Abstract
Endoplasmic reticulum (ER) stress activates three principal signaling pathways, collectively known as the unfolded protein response, leading to translational and transcriptional control mechanisms that dictate the cell’s response as adaptive or apoptotic. The present study illustrates that for HepG2 human hepatocellular carcinoma cells the signaling pathways triggered by ER stress extend beyond the three principal pathways to include mitogen-activated protein kinase (MAPK) signaling, leading to activation of transcription from the early growth response 1 (EGR1) gene. Analysis provided evidence for a SRC-RAS-RAF-MEK-ERK cascade mechanism that leads to enhanced phosphorylation of the transcription factor ELK1. ELK1 and serum response factor (SRF) are constitutively bound to the EGR1 promoter and are phosphorylated by nuclear localized ERK. The promoter abundance of both phospho-SRF and phopsho-ELK1 was increased by ER stress, but the SRF phosphorylation was transient. Knockdown of ELK1 had little effect on the basal EGR1 mRNA content, but completely blocked the increase in response to ER stress. Conversely, knockdown of SRF suppressed basal EGR1 mRNA content, but had only a small effect on the induction by ER stress. This research highlights the importance of MAPK signaling in response to ER stress and identifies ELK1 as a transcriptional mediator and the EGR1 gene as a target.
Keywords: unfolded protein response, thapsigargin, ELK1, RAS, RAF
1. Introduction
Endoplasmic reticulum (ER) stress occurs when there is a perturbation in ER-associated protein synthesis or folding, calcium flux, or other organelle malfunction. During ER stress, three membrane-associated sensors trigger individual signal transduction pathways collectively called the unfolded protein response (UPR) [1, reviewed in 2]. If the adaptation responses elicited by these three adaptive pathways are not sufficient to return to homoeostasis, apoptosis is triggered. Using mouse embryonic fibroblasts treated with thapsigargin (Tg), which triggers ER stress by perturbing calcium flux across the ER membrane [3], RNA-sequencing showed that ER stress activates hundreds of genes, including members of the early growth response (EGR) transcription factor family [4]. Separate studies with another cellular stress, amino acid deprivation, established that EGR1 is induced by a mechanism that involves the extracellular-regulated kinase (ERK) arm of mitogen-activated protein kinase (MAPK) signaling [5]. In HepG2 human hepatocellular carcinoma (HCC) cells, both the JNK and MEK arms of the MAPK pathways contribute to ATF4-independent amino acid stress signaling [5-9]. Furthermore, following amino acid deprivation, ERK-dependent signaling also leads to induction of Dickkopf homolog 1, a WNT signaling antagonist, [10] and JNK-dependent signaling promotes phosphorylation of ATF2, an important histone acetyltransferase [11]. In vivo, elevated levels of p-ERK localize in a specific group of neurons involved in aversion of diets deficient in an essential amino acids [12] and ERK signaling is involved in the aversion process [13, 14]. Depleting circulating asparagine by treatment of mice with the anti-leukemia drug asparaginase triggers induction of hepatic ERK signaling in the liver [15]. Collectively, these observations document that MAPK signaling is an underappreciated component of the over-lapping pathways triggered by amino acid limitation and ER stress.
A broad range of extracellular stimuli activate expression of the EGR1 gene, which encodes a transcription factor that functions as an immediate early response signal. EGR1 impacts cell growth, proliferation, differentiation, and apoptosis [reviewed in 16]. The signals that control EGR1 gene expression vary depending on the initial stimulus and target tissue. However, a common mechanism is ERK-dependent signaling that leads to phosphorylation of the E-twenty six-like transcription factor (ELK1) and the serum response factor (SRF), which are constitutively bound to E-twenty six-like (ETS) sequences and serum response element (SRE) sequences, respectively, within the EGR1 promoter [16, 17]. Once produced, EGR1 protein directly and indirectly regulates the expression of genes critical to cell proliferation [18]. Egr1 knockout mice, though viable, exhibit impaired liver regeneration following partial hepatectomy and Egr1 has been proposed as a central regulator of cell cycle progression during hepatocellular regeneration following injury [19]. Thus, control of hepatic EGR1 expression by ER stress may be a critical factor in liver physiology.
The present study documents that the ER stress-initiated induction of EGR1 transcription is mediated by SRC-RAS-RAF-MEK-ERK signaling leading to enhanced phosphorylation of EGR1 promoter-bound SRF and ELK1. Therefore, the results provide evidence that in some cells the ETS/SRF families of transcription factors are included within the umbrella of ER stress and that they enhance transcription through ETS/SRE genomic enhancer sequences. Furthermore, induction of immediate-early response genes, such as EGR1, in response to ER stress in tumor cells provides a possible link between cell stress and cell growth in the transformed state.
2. Materials and methods
2.1. Cell Culture
The cell lines used in these studies were cultured in Dulbecco’s modified Eagle’s medium (DMEM, pH 7.4, Corning, Manassas, VA), supplemented with 1X non-essential amino acids, 2 mM glutamine, 100 μg/ml streptomycin sulfate, 100 U/ml penicillin G, 0.25 μg/ml amphotericin B, and 10% v/v fetal bovine serum. Cells were maintained at 37°C in an atmosphere of 5% CO2 and 95% air and used while in growth phase at 60-70% confluence. Approximately 12 h prior to treatments, cells were replenished with fresh DMEM medium to ensure more complete nutrition when experiments were initiated.
2.2. Knockdown of Selected Proteins
The SMARTpool siRNA oligonucleotides for siH-RAS (#M-004142-00), siN-RAS (#M-003919-00), siK-RAS (#M-005069-00), siELK1 (#L-003885-00), siERK1 (#L-003592-00-0005), siERK2 (#L-003555-00-0005), siJNK1 (#L-003514-00-0005), siJNK2 (#L-003505-00-0005), siRAF1 (#L-003601-00-0005), and siSRF (#L-009800-00-0005) were purchased from Dharmacon/Thermo Fisher Scientific. The Silencer select siRNA for siSRC (#4390824) was purchased from Ambion/Thermo Fisher Scientific. At 72 h prior to activating ER stress, transient transfections were performed in 12-well plates using a total amount of 100 nM siRNA with DharmaFECT4 Transfection Reagent (#T-2004-01), according to the manufacturer’s protocol. The same amount of siRNA with a scrambled sequence (Dharmacon/Thermo Fisher Scientific, #D-001810-01-05) was used as the siControl.
2.3. RNA Isolation and Real-Time Quantitative PCR (RT-qPCR)
Total RNA was isolated with the TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s directions. The steady state mRNA levels were assayed by RT-qPCR, as described previously [20]. A 1 μg aliquot of total RNA was used to synthesize first-strand cDNA with the qScript cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD). For RT-qPCR, each cDNA sample was diluted 10X with TE buffer (10 mM TRIS, 1 mM EDTA, pH 8.0), 2 μl of this diluted solution was mixed with 10 μl of SYBR Green master mixture, and 5 pmol of forward and reverse primers were added in a total volume of 20 μl. The mixture was subjected to 40 cycles at 95°C for 15 s and then at 60°C for 60 s. The primers used are listed in Table 1. After RT-qPCR, melting curves were acquired by stepwise increase from 55°C to 95°C to ensure that only a single product was amplified in the reaction. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control and all calculations were based on the difference of threshold cycle number of the analyzed gene relative to the GAPDH mRNA content in the same sample. To measure the transcription activity, a pair of oligonucleotide primers (Table 1) corresponding the intron 1 and exon 2 of the EGR1 gene were used to measure the heterogeneous nuclear (hnRNA), as described in a previous report [21].
Table 1.
PCR Primers
| Primer Specificity | Primer Sequences |
|---|---|
| BiP/GRP78, mRNA | FP 5′-GCCATGGTTCTCACTAAAATGAAAGAAAC-3′ |
| RP 5′-TTGGGCATCATTAAAATAGGCTGGTAC-3′ | |
| GAPDH, mRNA | FP 5′-TTGGTATCGTGGAAGGACTC-3′ |
| RP 5′-ACAGTCTTCTGGGTGGCAGT-3′ | |
| EGR1, mRNA | FP 5′-AGAAGGACAAGAAAGCAGACAAAAGTGT-3′ |
| RP 5′-GGGGACGGGTAGGAAGAGAG-3′ | |
| EGR1, hnRNA | FP 5′-CTACGAGCACCTGACCGCAGG-3′ |
| RP 5′-ACAGGACGCCAGGATGGTGG-3′ | |
| K-RAS, mRNA | FP 5′-CTAGAACAGTAGACACAAAACAGG-3′ |
| RP 5′-CGAACTAATGTATAGAAGGCATC-3′ | |
| H-RAS, mRNA | FP 5′-TACGGCATCCCCTACATCGAGAC-3′ |
| RP 5′-CACCAACGTGTAGAAGGCATCCTC-3′ | |
| N-RAS, mRNA | FP 5′-GAGTTACGGGATTCCATTCATTGAAAC-3′ |
| RP 5′-TGGCGTATTTCTCTTACCAGTGTGTAAAA-3′ | |
| RAF1, mRNA | FP 5′-TATTGGGAAATAGAAGCCAGTGAAGTGA-3′ |
| RP 5′-AACATCTCCGTGCCATTTACCCTTATA-3′ | |
| ELK1, mRNA | FP 5′-CTGACCCCATCCCTGCTTCCTA-3′ |
| RP 5′-GAAGTGAATGCTAGGAGGCAGCG-3′ | |
| SRF, mRNA | FP 5′-AGTGGGGAGACCAAGGACACAC-3′ |
| FP 5′-TGGTGGTAGAGGTGCTAGGTGC-3′ | |
| SRC, mRNA | FP 5′-AGCGGCTCCAGATTGTCAACAA-3′ |
| RP 5′-GGATGTAGCCTGTCTGTCCTGTGC-3′ | |
| JNK1, mRNA | FP 5′-CCATTTCAGAATCAGACTCATGCCA-3′ |
| RP 5′-TGTGGTGTGAAAACATTCAAAAGGC-3′ | |
| JNK2, mRNA | FP 5′-GGGATTGTTGTGCTGCATTTGATAC-3′ |
| RP 5′-TGGTTCTGAAAAGGACGGCTTAGTTT-3′ | |
| ERK1, mRNA | FP 5′-CGCTTCCGCCATGAGAATGTC-3′ |
| RP 5′-CAGGTCAGTCTCCATCAGGTCCTG-3′ | |
| ERK2, mRNA | FP 5′-CGTGTTGCAGATCCAGACCATGAT-3′ |
| RP 5′-TGGACTTGGTGTAGCCCTTGGAA-3′ | |
| hEGR1 P1, ChIP Assay | FP 5′-CCCCGTCTCAGAAAGAATAAAAACATTA-3′ |
| RP 5′-CCTTGTGTCTGAATGTCCATTTTGC-3′ | |
| hEGR1 P2, ChIP Assay | FP 5′-CCTCTTTCGGATTCCCGCAG-3′ |
| RP 5′-GGTCCTTGTGGTGAGGGGTCA-3′ | |
| hEGR1 P3, ChIP Assay | FP 5′-GAGGGAGCGAGGGAGCAACC-3′ |
| RP 5′-CTCCAAATAAGGTGCTGCCCAAA-3′ | |
| hEGR1 P4, ChIP Assay | FP 5′-CATATTAGGGCTTCCTGCTTCCCATA-3′ |
| RP 5′-CCGCCTCTATTTGAAGGGTCTGG-3′ | |
| hEGR1 P5, ChIP Assay | FP 5′-CGCAGAGGACCGAGCTTTTGT-3′ |
| RP 5′-GCAGCCCCGCTCATCAAAA-3′ | |
| hEGR1 P6, ChIP Assay | FP 5′-GGGGATTCTCCGTATTTGCGTC-3′ |
| RP 5′-GGCTACCATTGACTCCCGAGGT-3′ | |
| hEGR1 P7, ChIP Assay | FP 5′-GTCCCAGCTCATCAAACCCAGC-3′ |
| RP 5′-AGAAGCGGCGATCACAGGACTC-3′ |
2.4. Protein Isolation and Immunoblotting
Whole cell protein was extracted with a RIPA buffer (50 mM Tris-HCl, 1% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, containing Pierce Protease and Phosphatase inhibitor mini-tablets (Thermo Fisher Scientific). Immunoblotting was performed as described previously [21]. The rabbit anti-EGR1 antibody (cat #4153) was obtained from Cell Signaling Technology (Danvers, MA) and the rabbit anti-β-actin polyclonal antibody (#A2066) was from Sigma-Aldrich. The bound secondary antibody was detected using an enhanced chemiluminescence kit (#32106, Thermo Fisher Scientific) and then exposing the blot to Classic Blue Autoradiography Film BX (MIDSCI, St. Louis, MO).
2.5. Chromatin Immunoprecipitation (ChIP)
ChIP analysis was performed according to a previously published protocol [21]. HepG2 cells were seeded at a density of 1.5 × 107 per 150 mm dish with DMEM medium and cultured for approximately 36 h, which included a transfer to fresh DMEM during the final 12 h prior to ER stress-induction. The rabbit anti-serum response factor (SRF, #sc-335) and, as a non-specific negative control a normal rabbit IgG (#sc-2027), were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The total ELK1 (#9182) and S103 phospho-SRF (#4261) antibodies were obtained from Cell Signaling, whereas the S383 phospho-ELK1 antibody was from Abcam (#32799, Boston, MA), a monoclonal antibody that recognizes both the phospho- and non-phosphorylated C-terminal domain of the largest subunit of DNA polymerase II was from Millipore (clone CTD4H8, #05-623). Immunoprecipitated DNA was analyzed with qPCR as described above, using primers listed in Table 1. The ChIP results are presented as the ratio to input DNA.
2.6. Statistical Analysis
Each experiment contained three or more replicate samples to detect experimental variation within an experiment and to ensure reproducibility each experiment was repeated one or more times with independent batches of cells. The data are expressed as the averages ± standard deviations within an individual experiment containing 3-4 replicates and the results analyzed using Student’s t test with a p ≤ 0.05 considered statistically significant.
3. Results
3.1. ER stress increases transcription from the EGR1 gene
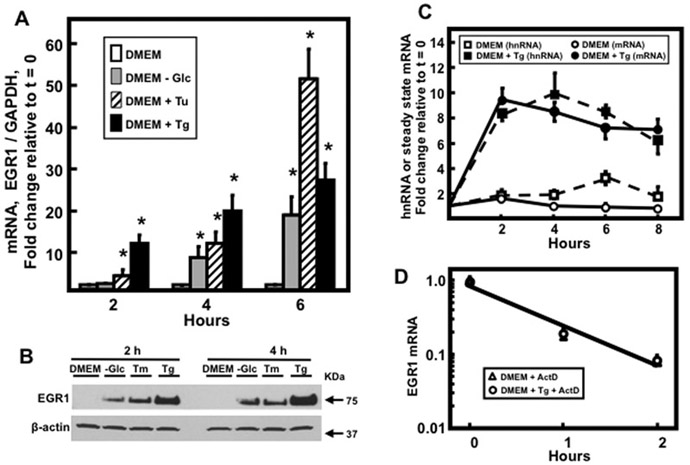
HepG2 human HCC cells were subjected to glucose starvation, tunicamycin (Tu), or thapsigargin (Tg), each of which causes ER stress [22]. Although the time course of each was different, all three stimuli resulted in an increase in EGR1 steady state mRNA (Fig.1A) and protein expression (Fig. 1B). To investigate the mechanism for the increased mRNA, cells were treated with Tg and the short-lived EGR1 hnRNA was assayed as a measure of transcription [21]. ER stress led to a significant induction of hnRNA that paralleled the rise in steady state mRNA (Fig. 1C). To determine if mRNA stabilization also contributed to the steady state mRNA level, EGR1 expression was induced by Tg treatment and then the cells were incubated with or without Tg in the presence of actinomycin D to block further transcription (Fig. 1D). The decay rate for EGR1 mRNA, about 30 min, was not significantly changed in the presence of Tg indicating that mRNA stabilization does not play a major role in the increased expression.
Fig. 1.
Regulation of EGR1 expression in response to ER stress. (Panel A) HepG2 human HCC cells were incubated in complete DMEM or DMEM lacking glucose, DMEM + 6 μM Tu, or DMEM + 50 nM Tg for the indicated time. EGR1 steady state mRNA content was assayed by RT-qPCR. GAPDH mRNA, which is not affected by ER stress, was used as an internal control and data shown are the means ± SD of at least triplicate samples within an experiment. The results shown are representative of multiple experiments. An asterisk indicates that the value is significantly different (p ≤ 0.05) from the DMEM control. (Panel B) HepG2 cells were incubated in complete DMEM or DMEM lacking glucose, DMEM + 6 μM Tu, or DMEM + 50 nM Tg for 2 and 4 hr and then EGR1 protein was measured in whole cell extracts by immunoblotting. The protein level of β-actin was used as the control. (Panel C) HepG2 human HCC cells were incubated in complete DMEM or DMEM + 50 nM Tg for the times indicated and EGR1 transcription activity, as measured by hnRNA, or steady state mRNA were assayed by RT-qPCR. (Panel D) The cells were incubated in DMEM + 50 nM Tg for 8 h and then transferred to complete DMEM or DMEM + Tg, which contained 5 μM actinomycin D (ActD), for an additional 2 h. EGR1 mRNA was measured by RT-qPCR and data were plotted as the logarithm of mRNA content versus time following incubation in ActD-containing medium. GAPDH mRNA, which is not affected by ER stress, was used as an internal control and results shown are the means ± SD of at least triplicate samples within an experiment. The results shown are representative of multiple experiments.
3.2. Cell Specificity of the EGR1 Induction
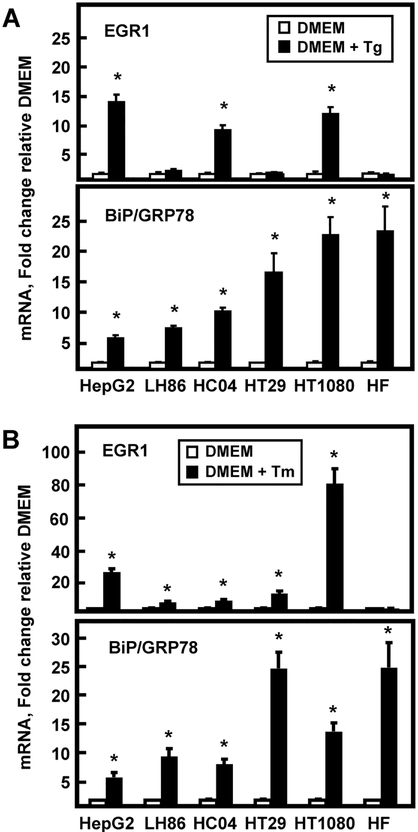
ER stress-associated induction of EGR1 mRNA expression by two different stimuli was monitored in several cell types other than HepG2 HCC to survey the breadth of the response (Fig. 2). For Tg treatment, LH86 human HCC, HT29 human colon adenocarcinoma, and human fibroblasts showed no induction of EGR1, whereas HepG2, HC-04 immortalized human hepatocytes, and HT1080 human fibrosarcoma cells were responsive (Fig. 2A). HepG2 and HT1080 cells also responded strongly to Tm as well, but the induction in Hc-04 cells was quantitatively smaller (Fig. 2B). Induction of BiP/GRP78, a known target of the UPR pathways, [23, 24], was used as a positive control for both stimuli and some degree of increase was observed in all cell types tested (Fig. 2). These results indicate that not all human hepatomas (LH86) respond the same as HepG2, that transformation is not a requirement for the response (HC-04), and that cell lineages (HT1080 colon adenocarcinoma) other than HCC exhibit ER stress-associated induction of EGR1. Interestingly, the results also document that the induction of EGR1 exhibits different cell specificity than does BiP/GRP78, a classic marker of ER stress and the UPR.
Fig. 2.
The cell specificity of the EGR1 ER stress response differs from that for BiP/GRP78. The indicated cell type was incubated in DMEM with either 50 nM Tg (Panel A) or 6 μM Tm (Panel B) for 6 h. Either EGR1 or BiP/GRP78 mRNA was assayed by RT-qPCR. GAPDH mRNA, which is not affected by ER stress, was used as an internal control and data shown are the means ± SD of at least triplicate samples within an experiment. The results shown are representative of multiple experiments. An asterisk indicates that the value is significantly different (p ≤ 0.05) from the DMEM control.
3.3. A MAPK Pathway is Required for ER Stress Activation of EGR1
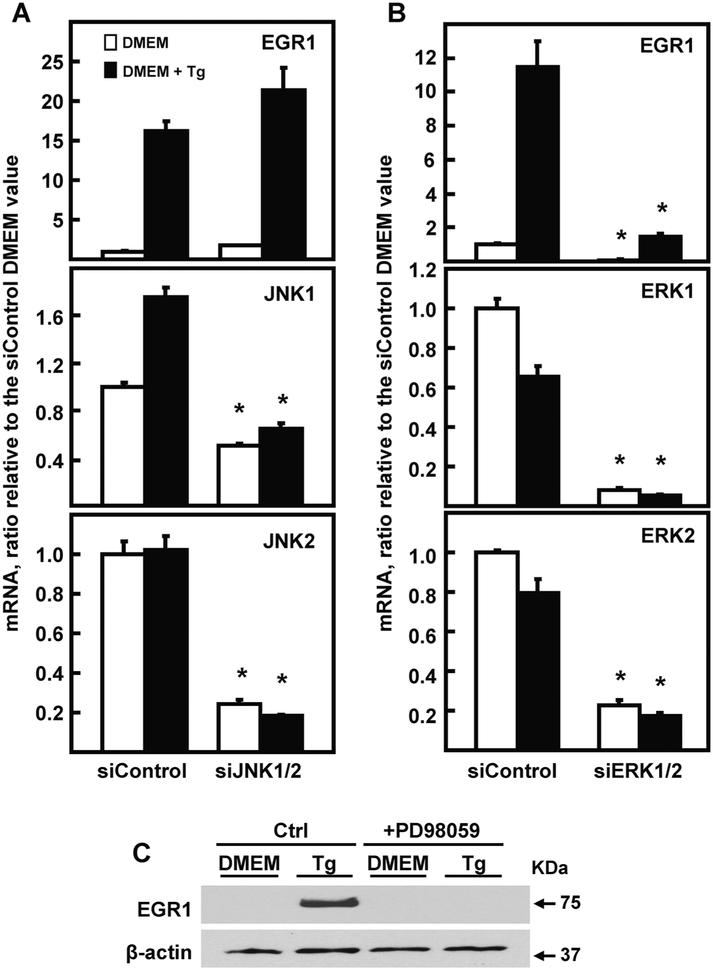
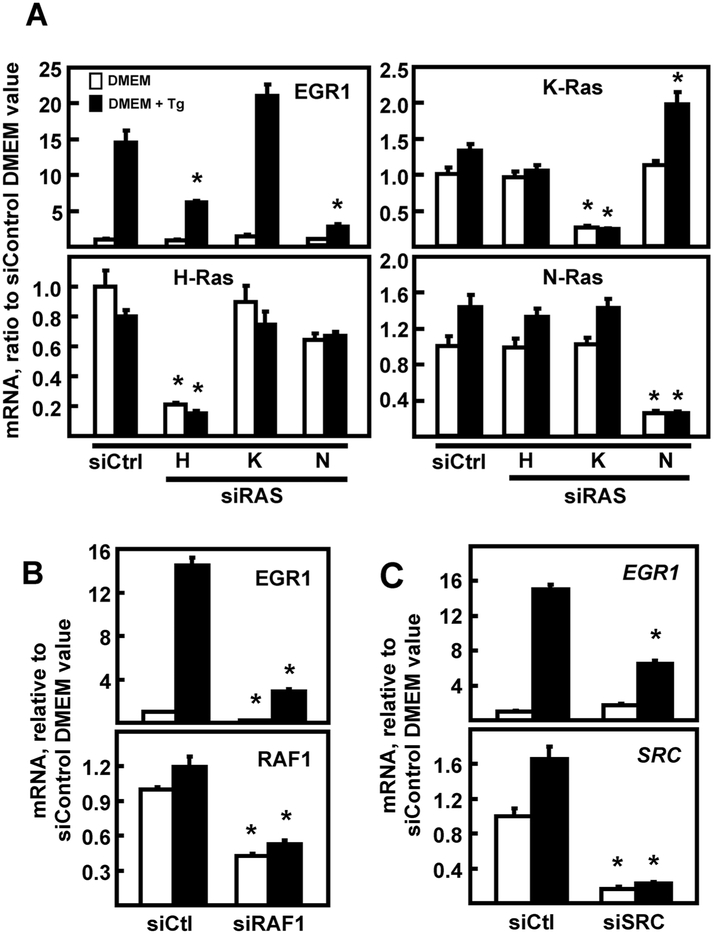
In HepG2 cells, both the JNK and MEK arms of the MAPK pathways contribute to ATF4-independent amino acid stress signaling [5-9]. It is known that there is cross talk between the UPR and the MAPK stress pathways [reviewed in 25]. Given these pathway interactions, as well as the partial overlap between amino acid and ER stress, the JNK and ERK pathways and their upstream effectors were investigated with regard to EGR1 induction by ER stress. Knockdown of JNK1/JNK2 did not inhibit EGR1 induction by ER stress (Fig. 3A), but the EGR1 up-regulation was largely dependent on ERK (Fig. 3B). Independent evidence for ERK involvement and confirmation of protein expression was obtained by documenting that the increase in EGR1 protein was blocked by the MEK inhibitor PD98059 (Fig. 3C). Activation of ERK is complex [26], but among the many possible upstream steps is SRC tyrosine kinase [27-29] and RAS-RAF [26]. Transient transfection of HepG2 cells with anti-sense oligonucleotides revealed a role for H-RAS and N-RAS, but not K-RAS in the EGR1 induction following Tg treatment (Fig. 4A). RAS often signals through RAF1, which is known to activate the MEK-ERK pathway [30]. Consistent with that hypothesis, RAF1 knockdown largely blocked the increase in EGR1 mRNA (Fig. 4B). Knockdown of SRC by siRNA also caused a significant suppression of the ER stress-enhanced EGR1 expression (Fig. 4C). These results suggest that activation of SRC, and possibly other tyrosine phosphorylation events triggered by ER stress, are upstream of the RAS-RAF-MEK-ERK cascade.
Fig. 3.
ERK, but not JNK, is required for ER stress induction of the EGR1 gene. HepG2 cells were transiently transfected with the indicated siRNAs, as described in the Methods section, and cultured for 48 h. Cells were incubated in DMEM ± Tg for 6 h and then steady state mRNA for the indicated gene was measured by RT-qPCR. Cells were transfected with 100 nM of an siRNA against a non-targeting control (siControl) or with a combination of two siRNAs used at 50 nM each: (Panel A) JNK1 and JNK2; (Panel B) ERK1 and ERK2. Steady state mRNA of EGR1, JNK1, JNK2, ERK1, or ERK2 was measured by RT-qPCR. The mRNA content of GAPDH was used as the internal control. For all panels, the target mRNA values were normalized to those for GAPDH within the same sample and the results shown are the means ± SD of triplicate samples within an experiment. The results shown are representative of multiple experiments. The data are graphed as relative values setting the siControl DMEM value to 1.0 and an asterisk indicates that the targeted siRNA value is significantly different (p ≤ 0.05) from the corresponding siControl value. (Panel C) HepG2 human HCC cells were incubated in complete DMEM ± 50 nM Tg and ± 20 μM PD98059 for 6 h. EGR1 protein was measured in whole cell extracts by immunoblotting. The protein level of β-actin was used as the control.
Fig. 4.
EGR1 induction by ER stress is dependent on RAS, RAF1, and SRC. HepG2 cells were transiently transfected with 100 nM siRNA, as described in the Methods section, and cultured for 48 h. Cells were incubated in DMEM ± Tg for 6 h and then steady state mRNA for the indicated gene was measured by RT-qPCR. (Panel A) Cells were transfected with an siRNA against a non-targeting control (Ctrl), or H, K, or N RAS, and then mRNA was measured for EGR1, each RAS form, and GAPDH as the internal control. (Panel B) HepG2 cells were transfected with an siRNA against a non-targeting control (Ctrl) or against RAF1 and then mRNA was measured for EGR1, RAF1, and GAPDH as the internal control. (Panel C) Cells were transfected with an siRNA against a non-targeting control (Ctrl) or siRNA against SRC and then mRNA was measured for EGR1, SRC, and GAPDH as the internal control. For all panels, the target mRNA values were normalized to those for GAPDH within the same sample and the results shown are the means ± SD of triplicate samples within an experiment. The results shown are representative of multiple experiments. The data are graphed as relative values setting the siControl DMEM value to 1.0. An asterisk indicates that the targeted siRNA value is significantly different (p ≤ 0.05) from the corresponding siControl.
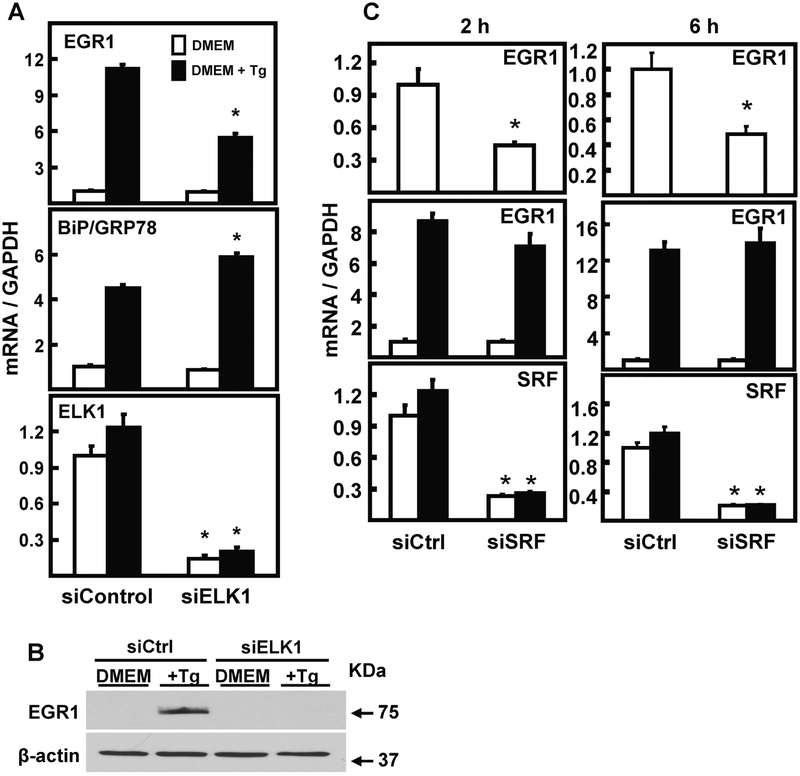
3.4. Induction of EGR1 is diminished in ELK1 knockdown HepG2 cells
ERK transduces cytoplasmic signaling to the nucleus by phosphorylation of transcription factors, including serum response factor (SRF) and ETS-like factor (ELK1), which are constitutively bound to target genes, but inactive prior to phosphorylation. SRF binding to genomic serum response elements (SRE) and ELK1 binding to E-twenty six (ETS) elements often exhibit cooperative binding [reviewed in 31, 32]. To determine the relative contribution of ELK1 and SRF to induction of EGR1 expression by ER stress, HepG2 cells were treated with siRNA specific for ELK1 and then treated with Tg for 6 h (Fig. 5). Knockdown of ELK1 expression caused a significant loss of stress-induced EGR1 mRNA (Fig. 5A) and protein (Fig. 5B) expression. To test the specificity of this reduction, it was observed that the level of BiP/GRP78 during ER stress was not suppressed in response to ELK1 knockdown (Fig. 5A). Initial experiments involving SRF knockdown did not show any significant effect on the induction of EGR1 mRNA after 6 h of ER stress. However, shortening the time of stress to 2 h revealed that although SRF knockdown still did not affect the stress-induced EGR1 mRNA level (Fig. 5C middle panels), loss of SRF did suppress the basal (DMEM only) expression of EGR1 mRNA (Fig. 5C, top panels). Collectively, the data indicate that SRF is required to maintain basal EGR1 expression, but only ELK1 is necessary for the gene’s response to ER stress.
Fig. 5.
EGR1 induction by ER stress is dependent on ELK1, but independent of SRF. (Panel A) HepG2 cells were transiently transfected with 100 nM siRNA against a non-targeting control (Ctrl) or Elk1 as described in the Methods section, and cultured for 48 h. Cells were incubated in DMEM ± Tg for 6 h and then steady state mRNA was measured by RT-qPCR for EGR1, GRP78, ELK1, and GAPDH as the internal control. (Panel B) HepG2 cells transfected with an siRNA against a non-targeting control (Ctrl) or Elk1 were cultured in DMEM ± Tg for 6 h. EGR1 protein was measured in whole cell extracts by immunoblotting. The protein level of β-actin was used as the control. (Panel C) HepG2 cells were transiently transfected with 100 nM siRNA against a non-targeting control (Ctrl) or SRF, as described in the Methods section, and cultured for 48 h. Cells were incubated in DMEM ± Tg for either 2 h or 6 h and then steady state mRNA for the indicated gene was measured by RT-qPCR. For Panels A and C, the target mRNA values were normalized to those for GAPDH within the same sample and the results shown are the means ± SD of triplicate samples within an experiment. The results shown are representative of multiple experiments. The data are graphed as relative values setting the DMEM value of either siControl, siELK1, or siSRF to 1.0. An asterisk indicates that the targeted siRNA value is significantly different (p ≤ 0.05) from the corresponding siControl.
3.5. ER Stress Causes Increased Association of p-ELK1 at the EGR1 Promoter
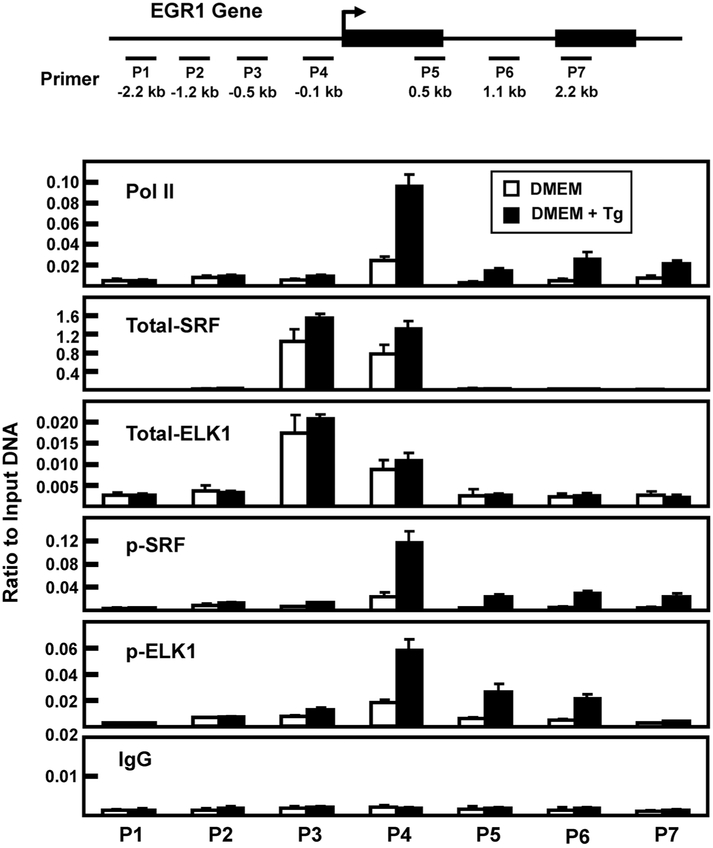
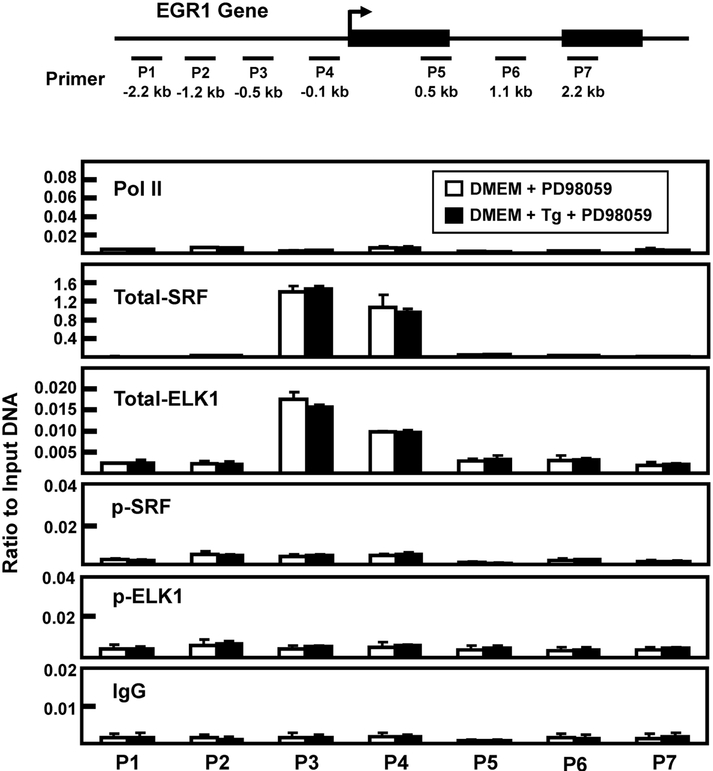
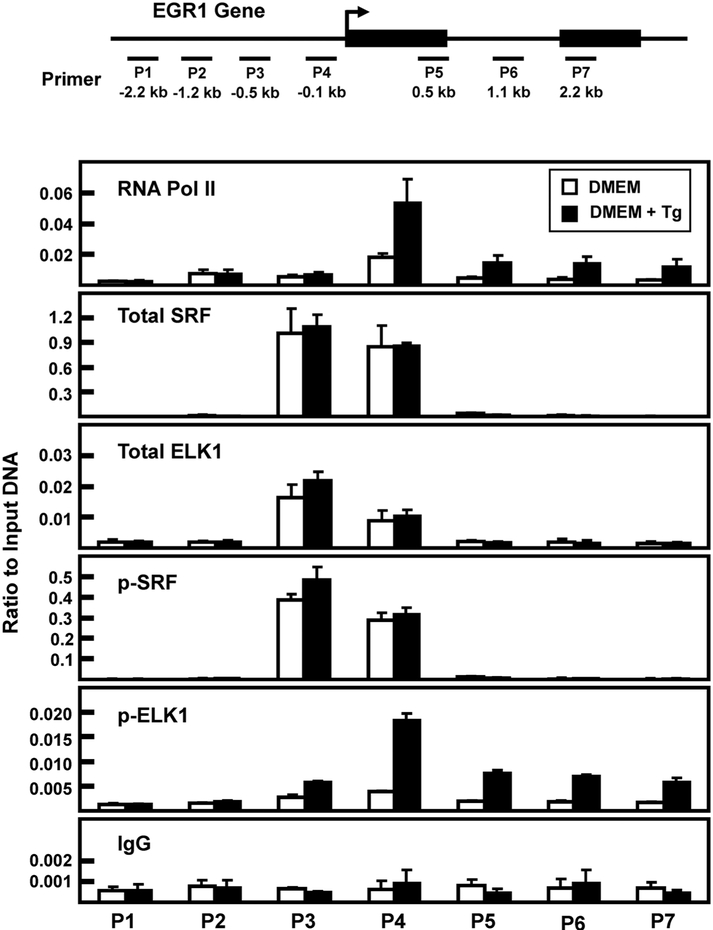
It has been documented that a series of SRE-ETS sequences upstream of the EGR1 transcription start site (TSS) mediate increased transcription in a stimuli- and cell-specific manner [16, 33]. Although the gene-associated abundance of total SRF and ELK1 does not typically change with gene activation, increased phosphorylation of ETS-bound ELK1 and SRE-bound SRF results in enhanced transcriptional activity [16, 33-35]. To investigate the gene association of total and phosphorylated ELK1 and SRF during ER stress, these factors were monitored by chromatin immunoprecipitation (ChIP) of the EGR1 promoter. Consistent with the hnRNA data indicating stress-induced transcription (Fig. 1C), following Tg treatment of HepG2 cells for 2 h there was an increase in RNA polymerase II (Pol II) association near the transcription start site (TSS) of the EGR1 gene (Fig. 6). There was constitutively bound SRF and ELK1 (“total”) within the proximal promoter region and there was a trend toward slightly higher amounts during ER stress. Conversely, the abundance of EGR1 promoter-bound p-SRF and p-ELK1 was much greater in response to the 2 h treatment with Tg. These results are consistent with previous studies concluding that ERK phosphorylates gene-associated SRF and ELK, but to confirm the role of ERK the ChIP analysis was repeated in cells treated with the MEK inhibitor PD98059 during the 2 h of ER stress. The results show that in the absence of ERK activity the amount of constitutive binding for SRF and ELK1 was largely unchanged, but the induction of gene-associated p-SRF and p-ERK was completely blocked (Fig. 7). Given that many of the experiments described above were performed after 6 h of Tg treatment, we monitored factor binding at this time point as well (Fig. 8). Once again, an increase in promoter-associated Pol II was observed, as was constitutive binding of both SRF and ELK1. However, in contrast to the 2 h data of Fig. 6, after 6 h of Tg treatment, there was little or no phosphorylation of SRF. These data, along with those of Fig. 5C, indicate that p-SRF may not be critical for induction of the EGR1 gene by ER stress.
Fig. 6.
Association of RNA polymerase II, SRF, and ELK1 at the EGR1 promoter after 2 h of Tg treatment. The locations of primers (labeled P1-P7) used to analyze the human EGR1 gene are illustrated relative to the transcription start site (arrow) and the two exons that comprise the protein-coding region of the gene are shown as black rectangles. The primer sequences are listed in Table 1. HepG2 cells were incubated in DMEM (Control) or DMEM + Tg for 2 h and then the cells were subjected to ChIP analysis with antibodies specific for RNA Pol II, total SRF, p-SRF, total ELK1, p-ELK1, and a non-specific IgG as a negative control. The data are plotted as the ratio to input DNA and are the averages ± SD for at least three samples within an experiment. The data shown are representative of multiple independent experiments.
Fig. 7.
Association of RNA polymerase II, SRF, and ELK1 at the EGR1 promoter after a 2 h treatment with Tg plus PD98059. The locations of primers (labeled P1-P7) used to analyze the human EGR1 gene are illustrated relative to the transcription start site (arrow) and the two exons that comprise the protein-coding region of the gene are shown as black rectangles. The primer sequences are listed in Table 1. HepG2 cells were incubated in DMEM (Control) or DMEM + Tg for 2 h, both conditions also included 20 μM PD98059. The cells were then subjected to ChIP analysis with antibodies specific for RNA Pol II, total SRF, p-SRF, total ELK1, p-ELK1, and a non-specific IgG as a negative control. The data are plotted as the ratio to input DNA and are the averages ± SD for at least three samples within an experiment. The data shown are representative of multiple independent experiments.
Fig. 8.
Association of RNA polymerase II, SRF, and ELK1 at the EGR1 promoter after 6 h of Tg treatment. The locations of primers (labeled P1-P7) used to analyze the human EGR1 gene are illustrated relative to the transcription start site (arrow) and the two exons that comprise the protein-coding region of the gene are shown as black rectangles. The primer sequences are listed in Table 1. HepG2 cells were incubated in DMEM (Control) or DMEM + Tg for 6 h and then the cells were subjected to ChIP analysis with antibodies specific for RNA Pol II, total SRF, p-SRF, total ELK1, p-ELK1, and a non-specific IgG as a negative control. The data are plotted as the ratio to input DNA and are the averages ± SD for at least three samples within an experiment. The data shown are representative of multiple independent experiments.
4. Discussion
Endoplasmic reticulum stress triggers a number of adaptive signaling pathways to alter cell homeostasis. The most widely studied ER stress pathways are triggered by three sensors located in the ER membrane. To our knowledge, there is no published evidence that any of these three pathways lead to modulation of the EGR1 gene. However, the present data illustrate the link between ER stress and the ERK arm of the MAPK pathways. The results also document that the list of ER stress-associated transcription factors and their respective responsive genomic sequences should include SRF, ELK1, and EGR1. These conclusions are supported by the following novel observations. 1) The results extend to HepG2 human HCC cells our previously published RNA-sequencing data indicating that EGR1 expression is highly induced in mouse embryonic fibroblasts in response to ER stress [4]. 2) The ER stress-dependent increase in EGR1 expression is largely the result of enhanced transcription. 3) Activation of EGR1 transcription requires SRC-RAS-RAF1-MEK-ERK-ELK1 signaling. 4) The induction of EGR1 transcription by ER stress is associated with increased phosphorylation of constitutively bound SRF and ELK1. 5) Consistent with the DNA binding studies, suppression of ELK1 expression in the cell adversely affects the induction of EGR1 by ER stress, whereas SRF is dispensable for induced EGR1 expression but contributes to basal expression. Collectively, the results are similar to those reported previously for activation of the EGR1 gene in response to amino acid deprivation [5]. Thus, both amino acid limitation and ER stress must converge at a common point and we believe this to be an unidentified step upstream of RAS.
Activation of the MAPK pathways is often associated with the pro-survival characteristics of transformed cells, either through suppression of apoptosis or enhanced proliferation. Likewise, the three traditional arms of the UPR that are triggered by ER stress are generally increased in activity following transformation and permit cells to adapt to the stringent conditions within a rapidly growing tumor. Therefore, it is not surprising that there is evidence for cross talk between these critical signaling processes [reviewed in 25]. Although there have been reports that ER stress activates MEK in hepatoma cells [36], the mechanism by which this occurs and downstream targets remain largely unknown. Although future studies will be required to determine if one or more of the traditional arms of the UPR contributes to activation of the EGR1 gene following ER stress, the present results document that ER stress in HepG2 cells increases p-ERK production and we also provide evidence for both upstream and downstream signaling steps. These data illustrate that SRC, RAS, and RAF are contributing components upstream of MEK-mediated EGR1 induction. It has been reported that SRC can mediate activation of RAS, but the cellular signaling circumstances that lead to this activation are not fully understood [27-29].
Although partial functional redundancy is recognized among the three members of the RAS protein family, independent and unique activities by each one also exist [37]. The present results following ER stress activation illustrate that both H-RAS and N-RAS are required for induction of the EGR1 gene. Interestingly, K-RAS is not essential. Mutations in K-RAS are by far the most common in human cancers, compared to H-RAS or N-RAS, but RAS mutations are not highly associated with hepatocellular carcinomas [37]. The basis for this selectivity among the RAS family members with regard to ER stress is not known, but there are many other reports that illustrate differences between K-RAS and the other two members. For example, K-RAS deficiency is embryonically lethal, yet mice with single or double knockout of H-RAS and N-RAS are viable [37]. H-RAS and N-RAS undergo palmitoylation that contributes to the localization within the plasma membrane, whereas this modification and plasma membrane sub-domain targeting does not appear to occur for K-RAS [38]. K-RAS complexes with calmodulin, whereas H-RAS and N-RAS do not [37]. Distribution of calcium levels within sub-cellular organelles is intimately linked to ER stress, indeed, perturbation of ER calcium content is the molecular basis for Tg-induced ER stress [3]. Perhaps the lack of H-RAS or N-RAS association with calmodulin influences the calcium-dependent ER signaling in some unidentified manner. Additional experimentation is required to understand the specific contribution of H-RAS and N-RAS to the cellular response to ER stress.
Phosphorylation of ERK coincides with its translocation to the nucleus where it is recruited to and phosphorylates constitutively bound transcription factors, which in turn increases transcription of the selected target genes. Among this collection of ERK transcription factor targets are members of the ETS family, including ELK1 [32, 34, 35]. ELK1 can act in a combinatorial manner with SRF [32] and the EGR1 promoter is known to contain multiple SRF and ELK1 binding sites in close proximity [39]. The present data show that ELK1 is constitutively bound to the EGR1 gene in HepG2 cells. Induction of ER stress for either 2 or 6 h did not alter the total abundance of the protein bound, but the amount of gene-associated p-ELK1 was increased in an ERK-dependent manner. Furthermore, knockdown of ELK1 by siRNA prevented the induction of EGR1 mRNA and protein. These data document that increased phosphorylation of ELK1 is a critical step during activation of EGR1 expression in response to ER stress. Conversely, although ER stress did result in a transient phosphorylation of EGR1 promoter-bound SRF, present at 2 h but gone by 6 h, based on the knockdown results SRF appears to play a role in maintenance of basal EGR1 expression with little or no contribution to the induction following ER stress. The exact role of the transient phosphorylation is unclear.
EGR1 is most often considered a tumor suppressor, but has been reported to function as a tumor promoter in some contexts. Krones-Herzig et al. [18] used Egr1-deficient mouse embryonic fibroblasts to document Egr1-dependent regulation of 266 genes and four major downstream nodes mediated by TGFβ, IL6, IGF1, and p53. With regard to tumor growth and ER stress, hypoxia-induced UPR signaling is a hallmark of transformed cells and tumor tissue, including HCC cells [40]. One of the outcomes of UPR signaling in many chronic liver diseases is increased angiogenesis [41]. Promotion of angiogenesis and tumor-associated vascularization is certainly required during tumor growth and it is recognized that HCC progression is dependent on VEGF-A driven endothelial cell proliferation [42]. EGR1 binds to the VEGF-A proximal promoter during hepatocyte growth factor mediated proliferation of HepG2 and Hep3B hepatoma cells [43] and EGR1 promotes the invasion of hepatocellular carcinoma cells [44]. Based on our results in cultured HepG2 cells, it is tempting to speculate that increased EGR1 expression in response to hypoxia or other ER stress modulators would promote HCC tumor growth in vivo.
In summary, induction of the EGR1 gene in response to ER stress in HepG2 human HCC cells was dependent on a SRC-RAS-RAF-MEK-ERK-ELK1 signaling cascade. These results contribute to the mounting evidence that the Unfolded Protein Response extends beyond the ER localized transducers of ATF6, IRE1, and PERK, to include the MAPK pathways. As a consequence, the ERK-phosphorylated ETS family member ELK1 can be added to the list of transcription factors that mediate the regulatory transcription network of the UPR. Furthermore, the increased expression of EGR1 implies that EGR1 responsive genes must be evaluated for their contribution to the adaptive and/or apoptotic ER stress response.
HIGHLIGHTS:
In certain cell types, EGR1 expression is highly induced in response to ER stress
The enhancement of EGR1 abundance results from increased transcription
The signaling that leads to increased EGR1 transcription is the MEK-ERK pathway
The transcription factors ELK1 and EGR1 contribute to the cellular ER stress response
Acknowledgments
Acknowledgements: This research was supported by a grant to MSK from the National Cancer Institute, the National Institutes of Health (CA203565).
Abbreviations:
- ATF6
activating transcription factor 6
- BiP/GRP78
binding immunoglobulin protein/glucose regulated protein 78
- ChIP
chromatin immunoprecipitation
- ELK1
E-twenty six (ETS)-like factor 1
- ERK
extracellular-signal regulated kinase
- ETS
E-twenty six; GAPDH, glyceraldehyde-3-phosphate dehydrogenase gene
- HCC
hepatocellular carcinoma
- IRE1
inositol-requiring endonuclease/kinase 1
- JNK
JUN N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/ERK kinase
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- RT-PCR
reverse transcriptase-polymerase chain reaction
- RT-qPCR
quantitative RT-PCR
- SRF
serum response factor
- SRE
serum response element
- Tg
thapsigargin
- Tu
tunicamycin
- UPR
unfolded protein response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Shan, Dudenhausen, and Kilberg
Induction of Early Growth Response Gene 1 (EGR1) by Endoplasmic Reticulum Stress is Mediated by the Extracellular Regulated Kinase (ERK) Arm of the MAPK Pathways
References
- [1].Han J, Kaufman RJ, Physiological/pathological ramifications of transcription factors in the unfolded protein response, Genes Dev, 31 (2017) 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baird TD, Wek RC, Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism, Adv Nutr, 3 (2012) 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Carreras-Sureda A, Pihan P, Hetz C, Calcium signaling at the endoplasmic reticulum: fine-tuning stress responses, Cell Calcium, 70 (2018) 24–31. [DOI] [PubMed] [Google Scholar]
- [4].Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, Kilberg MS, Sartor MA, Kaufman RJ, ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death, Nature Cell Biol, 15 (2013) 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shan J, Balasubramanian MN, Donelan W, Fu L, Hayner J, Lopez MC, Baker HV, Kilberg MS, A MEK-Dependent Transcriptional Program Controls Activation of the Early Growth Response 1 (EGR1) Gene During Amino Acid Limitation, J. Biol. Chem, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thiaville MM, Pan YX, Gjymishka A, Zhong C, Kaufman RJ, Kilberg MS, MEK signaling is required for phosphorylation of eIF1Clpha following amino acid limitation of HepG2 human hepatoma cells, J. Biol. Chem, 283 (2008) 10848–10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fu L, Balasubramanian M, Shan J, Dudenhausen EE, Kilberg MS, Auto-activation of c-JUN gene by amino acid deprivation of hepatocellular carcinoma cells reveals a novel c-JUN-mediated signaling pathway, J. Biol. Chem, 286 (2011) 36724–36738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fu L, Kilberg MS, Elevated cJUN expression and an ATF/CRE site within the ATF3 promoter contribute to activation of ATF3 transcription by the amino acid response, Physiol. Genomics, 45 (2013) 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shan J, Donelan W, Hayner JN, Zhang F, Dudenhausen EE, Kilberg MS, MAPK signaling triggers transcriptional induction of cFOS during amino acid limitation of HepG2 cells, Biochim. Biophys. Acta, 1854 (2015) 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhou D, Zhang Y, Pan YX, Chen H, Dickkopf homolog 1, a Wnt signaling antagonist, is transcriptionally up-regulated via an ATF4-independent and MAPK/ERK-dependent pathway following amino acid deprivation, Biochim. Biophys. Acta, 1809 (2011) 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chaveroux C, Jousse C, Cherasse Y, Maurin AC, Parry L, Carraro V, Derijard B, Bruhat A, Fafournoux P, Identification of a novel amino acid response pathway triggering ATF2 phosphorylation in mammals, Mol. Cell Biol, 29 (2009) 6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sharp JW, Ross-Inta CM, Hao S, Rudell JB, Gietzen DW, Co-localization of phosphorylated extracellular signal-regulated protein kinases 1/2 (ERK1/2) and phosphorylated eukaryotic initiation factor 1Clpha (eIF1Clpha) in response to a threonine-devoid diet, J.Comp Neurol, 494 (2006) 485–494. [DOI] [PubMed] [Google Scholar]
- [13].Hao S, Ross-Inta CM, Gietzen DW, The sensing of essential amino acid deficiency in the anterior piriform cortex, that requires the uncharged tRNA/GCN2 pathway, is sensitive to wortmannin but not rapamycin, Pharmacol. Biochem. Behav, 94 (2010) 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Anthony TG, Gietzen DW, Detection of amino acid deprivation in the central nervous system, Current opinion in clinical nutrition and metabolic care, 16 (2013) 96–101. [DOI] [PubMed] [Google Scholar]
- [15].Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG, The GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent, L-asparaginase, J. Biol. Chem, 284 (2009) 32742–32749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pagel JI, Deindl E, Early growth response 1--a transcription factor in the crossfire of signal transduction cascades, Indian J. Biochem. Biophys, 48 (2011) 226–235. [PubMed] [Google Scholar]
- [17].Hodge C, Liao J, Stofega M, Guan K, Carter-Su C, Schwartz J, Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2, J. Biol. Chem, 273 (1998) 31327–31336. [DOI] [PubMed] [Google Scholar]
- [18].Krones-Herzig A, Mittal S, Yule K, Liang H, English C, Urcis R, Soni T, Adamson ED, Mercola D, Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53, Cancer Res, 65 (2005) 5133–5143. [DOI] [PubMed] [Google Scholar]
- [19].Liao Y, Shikapwashya ON, Shteyer E, Dieckgraefe BK, Hruz PW, Rudnick DA, Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1-deficient mice, J. Biol. Chem, 279 (2004) 43107–43116. [DOI] [PubMed] [Google Scholar]
- [20].Shan J, Fu L, Balasubramanian MN, Anthony T, Kilberg MS, ATF4-Dependent Regulation of the JMJD3 Gene During Amino Acid Deprivation Can be Rescued in Atf4-Deficient Cells by Inhibition of Deacetylation, J. Biol. Chem, 287 (2012) 36393–36403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen H, Pan YX, Dudenhausen EE, Kilberg MS, Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation, J. Biol. Chem, 279 (2004) 50829–50839. [DOI] [PubMed] [Google Scholar]
- [22].Kaufman RJ, Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls, Genes Dev, 13 (1999) 1211–1233. [DOI] [PubMed] [Google Scholar]
- [23].Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K, Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1, Dev Cell, 13 (2007) 365–376. [DOI] [PubMed] [Google Scholar]
- [24].Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, Yates JR 3rd, Su AI, Kelly JW, Wiseman RL, Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments, Cell reports, 3 (2013) 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Darling NJ, Cook SJ, The role of MAPK signalling pathways in the response to endoplasmic reticulum stress, Biochim. Biophys. Acta, 1843 (2014) 2150–2163. [DOI] [PubMed] [Google Scholar]
- [26].Delire B, Starkel P, The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications, Eur. J. Clin. Invest, 45 (2015) 609–623. [DOI] [PubMed] [Google Scholar]
- [27].Williams NG, Roberts TM, Li P, Both p21ras and pp60v-src are required, but neither alone is sufficient, to activate the Raf-1 kinase, Proc. Natl. Acad. Sci. U. S. A, 89 (1992) 2922–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van der Geer P, Wiley S, Gish GD, Pawson T, The Shc adaptor protein is highly phosphorylated at conserved, twin tyrosine residues (Y239/240) that mediate protein-protein interactions, Curr. Biol, 6 (1996) 1435–1444. [DOI] [PubMed] [Google Scholar]
- [29].Bunda S, Heir P, Srikumar T, Cook JD, Burrell K, Kano Y, Lee JE, Zadeh G, Raught B, Ohh M, Src promotes GTPase activity of Ras via tyrosine 32 phosphorylation, Proc. Natl. Acad. Sci. U. S. A, 111 (2014) E3785–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Morrison DK, MAP kinase pathways, Cold Spring Harbor perspectives in biology, 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hollenhorst PC, McIntosh LP, Graves BJ, Genomic and biochemical insights into the specificity of ETS transcription factors, Annu. Rev. Biochem, 80 (2011) 437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Odrowaz Z, Sharrocks AD, ELK1 uses different DNA binding modes to regulate functionally distinct classes of target genes, PLoS genetics, 8 (2012) e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen CC, Lee WR, Safe S, Egr-1 is activated by 17beta-estradiol in MCF-7 cells by mitogen-activated protein kinase-dependent phosphorylation of ELK-1, J. Cell. Biochem, 93 (2004) 1063–1074. [DOI] [PubMed] [Google Scholar]
- [34].O'Donnell A, Odrowaz Z, Sharrocks AD, Immediate-early gene activation by the MAPK pathways: what do and don’t we know?, Biochem. Soc. Trans, 40 (2012) 58–66. [DOI] [PubMed] [Google Scholar]
- [35].Yang SH, Sharrocks AD, Whitmarsh AJ, MAP kinase signalling cascades and transcriptional regulation, Gene, 513 (2013) 1–13. [DOI] [PubMed] [Google Scholar]
- [36].Dai R, Chen R, Li H, Cross-talk between PI3K/Akt and MEK/ERK pathways mediates endoplasmic reticulum stress-induced cell cycle progression and cell death in human hepatocellular carcinoma cells, Int. J. Oncol, 34 (2009) 1749–1757. [DOI] [PubMed] [Google Scholar]
- [37].Stephen AG, Esposito D, Bagni RK, McCormick F, Dragging ras back in the ring, Cancer cell, 25 (2014) 272–281. [DOI] [PubMed] [Google Scholar]
- [38].McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin RA, Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance, Biochim. Biophys. Acta, 1773 (2007) 1263–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gregg J, Fraizer G, Transcriptional Regulation of EGR1 by EGF and the ERK Signaling Pathway in Prostate Cancer Cells, Genes & cancer, 2 (2011) 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Vandewynckel YP, Laukens D, Devisscher L, Bogaerts E, Paridaens A, Van den Bussche A, Raevens S, Verhelst X, Van Steenkiste C, Jonckx B, Libbrecht L, Geerts A, Carmeliet P, Van Vlierberghe H, Placental growth factor inhibition modulates the interplay between hypoxia and unfolded protein response in hepatocellular carcinoma, BMC Cancer, 16 (2016) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Paridaens A, Laukens D, Vandewynckel YP, Coulon S, Van Vlierberghe H, Geerts A, Colle I, Endoplasmic reticulum stress and angiogenesis: is there an interaction between them?, Liver international : official journal of the International Association for the Study of the Liver, 34 (2014) e10–18. [DOI] [PubMed] [Google Scholar]
- [42].Liu LP, Liang HF, Chen XP, Zhang WG, Yang SL, Xu T, Ren L, The role of NF-kappaB in Hepatitis b virus X protein-mediated upregulation of VEGF and MMPs, Cancer Invest, 28 (2010) 443–451. [DOI] [PubMed] [Google Scholar]
- [43].Lee KH, Kim JR, Hepatocyte growth factor induced up-regulations of VEGF through Egr-1 in hepatocellular carcinoma cells, Clin. Exp. Metastasis, 26 (2009) 685–692. [DOI] [PubMed] [Google Scholar]
- [44].Ozen E, Gozukizil A, Erdal E, Uren A, Bottaro DP, Atabey N, Heparin inhibits Hepatocyte Growth Factor induced motility and invasion of hepatocellular carcinoma cells through early growth response protein 1, PloS one, 7 (2012) e42717. [DOI] [PMC free article] [PubMed] [Google Scholar]