n ≥3) or S.E.M for in vivo studies. Data was analyzed by Student’s t-test. Differences were considered statistically significant at p<0.05.
3. Results
3.1. HER2 inhibits anoikis
First we wanted to test anoikis resistance in MDA-MB-231 cells relative to HH cells. For this we compared the survival of MDA-MB-231 and HH cells under anchorage-independent conditions using anoikis assay. We observed that only about 60% of MDA-MB-231 cells survived under anchorage independent conditions (Fig. 1A-i). On the other hand, about 90% HH cells survived under anchorage-independent conditions (Fig. 1A-ii). These results indicated that HER2 expression enhanced anoikis resistance in MDA-MB-231 cells. To confirm these observations, we silenced HER2 using shRNA, while the control cells were transfected with a scrambled shRNA. After HER2 silencing in HH cells, only about 50% cell survival was observed under anchorage independent conditions (Fig. 1A-iii). Next, we wanted to test the role of HER2 in anoikis resistance in SKBR3 cell line with constitutive high expression of HER2. Nonetheless, expression of HER2 was several folds higher in HH cells as compared to SKBR3 cells as shown in supplementary figure 1. For this we compared the survival of SKBR3 cells under anchorage-independent conditions. We observed that there was no significant change in cell survival under anchorage-independent conditions (Fig. 1B-i). Similar to HH cells, we also silenced HER2 in SKBR3 cells using shRNA and observed that only 40% cell survived under anchorage-independent conditions after knocking down HER2 as compared to adherent cells (Fig. 1B-ii). Furthermore, the cells that attached from anchorage independent cultures were also stained and imaged. The bright field images of the anoikis resistant cells showed drastic differences between the patterns of cell attachment. The MDA-MB-231 cells reattached to the plates in a diffused manner, whereas HH cells reattached as aggregates (Fig. 1C). The aggregates formed by HH cells were breast cancer spheroids, which tends to have cancer stem cell like properties and contribute to tumor progression [31]. However, the cells with HER2 knockdown attached mostly as diffused cells and aggregates were not as distinct as with HH cells (Fig. 1C). These results suggested that HER2 plays important role in forming spheroids and imparts anoikis resistance in breast cancer cells.
Figure 1– HER2 promotes anoikis resistance.
Cell survival was analyzed in cells after 48h of anchorage independent conditions, using anoikis assay. A) Comparison of anoikis resistance in MDA-MB-231 and HH cells and in HH cells after silencing HER2 using shRNA transfection. B) Analysis of Anoikis resistance in SKBR3 and HER2 shRNA transfected SKBR3 cells. C) Bright field images of sulforhodamine B stained MDA-MB-231, HH and HH with HER2shRNA cells, re-attached after 48h of anchorage independent conditions. Values were plotted as mean ± SD (n=3).
3.2. HER2 increases proliferation of anoikis resistant cells
Next we sought to determine the proliferation rate of the cells cultured under anchorage-independent conditions. Hence we compared the proliferation rates of anoikis resistant MDA-MB-231 and HH cells and SKBR3 cells by culturing equal number of anoikis resistant cells for 24h. This was done by comparing the ratio of cell densities of adhered vs anchorage independent cultured MDA-MB-231 and HH cells. Our results showed that the ratio of cell densities from anoikis resistant vs adherent cells was about 50% for MDA-MB-231 cells and 145% for HH cells, suggesting that HER2 not only imparts resistance to anoikis but also accelerates the proliferation of anoikis resistant cells (Fig. 2A-i&ii). Furthermore, to confirm the role of HER2, we knockdown HER2 using shRNA and performed the experiment under similar conditions. HER2 silencing significantly reduced the ratio of cell densities of anoikis resistant HH cells (Fig. 2A-iii). On the other hand, we found that the ratio of cell densities for anoikis resistant vs. adherent control cells was about 0.92 for SKBR3 cells which has constitutively high HER2 levels (Fig. 2B-i). However, the ratio of cell densities significantly reduced to 0.5 after knocking down HER2 with shRNA (Fig. 2B-ii). In addition, we evaluated the level of proliferation markers such as PCNA in MDA-MB-231 and SKBR3 cells under anchorage independent and adherent condition. We observed 0.5 fold reduction in the level of PCNA under anchorage dependent vs adhered cells in MDA-MB-231 cells. However, PCNA was slightly elevated (1.2 folds) in the SKBR3 under anchorage independent conditions confirming the role of HER2 in proliferation of anoikis resistant cells (Fig. 2C). Furthermore, to evaluate the role of HER2 in cell invasion, we performed cell invasion assay under anchorage dependent and independent conditions in MDA-MB-231 as well as in HH cells. Our results indicated that percent cell invasion under anchorage-independent conditions in HH cells was significantly higher as compared to anchorage dependent conditions in the same cells. However, we did not observed any significant difference in percent cell invasion in MDA-MB-231 cells under anchorage dependent vs independent conditions. These results further validate the involvement of HER2 in anoikis resistance (Supplementary Figure 2).
Figure 2– Proliferation of anoikis resistant cells.
Proliferation rate of anoikis resistant MDA-MB-231, HH and SKBR3 cells was analyzed by comparing cell densities after 24h incubation. Proliferation rates of anoikis resistant MDA-MB-231, HH and HH with HER2shRNA cells. Proliferation rates of anoikis resistant SKBR3 cells and SKBR3 transfected with shRNA HER2. C) Western blot analysis for proliferation marker PCNA under adherent vs anchorage independent condition in MDA-MB-231 (i) cells and SKBR3 (ii) cells. D) Mammosphere formation assay in MDA-MB-231, HH and HH with HER2shRNA in agar gel for 30 days; Multiphoton image of mammosphere formed in HH cell culture, Green fluorescence indicates HER2 staining. Values were plotted as mean ± SD (n=3).
3.3. HER2 promotes mammosphere formation
Mammosphere formation is a characteristic of mammary stem/progenitor cells [32]. Hence we tested mammosphere formation capability of MDA-MB-231 cells in agar and compared with HH cells. To achieve this, cells cultured under anchorage independent condition for 48h were resuspended in agar solution and cultured further for about 30 days. No MDA-MB-231 mammospheres could be observed in agar (Fig. 2D-i). On the other hand, HH cells in agar developed numerous mammospheres of substantial sizes (Fig. 2D-ii). The green fluorescence in enlarged image indicates presence of HER2 in mammospheres. However, when HER2 was knocked down in HH cells, the number and size of mammospheres was notably reduced (Fig. 2D-iii). These observations reinforced a direct role of HER2 in promoting anoikis resistance and tumor growth in anoikis resistant cells.
3.4. Anoikis resistant cells show induced sonic hedgehog signaling
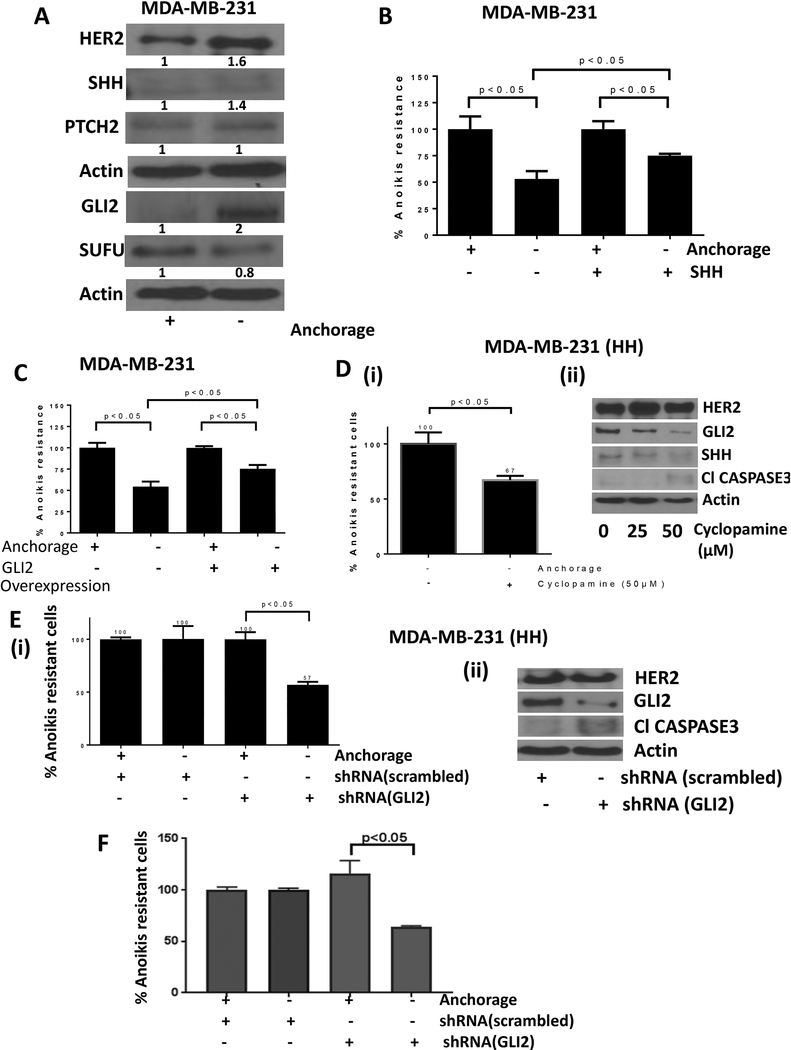
To delineate the underlying mechanisms of HER2-mediated anoikis resistance, we performed western blot analysis of the cell lysates from adherent and anchorage independent cultured cells. We observed that anchorage independent cells showed 1.6 fold increased expression of HER2 compared to adherent control cells (Fig. 3A). The anoikis resistant cells also showed a 1.4 fold modest increase in the expression of SHH, a ligand for sonic hedgehog signaling (SHH) (Fig. 3A). The expression of PTCH2, a receptor for SHH pathway remained unchanged, but the anoikis resistant cells showed a 2 fold increase in the expression of GLI2, a transcription factor of SHH pathway. A modest and statistically insignificant change was observed in the expression of GLI1 (data not shown). SUFU, an intrinsic inhibitor of SHH pathway, was reduced by 20% under anchorage independent conditions (Fig. 3A). These results suggested the activation of HER2 as well as SHH signaling in anoikis resistant cells.
Figure 3– SHH signaling promote anoikis resistance.
A) Western blot analysis for protein modulations in adherent vs anchorage independent MDA-MB-231 cells. B) Anoikis assay for MDA-MB-231 cells treated with SHH ligand and respective control cells. C) Anoikis assay for MDA-MB-231 cells transfected with empty vector or GLI2 overexpression plasmid. D) Anoikis assay (i) and western blot analysis (ii) for HH cells treated with cyclopamine, a pharmacological inhibitor of SHH signaling. E) Anoikis assay (i) and western blot analysis (ii) for HH cells transfected with scrambled shRNA or GLI2 shRNA to compare anoikis resistance. F) Anoikis assay in SKBR3 cells transfected with GLI2 shRNA. Values were plotted as mean ± SD (n=3).
3.5. SHH signaling regulates anoikis resistance
Since our experiments showed enhanced SHH signaling in anoikis resistant cells, next step was to elucidate the role of SHH pathway in anoikis resistance in breast cancer cells. MDA-MB-231 cells were treated with recombinant SHH and analyzed for anoikis resistance. Our results showed that SHH treatment increased anoikis resistance from about 50% to 80% in MDA-MB-231 cells (Fig. 3B). In order to confirm the involvement of GLI2, we overexpressed or knocked down GLI2. Our results showed that GLI2 overexpression by transient transfection enhanced the anoikis resistance in MDA-MB-231 cells by about 25% (Fig. 3C). In fact, GLI2 overexpressing MDA-MB-231 cells showed 75% anoikis resistance relative to 50% in cells with constitutive levels of GLI2 (Fig. 3C). Furthermore, to confirm the role of SHH in anoikis resistance, we used cyclopamine, a pharmacological inhibitor of SHH signaling. HH cells were treated with 50μM cyclopamine and anoikis resistance of the cells was analyzed. The results showed a significant suppression of survival of anoikis resistant cells treated with cyclopamine. Cyclopamine treatment reduced anoikis resistance in HH cells by about 33% (Fig. 3D-i). Our results also showed that cyclopamine treatment inhibited GLI2 and SHH expression along with inducing apoptosis through cleavage of caspase3 in MDA-MB-231 cells (Fig. 3D-ii). In addition, HH cells and SKBR3 cells were transfected with GLI2 shRNA and anoikis resistance was evaluated. We observed that silencing of GLI2 suppressed anoikis resistance by approximately 50% in HH cells (Fig. 3E-i) and about 35% in SKBR3 cells (Fig. 3F). Western blot data showed that GLI2 suppression by shRNA also caused cleavage of caspase 3 suggesting apoptosis in HH (Fig. 3E-ii). These findings clearly showed a significant role of GLI2 in anoikis resistance in breast cancer cells.
3.6. HER2 mediates activation of GLI2
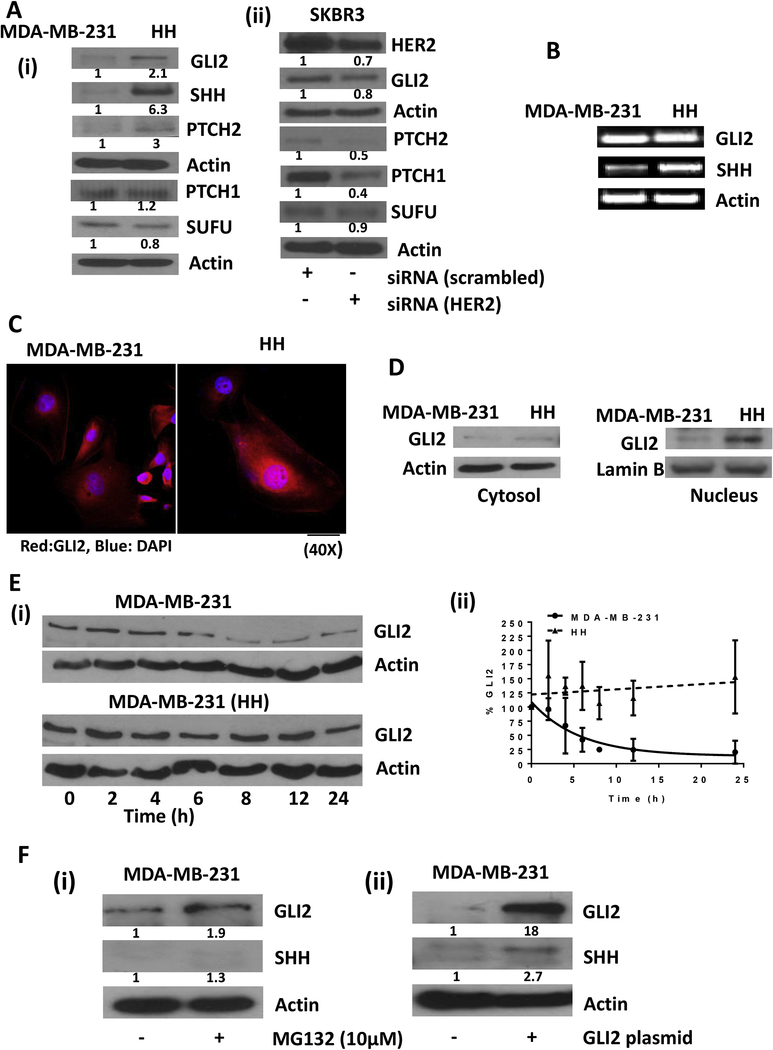
To evaluate a link between HER2 and GLI2, we first compared the constitutive expression of proteins belonging to sonic hedgehog signaling in MDA-MB-231 and HH cells. Western blot analysis from whole cell lysates showed a 6 fold increase in SHH expression in HH cells as compared to parent MDA-MB-231 cells (Fig. 4A-i). Although PTCH2 was increased by 3 fold in HH cells, the PTCH1 expression remained unchanged. Furthermore, HH cells showed a 2.1 fold increased expression of transcription factor GLI2. The expression of SUFU, which is an intrinsic inhibitor of SHH pathway, was reduced by 20% in HH cells (Fig. 4A-i). We also compared the constitutive expression of sonic hedgehog signaling molecules in parent SKBR3 cells and SKBR3 cells transfected with HER2 siRNA by western blot. We observed that GLI2 was significantly downregulated in cells silenced with HER2. Reduction in expression of PTCH1 and PTCH2 was also observed in HER2 silenced SKBR3 cells indicating a cross talk between HER2 and sonic hedgehog signaling (Fig. 4A-ii). These results indicate the cross-talk between HER2 and SHH signaling under anchorage dependent conditions. We also looked at the mRNA levels of GLI2 and SHH in MDA-MB-231 and MDA-MB-231 (HH) cells. The results from this experiment showed increased mRNA levels of SHH, however, surprisingly, GLI2 mRNA levels were found to be unchanged in HH cells (Fig. 4B). The immunofluorescence showed an increased expression of GLI2 in HH cells (Fig. 4C). Interestingly, the expression of GLI2 was found to be more in the nucleus of HH cells as compared to MDA-MB-231 cells (Fig. 4C). These results were further confirmed by western blot analysis of nuclear lysates from both the cell lines. The results showed significantly higher levels of GLI2 in the nuclear fraction of HH cells relative to MDA-MB-231 cells (Fig. 4D). These observations indicated enhanced SHH signaling with HER2 expression.
Figure 4– HER2 regulates SHH signaling.
A) Western blot analysis for comparison of constitutive protein expressions of SHH signaling in MDA-MB-231 vs HH cells (i) and in SKBR3 parent cells vs SKBR3 cells transfected with siRNA HER2 (ii). B) PCR assay for analysis of transcript levels of GLI2 and SHH in MDA-MB-231 and HH cells. C) Immunofluorescence for GLI2 (indicated by red color) in MDA-MB-231 and HH cells. D) Western blot analysis for GLI2 in cytosolic and nuclear extracts of MDA-MB-231 and HH cells. E) Western blot analysis for GLI2 in MDA-MB-231 and HH cells treated with cycloheximide at different time points (i). The GLI2 expression was quantitated and values from three independent experiments were plotted against time (ii). F) Western blot analysis for GLI2 and SHH in MDA-MB-231 cells treated with proteasome inhibitor MG-132 (i) and GLI2 overexpression plasmid transfection (ii). Values were plotted as mean ± SD (n=3).
3.7. GLI2 stability regulates SHH signaling
Although, we observed increased expression of GLI2 in HH cells, there was no change at transcriptional level. Hence we assessed the stability of GLI2 protein in MDA-MB-231 vs HH cells by inhibiting protein synthesis using 1μg/ml cycloheximide. Western blot analysis of cell lysates from a time-dependent treatment with cycloheximide showed that GLI2 was stable for a longer duration in HH cells as compared to parent cells (Fig. 4E-i). GLI2 was reduced by 50% in MDA-MB-231 cells within 5h of cycloheximide treatment, whereas the expression of GLI2 was maintained in HH cells for at least 12h (Fig. 4E-ii). To further confirm the degradation of GLI2, MDA-MB-231 cells were treated with 10μM MG-132, a proteasome inhibitor for 20h and whole cell lysates were analyzed using western blot analysis. Our results showed that GLI2 expression was increased by 1.9 fold in MDA-MB-231 cells treated with proteasome inhibitor (Fig. 4F-i). In addition, proteasome inhibitor treatment also increased the expression of SHH by 1.3 fold. To delineate the mechanism of SHH induction, MDA-MB-231 cells were transfected with GLI2. We observed about 18 fold increase in GLI2 expression in the cells transfected with GLI2 plasmid. Furthermore, GLI2 overexpression increased SHH expression by about 2.7 fold (Fig. 4F-ii). Our results suggested that HH cells have higher GLI2 expression due to reduced proteasomal degradation of GLI2, which further leads to increased SHH signaling.
3.8. HER2 inhibits GSK-mediated GLI2 degradation
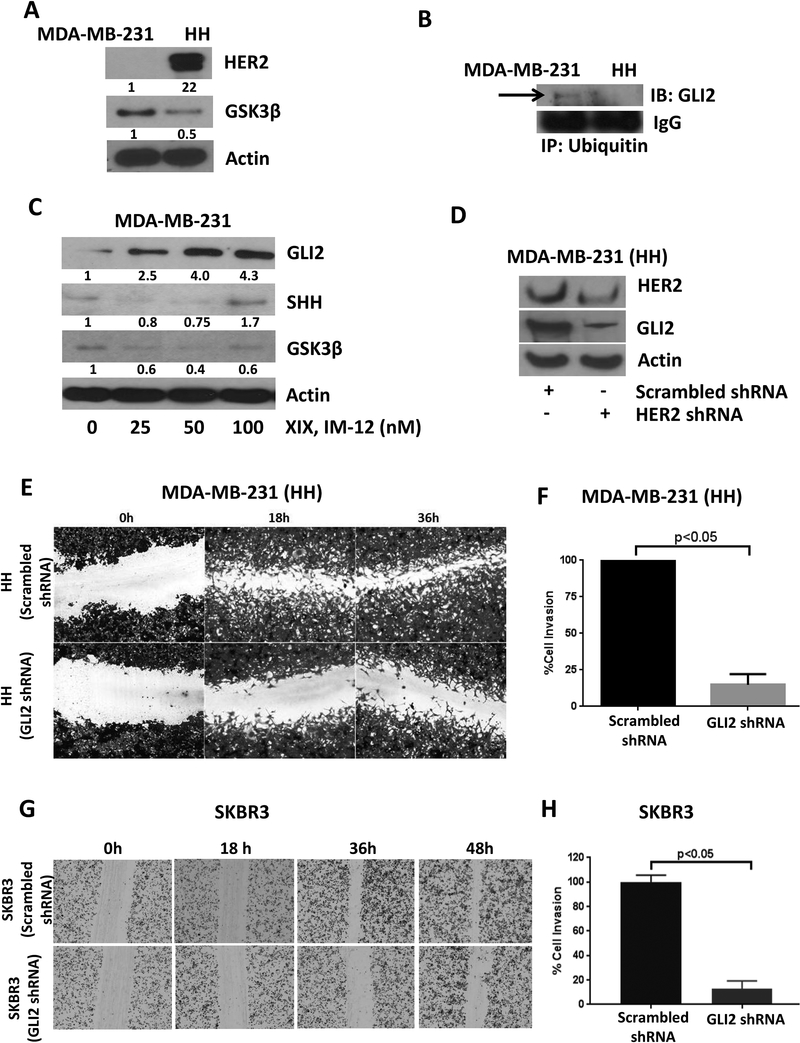
To further delineate the mechanism of reduced proteasomal degradation of GLI2 in HH cells, we determined GSK3β expression, which is known to negatively regulate GLI2. The western blot analysis showed a 50% reduced expression of GSK3β in HH cells as compared to MDA-MB-231 cells (Fig. 5A). However, phosphorylation of GSK3β at Ser9 did not change in HH cells (data now shown). Immunoprecipitation analysis showed decreased association of ubiquitin with GLI2 in HH cells as compared to parent MDA-MB-231 cells, further confirming reduced proteasomal degradation of GLI2 in HH cells (Fig. 5B). To elucidate the role of GSK3β in GLI2 degradation, MDA-MB-231 cells were treated with various concentrations of XIX IM-12, a GSK3β inhibitor, for 24h. Inhibition of GSK3β caused an increase in the expression of GLI2 in a concentration-dependent manner in MDA-MB-231 cells, where constitutive level of GLI2 was low. We also observed increase in the expression of SHH, which could be because of some feedback mechanism, as activation of the GLI2 leads to the transcription of Shh [33] (Fig. 5C). We also observed significantly reduced expression of GLI2 when HER2 was silenced in HH cells using shRNA (Fig. 5D).
Figure 5– GSK3β mediates GLI2 stability in HH cells.
A) Constitutive expression of GSK3β in MDA-MB-231 and HH cells. B) Immunoprecipitation of ubiquitin in MDA-MB-231 and HH cells and immunoblotting for GLI2. C) Western blot analysis for GLI2 signaling in MDA-MB-231 cells after 24h treatment with GSK3β inhibitor, XIX, IM-12. D) Effect of HER2 silencing on GLI2 expression in HH cells. HH cells were transfected with scrambled or GLI2 shRNA. The control and GLI2 silenced cells were plated and analyzed for cell migration using wound healing and cell invasion using Boyden’s chamber assay in HH cells (E&F) and in SKBR3 cells (G&H). Values were plotted as mean ± SD (n=3).
3.9. GLI2 silencing suppresses cell migration and invasion
To confirm the role of GLI2 in metastasis of cells with high HER2 expression, the HH and SKBR3 cells were transfected with scrambled or GLI2 shRNA. Wound healing and cell invasion capability of GLI2 silenced cells was compared with control cells. We observed a significant reduction in cell migration and invasion by GLI2 silencing. Our results showed approximately 75–80% reduction in wound healing of the HH cells transfected with GLI2 shRNA and about 50–60% reduction in SKBR3 cells transfected with GLI2 shRNA (Fig. 5E&5G). The HH and SKBR3 cells with GLI2 silencing also showed approximately 80% and 90% reduced cell invasion respectively (Fig. 5F&H). These results clearly indicated that GLI2 is required by cells expressing high HER2 for their migration and invasion ability.
3.10. HER2 overexpression causes enhanced metastasis of anoikis resistant cells in vivo
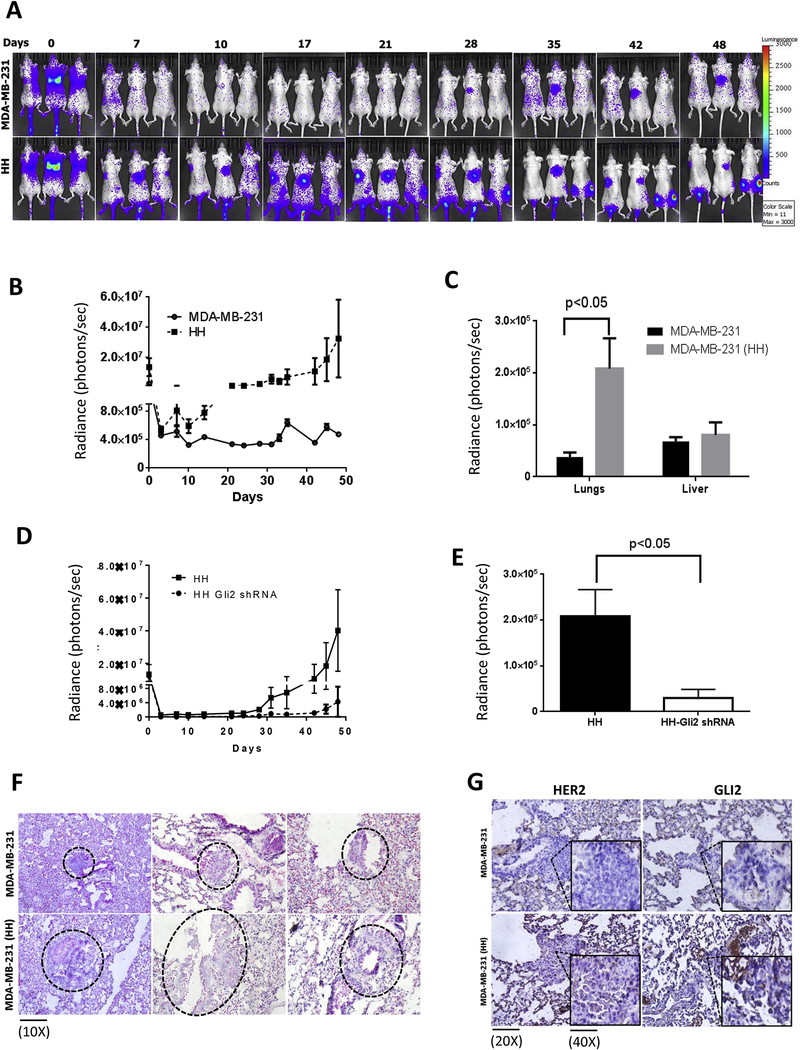
The in vitro observations were further confirmed in an in vivo metastasis model. Equal number of viable luciferase transfected and anoikis resistant MDA-MB-231 and HH cells were injected by tail vein route in athymic nude mice. In addition, we also injected anoikis resistant HH cells transfected with GLI2 shRNA, to confirm the role of SHH signaling in anoikis resistance. The metastasis was monitored periodically by in vivo imaging. The imaging data showed enhanced rate and extent of metastasis in mice injected with anoikis resistant HH cells as compared to MDA-MB-231 cells (Fig. 6A). At the end of experiment, mice were euthanized humanely and livers and lungs were collected for imaging. A 5.5 fold increase in luminescence was observed in the lungs of mice injected with HH cells (Fig. 6C). We also observed a minor increase of about 1.2 fold in the luminescence in the livers of mice injected with HH cells (Fig. 6C). However, metastasis of anoikis resistant HH cells that were transfected with GLI2 shRNA was significantly suppressed as suggested by luminescence curve (Fig. 6D). Our results showed a significant reduction in luminescence 24h after the cell injection in HH cells with GLI2 shRNA (Fig. 6D). Interestingly, luminescence signal did not show any significant increase till day 40 (Fig. 6D). The luminescence from HH GLI2 shRNA cells was about 10 fold less than luminescence from HH cells at the end of the experiment (Fig. 6E). These results indicated enhanced metastasis of HH cells when compared with MDA-MB-231 cells. In addition, GLI2 was also shown to play a crucial role in metastasis of HH cells.
Figure 6– Increased metastasis of HH cells in vivo and increased expression of HER2 and GLI2 in tumors from anoikis resistant MDA-MB-231 and HH cells.
About 0.5 × 106 MDA-MB-231 or HH anoikis resistant cells were injected by tail vein route in athymic nude female mice (n=6). The mice were imaged periodically for bioluminescence after luciferin injections. A&B) Time dependent luminescence imaging data from athymic nude mice for MDA-MB-231 and HH cells. C) Luminescence imaging data from isolated lungs and livers of from MDA-MB-231 and HH injected mice at the end of experiment. D) Time dependent luminescence signal from mice injected with HH and HH with GLI2shRNA cells. E) Average luminescence of the lungs from HH and HH-GLI2 shRNA group, which were imaged ex vivo at the end of the experiment. Values were plotted as means ± SEM (n=6). The lungs from mice of both the groups were fixed in formalin and processed for microscopic evaluation of tumors. F) H&E staining for tumors from different mouse lung samples of MDA-MB-231 and HH injected mice. G) immunohistochemical staining for HER2 and GLI2 in tumors from MDA-MB-231 and HH injected mice lungs. The inset in all panel shows enlarged view of tumor cells. Values were plotted as mean ± SEM (n=6).
3.11. HER2 overexpressing tumors show increased expression of nuclear GLI2
The metastatic tumors formed by HH anoikis resistant cells were compared with MDA-MB-231 anoikis resistant cells by histology and immunohistochemistry for GLI2 and HER2 expression. H&E staining showed a drastic difference in the histology of tumors formed by MDA-MB-231 cells and HH cells (Fig. 6F). The HH tumors were highly invasive and larger, whereas MDA-MB-231 tumors were more localized and smaller in size (Fig. 6F). Furthermore, immunostaining showed significantly increased HER2 expression in tumors of HH as compared to tumors formed by MDA-MB-231 (Fig. 6G). Moreover, expression of GLI2 was more in the nucleus of cells of tumors induced by HH cells as compared to MDA-MB-231 tumors (Fig. 6G). These observations suggest that GLI2 expression was significantly higher in HH tumors, which may account for increased metastatic potential of breast cancer cells.
4. Discussion
Anchorage-independent growth of cells is an important hallmark of cancer metastasis [2]. Few molecular pathways have been implicated in anoikis resistance leading to anchorage-independent cell growth [3]. It has been shown that HER2 positive tumors have more metastatic potential, however, exact mechanism behind HER2-mediated metastasis is not known [34]. Therefore, it is important to understand the underlying mechanisms of higher metastatic rates in HER2 overexpressing breast tumors in patients. In the current study we studied the role of HER2 in anoikis resistance through hedgehog signaling as a possible mechanism of enhanced breast cancer metastasis.
HER2 is a receptor tyrosine kinase overexpressed in about 25% of breast cancer patients and known to be associated with poor prognosis [4, 35]. Few studies indicated higher anoikis resistance in HER2 positive breast cancer cells that were dependent on HIF1α and integrinα5 signaling [6, 7]. In line with these studies, we observed increased anoikis resistance in HER2 overexpressing breast cancer cells relative to cells with basal HER2. We also observed that HER2 inhibition significantly suppressed survival of anoikis-resistant cells. In addition we observed an increased proliferation and mammosphere formation by anoikis resistant HER2 overexpressing breast cancer cells as compared to anoikis resistant cells with constitutive HER2 expression. These observations clearly implicated HER2 in anoikis resistance.
Sonic hedgehog (SHH) signaling has recently gained significant attention due to its role in various cancers including breast cancer [11, 12, 16]. GLI1 and GLI2 are major transcription factors known to mediate the action of SHH ligand [36]. Previously, we have shown the important role of sonic hedgehog signaling in anoikis resistance in ovarian cancer cells [17]. We observed that GLI1 inhibition suppressed anoikis resistance in ovarian cancer cells [17]. In agreement with our previous study, we observed sonic hedgehog signaling mediated anoikis resistance in breast cancer cells in our current study. We observed that inhibition of sonic hedgehog signaling by small molecule inhibitor cyclopamine or silencing GLI2 by shRNA significantly reduced anoikis resistance in breast cancer cells. Interestingly, GLI2 is known to play a significant role in breast cancer progression [16]. In contrast to our previous study, we did not observe modulation of GLI1 in the present study. However, this variation could be attributed to the difference in cancer models. Our present results also showed activation of sonic hedgehog signaling with HER2 overexpression and inhibition of sonic hedgehog signaling with HER2 silencing, suggesting a direct link between HER2 and sonic hedgehog signaling. Interestingly, we also observed that HER2-SHH interaction was not only specific to anoikis resistance (anchorage independent) but also observed under anchorage-dependent conditions. To the best of our knowledge, ours is the first study to identify the cross-talk between HER2 and SHH signaling in breast cancer and its direct implication in metastasis.
A study by Pan et. al. has shown regulation of GLI2 activity by GSK3β mediated proteasomal degradation [37]. In our current study, we observed higher expression of SHH and GLI2 in HH cells as compared to parent MDA-MB-231 cells. In addition, we also observed reduced expression of GLI2 and other molecules of sonic hedgehog signaling in SKBR3 cells when HER2 was silenced as compared to parent SKBR3 cells. Furthermore, there was increased nuclear localization of GLI2 in HH cells, suggesting increased GLI2 activity with HER2 overexpression. Interestingly, when protein synthesis was inhibited, GLI2 was stable for a longer duration in HH cells as compared to MDA-MB-231 cells. We also observed reduced expression of GSK3β in HH cells relative to parent cells with low HER2. Interestingly, GLI2 expression was increased in MDA-MB-231 cells by treatment with GSK3β or proteasome inhibitors. Hence, it can be concluded that HER2 inhibits degradation of GLI2 by suppressing GSK3β activity, resulting in the increased transcriptional activity of GLI2. This was further confirmed by reduced association of GLI2 to ubiquitins in HH cells relative to MDA-MB-231 cells with low HER2. These results clearly suggest a direct role of GSK3β in regulating HER2 mediated proteasomal degradation of GLI2 in our model.
The metastasis of anoikis resistant HH cells to lungs was increased by 5 fold as compared to parent MDA-MB-231 cells. The metastatic tumors in the lungs induced by HH cells were bigger and invasive as compared to small and localized tumors induced by MDA-MB-231 cells. Immunohistochemical staining demonstrated increased expression of HER2 and GLI2 in the metastatic tumor lesions in the lungs.
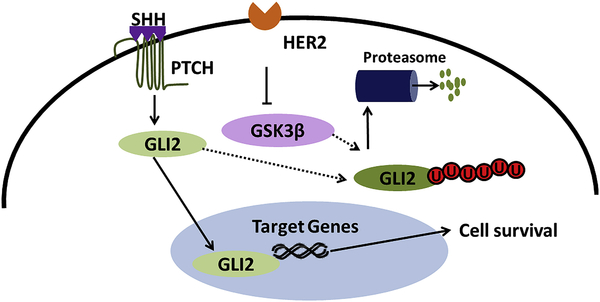
Taken together our study provides a novel insight into the mechanism of anoikis resistance in breast cancer. Furthermore, our study also demonstrated a novel cross-talk between HER2 and SHH signaling mediated by GLI2 (Figure 7). This study also raises a possibility of the role of SHH signaling in drug resistance due to its cross-talk with HER2. To the best of our knowledge, this is the first report to identify the novel cross-talk of HER2 and SHH signaling and their role in anoikis resistance and implications in metastasis. The outcomes from this study purports SHH signaling as a potential target in HER2 positive cancer patients.
Figure 7–
Schematic molecular mechanism of SHH pathway regulation by HER2 in breast cancer cells
Supplementary Material
Highlights:
Metastastic breast cancer cells acquire resistance to anoikis
Increased anoikis resistance has been observed in HER2 overexpression breast cancer cells
HER2 overexpression was also associated with increased sonic hedgehog signaling
Acknowledgements
This work was supported in part by R01 grant CA129038 (to Sanjay K. Srivastava) awarded by the National Cancer Institute, NIH. Instrument grant RP110786 awarded by CPRIT (Cancer Prevention and Research Institute of Texas) for multiphoton microscope is also acknowledged. Kind gift of MDA-MB-231 (HH) cells by Dr. Patricia S. Steeg (National Cancer Institute, Maryland) and Dr. Quentin Smith (Texas Tech University Health Sciences Centre, Amarillo, Texas) are greatly appreciated.
Abbreviations:
- HER2
Human epidermal growth receptor 2
- HH
MDA-MB-231 HER2 overexpressing cells
- SHH
Sonic hedgehog
- PCR
Polymerase chain reaction
- IHC
Immunohistochemistry
Footnotes
Conflict of interest
Authors declare that there are no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Frisch SM, Francis H, Disruption of epithelial cell-matrix interactions induces apoptosis, The Journal of Cell Biology, 124 (1994) 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell, 144 (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [3].Guadamillas MC, Cerezo A, Del Pozo MA, Overcoming anoikis--pathways to anchorage-independent growth in cancer, Journal of Cell Science, 124 (2011) 3189–3197. [DOI] [PubMed] [Google Scholar]
- [4].Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL, Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene, Science, 235 (1987) 177–182. [DOI] [PubMed] [Google Scholar]
- [5].Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, Shak S, Stewart SJ, Press M, Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer, Journal of Clinical Oncology, 20 (2002) 719–726. [DOI] [PubMed] [Google Scholar]
- [6].Haenssen KK, Caldwell SA, Shahriari KS, Jackson SR, Whelan KA, Klein-Szanto AJ, Reginato MJ, ErbB2 requires integrin alpha5 for anoikis resistance via Src regulation of receptor activity in human mammary epithelial cells, Journal of Cell Science, 123 (2010) 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Whelan KA, Schwab LP, Karakashev SV, Franchetti L, Johannes GJ, Seagroves TN, Reginato MJ, The oncogene HER2/neu (ERBB2) requires the hypoxia-inducible factor HIF-1 for mammary tumor growth and anoikis resistance, The Journal of Biological Chemistry, 288 (2013) 15865–15877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Grassian AR, Schafer ZT, Brugge JS, ErbB2 stabilizes epidermal growth factor receptor (EGFR) expression via Erk and Sprouty2 in extracellular matrix-detached cells, The Journal of Biological Chemistry, 286 (2011) 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, Muthuswamy SK, Brugge JS, Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis, Nature cell biology, 5 (2003) 733. [DOI] [PubMed] [Google Scholar]
- [10].Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG, The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells, Cell, 138 (2009) 1083–1095. [DOI] [PubMed] [Google Scholar]
- [11].Kasper M, Jaks V, Fiaschi M, Toftgard R, Hedgehog signalling in breast cancer, Carcinogenesis, 30 (2009) 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hui M, Cazet A, Nair R, Watkins DN, O’Toole SA, Swarbrick A, The Hedgehog signalling pathway in breast development, carcinogenesis and cancer therapy, Breast Cancer Research, 15 (2013) 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Javelaud D, Alexaki VI, Dennler S, Mohammad KS, Guise TA, Mauviel A, TGF-β/SMAD/GLI2 signaling axis in cancer progression and metastasis, Cancer research, 71 (2011) 5606–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Miyagawa S, Matsumaru D, Murashima A, Omori A, Satoh Y, Haraguchi R, Motoyama J, Iguchi T, Nakagata N, Hui C.-c., The role of sonic hedgehog-Gli2 pathway in the masculinization of external genitalia, Endocrinology, 152 (2011) 2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pan Y, Gong Y, Ruan H, Pan L, Wu X, Tang C, Wang C, Zhu H, Zhang Z, Tang L, Sonic hedgehog through Gli2 and Gli3 is required for the proper development of placental labyrinth, Cell death & disease, 6 (2016) e1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Im S, Choi HJ, Yoo C, Jung JH, Jeon YW, Suh YJ, Kang CS, Hedgehog related protein expression in breast cancer: gli-2 is associated with poor overall survival, Korean Journal of Pathology, 47 (2013) 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kandala PK, Srivastava SK, Diindolylmethane-mediated Gli1 protein suppression induces anoikis in ovarian cancer cells in vitro and blocks tumor formation ability in vivo, The Journal of Biological Chemistry, 287 (2012) 28745–28754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fofaria N, Srivastava SK, Critical role of STAT-3 in melanoma metastasis through anoikis resistance, Oncotarget, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fofaria NM, Srivastava SK, STAT3 induces anoikis resistance, promotes cell invasion and metastatic potential in pancreatic cancer cells, Carcinogenesis, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jeng K.-s., Sheen I-S, Jeng W-J, Yu M-C, Hsiau H-I, Chang F-Y, High expression of Sonic Hedgehog signaling pathway genes indicates a risk of recurrence of breast carcinoma, OncoTargets and therapy, 7 (2014) 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sahu RP, Zhang R, Batra S, Shi Y, Srivastava SK, Benzyl isothiocyanate-mediated generation of reactive oxygen species causes cell cycle arrest and induces apoptosis via activation of MAPK in human pancreatic cancer cells, Carcinogenesis, 30 (2009) 1744–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gupta P, Srivastava SK, Antitumor activity of phenethyl isothiocyanate in HER2-positive breast cancer models, BMC Medicine, 10 (2012) 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gupta P, Adkins C, Lockman P, Srivastava SK, Metastasis of breast tumor cells to brain is suppressed by phenethyl isothiocyanate in a novel in vivo metastasis model, PLoS One, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sahu RP, Srivastava SK, The role of STAT-3 in the induction of apoptosis in pancreatic cancer cells by benzyl isothiocyanate, Journal of National Cancer Institute, 101 (2009) 176–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M, A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2, Human Molecular Genetics, 14 (2005) 2181–2188. [DOI] [PubMed] [Google Scholar]
- [26].Ranjan A, Srivastava SK, Penfluridol suppresses pancreatic tumor growth by autophagy-mediated apoptosis, Sci Rep, 6 (2016) 26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gupta P, Srivastava SK, Inhibition of HER2-integrin signaling by Cucurbitacin B leads to in vitro and in vivo breast tumor growth suppression, Oncotarget, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ranjan A, Gupta P, Srivastava SK, Penfluridol: An Antipsychotic Agent Suppresses Metastatic Tumor Growth in Triple-Negative Breast Cancer by Inhibiting Integrin Signaling Axis, Cancer Res, 76 (2016) 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pramanik KC, Fofaria NM, Gupta P, Ranjan A, Kim SH, Srivastava SK, Inhibition of beta-catenin signaling suppresses pancreatic tumor growth by disrupting nuclear beta-catenin/TCF-1 complex: critical role of STAT-3, Oncotarget, 6 (2015) 11561–11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gupta P, Srivastava SK, HER2 mediated de novo production of TGFbeta leads to SNAIL driven epithelial-to-mesenchymal transition and metastasis of breast cancer, Molecular oncology, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Reynolds DS, Tevis KM, Blessing WA, Colson YL, Zaman MH, Grinstaff MW, Breast Cancer Spheroids Reveal a Differential Cancer Stem Cell Response to Chemotherapeutic Treatment, Scientific reports, 7 (2017) 10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS, In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells, Genes & development, 17 (2003) 1253–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xu X, Lu Y, Li Y, Prinz RA, Sonic Hedgehog signaling in thyroid cancer, Frontiers in endocrinology, 8 (2017) 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Eccles SA, The role of c-erbB-2/HER2/neu in breast cancer progression and metastasis, Journal of Mammary Gland Biology and Neoplasia, 6 (2001) 393–406. [DOI] [PubMed] [Google Scholar]
- [35].Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. , Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer, Science, 244 (1989) 707–712. [DOI] [PubMed] [Google Scholar]
- [36].Katoh Y, Katoh M, Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review), International Journal of Molecular Medicine, 22 (2008) 271–275. [PubMed] [Google Scholar]
- [37].Pan Y, Bai CB, Joyner AL, Wang B, Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation, Molecular and Cellular Biology, 26 (2006) 3365–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.