Abstract
N-methyl-D-aspartate receptors (NMDARs) are glutamatergic receptors that take part in excitatory synaptic transmission and drive functional and structural neuronal plasticity, including activity-dependent changes in dendritic morphology. Forebrain NMDARs contribute to neuronal plasticity in at least two ways: through calcium-mediated processes or via direct intracellular postsynaptic signaling. Both properties are regulated by the GluN2 subunits. However, the separate contributions of these properties to the regulation of dendritic morphology are unknown. We created transgenic mice that express chimeric GluN2 subunits and examined the impact on pyramidal cell dendritic morphology in hippocampal region CA1. Golgi-Cox impregnation and transgenic expression of green fluorescent protein were employed to visualize dendritic arbors. In adult mice with a predominantly native GluN2A background, overexpression of the GluN2B carboxy terminus increased the total path of the dendritic arbor without affecting branch number or tortuosity. Overexpressing the amino terminus and transmembrane domains of GluN2B had little effect. It may be inferred from these results that NMDAR-dependent intracellular signaling regulates dendritic morphology of hippocampal pyramidal cells more so than calcium conductance dynamics. The findings add to the understanding of NMDAR-mediated signaling in hippocampal neurons and support re-investigation of the molecular underpinnings of NMDAR involvement in postnatal dendrite maturation.
Keywords: NMDA, dendritic morphology, protein signaling, GluN2, transgenic, development
Introduction
N-methyl-D-aspartate receptors (NMDARs), a subclass of ionotropic glutamate receptors at excitatory synapses, are the primary inducers of activity-dependent plasticity in the central nervous system. When activated, NMDARs initiate changes in dendritic structure and function including modifications in dendritic branching (Jan & Jan, 2001) and in synaptic efficacy (Luscher & Malenka, 2012). In an inverted-U fashion, acute or moderate activation of NMDARs leads to dendritic growth while activity blockade or excessive activity triggers dendritic retraction (Hardingham & Bading, 2010). The complete chain of events from activation of NMDARs to dendritic plasticity, especially structural modifications, is not fully understood.
NMDARs consist of four protein subunits, two of which are obligatory GluN1 subunits; the remaining two subunits vary per neuron type and age of the animal (Dingledine, Borges, Bowie, & Traynelis, 1999). In the forebrain, the two accessory subunits are GluN2 subunits, either GluN2A or GluN2B (or one of each), though substantial levels of GluN3 have been observed in neonatal brains (Pachernegg, Strutz-Seebohm & Hollmann, 2012). In adult animals, both GluN2A and GluN2B subunits may be incorporated into NMDARs with total GluN2A content exceeding GluN2B (Dumas, 2005; Ferreria, 2015; Monyer, Burnashev, Laurie, Sakmann & Seeburg, 1994; Sanders et al., 2013). Chronic application of GluN2B antagonists reduced filopodia formation in cultured hippocampal neurons, while antagonists of GluN2A inhibit secondary dendrite formation (Henle, Dehmel, Leemhuis, Fischer, & Meyer, 2012). In genetically modified mice lacking the GluN2A subunit, total dendritic length and arbor complexity were reduced (Kannangara et al., 2014). Thus, GluN2A and GluN2B subunits appear to regulate dendritic morphology differentially. Further molecular investigation is warranted to gain a greater understanding of how NMDARs with GluN2A or GluN2B impact dendritic morphology.
Numerous functional properties of NMDARs vary with the inclusion of GluN2A or GluN2B. For decades, calcium entry into the postsynaptic neuron and its impact on second messenger systems were considered the focal mechanism by which NMDAR activation altered neuron structure and function (Hardingham & Bading, 2010; Konur & Ghosh, 2005; Malenka & Bear, 2004). When activated, NMDARs containing GluN2B display a reduced open probability, but also exhibit reduced deactivation time (Sanz-Clemente, Nicoll,& Roche, 2013; Shipton & Paulsen, 2014), leading to prolonged calcium entry and greater charge transfer compared to NMDARs containing GluN2A (Berberich, Jensen, Hvalby, Seeburg & Kohr, 2007). These parameters of ion conductance are largely regulated by amino acid signaling motifs in the aminoterminal domain (NTD) and transmembrane domains (TMDs). However, NMDARs also operate in calcium-independent ways to regulate neuronal plasticity (Birnbaum, Bali, Rajendran, Nitsch, & Tackenberg, 2015; Chung, 2013; Nabavi et al., 2013; Vissel, Krupp, Heinemann & Westbrook, 2001; Li et al., 2016; Stein, Gray, & Zito, 2015; Dore et al., 2017). Glutamate binding to NMDARs can produce signals in the postsynaptic neuron in the absence of calcium conductance, via direct protein-protein interactions mediated by the carboxy-terminal domain (CTD) of the GluN2 subunits (Sanz-Clemente, Nicoll, & Roche, 2013; Blanke & VanDongen, 2009; Maki, Aman, Amico-Ruvio, Kussius & Popescru, 2012; Dore & Malinow, 2015; Dore, Aow, & Malinow, 2016; Aow, Dore & Malinow, 2015). Given this additional level of complexity in NMDAR function, molecular experiments are necessary to tease out the individual contributions of distinct GluN2-dependent properties to dendritic morphology.
We applied molecular subcloning to replace the CTD of the GluN2A subunit with the CTD of GluN2B (termed ABc, GluN2A receptor with the GluN2B carboxy terminus) and vice versa, we replaced the CTD of the GluN2B subunit with the CTD of GluN2A (termed BAc) and expressed these constructs in separate lines of transgenic mice (Sanders et al., 2013). We performed Golgi-Cox neuronal impregnations and separately crossed the ABc and BAc lines with mice that express enhanced green fluorescent protein (eGFP) sparsely throughout the forebrain to label hippocampal pyramidal neurons. In hippocampal pyramidal neurons from ABc mice, average branch length increased across most of the dendritic arbor and the number of apical branch intersections, most especially at later annuli, compared to BAc and WT animals. Also, the total arbor path of the pyramidal neuron’s apical and basal dendrites was increased in ABc mice compared to BAc and wild-type mice. These results suggest that increasing the GluN2B CTD content results in more expansive dendrites in adult hippocampal neurons and help clarify the different roles of the GluN2A and GluN2B CTDs in neuronal plasticity.
Experimental Procedures
Subjects
All animals (n = 31) used in this project were C57BL6J transgenic mice perfused between the ages of 30–60 days after birth. Male (n = 13) and female (n = 18) mice were bred in the Krasnow Institute Animal Facility (accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International, AAALAC). Animals were maintained in individually ventilated cages (Animal Care Systems) on a 12:12 h light/dark cycle. Food and water were available ad libitum. All the following procedures were performed in accordance with the regulations stated in the Guide for Care and Use of Laboratory Animals by the National Research Council and approved by the George Mason University Institutional Animal Care and Use Committee (IACUC).
TRE_ABc and TRE_BAc mice were generated in-house (Sanders et al., 2013). Driver mice expressing tTA under the transcriptional regulation of the CamKII minimal promoter (Mayford, Wang, Kandel & O’Dell, 1995) were obtained as a generous gift from Dr. Clifford Kentros (“Line-84,” Kavli Institute, Norway) and from Jackson Laboratories [Tg(Camk2atTA)1Mmay]. Thy1-eGFP mice were obtained from Jackson Laboratories [Tg(Thy1EGFP)MJrs/J]. Double positive ABc or BAc mice were crossed with the Thy1-eGFP mice for the fluorescent imaging experiments. All animals were tail clipped and ear tagged under Isoflurane anesthetic (3% vapor, Phoenix Pharmaceuticals, LLC). Tail clips were placed in wellplates from Transnetyx Inc. and shipped out for commercial processing. Genotyping results were received 24–72 hours later. All CA1 neurons were collected with neuronal somas located in the following coordinates: from Bregma (in mm): −2.06, (Medial-Lateral: 1.25–1.75, Dorsal-Ventral: 4.25).
Tissue processing
Golgi-Cox impregnation:
ABc (n = 5), BAc (n = 4) and WT (n = 6) mice were overdosed with Isoflurane anesthetic (> 5% vapor, Phoenix Pharmaceuticals, LLC) and perfused with ice-cold 0.9% sterile saline for ten minutes. Brains were quickly extracted, immediately placed in Golgi-Cox solution (5% potassium dichromate, 5% mercuric chloride, and 5% potassium chromate), and placed in the dark for 12 to 14 days. The brains were then transferred to a 30% sucrose solution for 48 hours and sectioned to 75 μm thickness using a vibratome. Sections were briefly maintained in phosphate buffered saline (PBS) in 24-well culture plates and then mounted onto microscope slides (Superfrost Plus Microscope Slides, Fisher Scientific) coated in 1% gelatin (Type B powdered gelatin, BioReagent, Sigma-Aldrich). During mounting, the sections were temporarily covered with parafilm and gently pressed into the gelatin to optimize the bond between the tissue and the slide and then incubated at 60° C for 24 hours. After incubation, mounted sections were alternately bathed in Omnipure H2O for 1 minute, in ammonium hydroxide for 30 minutes, and Kodak Fix solution (5160353, Kodak) for 30 minutes at room temperature. The slides were then run through ascending concentrations of ethanol (in water) to dehydrate the tissue and cleared of excess lipids with xylene before being coverslipped.
Thy1_eGFP:
The Thy-1 eGFP mice were bred with double positive ABc and BAc mice to produce animal subjects. ABc (n = 5), BAc (n = 3), and WT (n = 8) mice were overdosed with Isoflurane anesthetic and perfused with ice-cold 4% paraformaldehyde for ten minutes. Brains were rapidly extracted, left in PFA for 24 hours, and then transferred to 30% sucrose for 48 hours. These brains were then sectioned in a vibratome submerged in PBS at 75μm thickness and stored in 24-well culture plates. Slices were then mounted onto charged slides (Colorfrost Plus, Fisher Scientific) and coverslipped with antifade mounting medium (Vectashield, Vector Laboratories, Inc.).
Imaging, Tiling, and Tracing
Brain slices from Thy-1_eGFP mice were imaged with a Nikon Eclipse 80i confocal microscope. EZ-C1 3.91 software was used to collect images (40X magnification), acquire z-stacks, and render. Z-stacks were imported and then were tiled in Neurolucida to create a complete 2D neuron image (Fig. 1A). Golgi-Cox labeled brain sections were imaged on an Olympus AX70 Brightfield microscope. Neurolucida software was used to capture z-stack images and render a single 2D image for tracing. Z-stacks were gathered at 40X (Fig. 1B).
Figure 1:
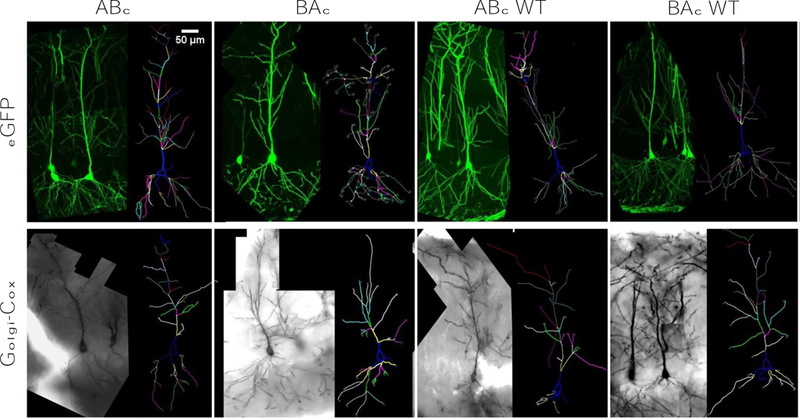
Representative images and traces from eGFP and Golgi-Cox labeled neurons. A) eGFP labeled neurons and their corresponding traces for each genotype. B) Golgi-Cox labeled neurons and their corresponding traces for each genotype. Scale bars are equal to 50µm for each respective image. Branch order is color-coded in each trace.
Neurolucida was used to trace completely tiled z-stacked images (Fig. 1). Three raters, blinded to genotype, then individually traced each neuron to ensure accuracy and reduce interrater variability. Since eGFP neurons (Fig. 1A) and Golgi-Cox stained neurons (Fig. 1B) were gathered on different microscopes with different software, traces from eGFP labeled neuron were proportionally scaled on the x- and y-axis to match the Golgi-Cox labeled cells. Three neurons per animal from three to five animals per genotype were analyzed blind (n = 9 – 15 cells per genotype group). Each neuron was chosen based on its relative isolation from other labeled cells. If a target cell demonstrated substantial dendritic overlap with other cells, it was passed over for selection. Isolated cells with obvious dendritic truncations due to tissue sectioning were eliminated from consideration. Selection criteria were applied equally across genotypes and across methodologies.
Data analysis
The same three cells per animal were traced by three independent raters, with completed traces imported into Neurolucida Explorer (Version 11.01.1, MBF Bioscience) for analysis with the Branch Structure Analysis and Sholl Analysis data settings. Collecting images blind with respect to labeling method was not possible. All imaged cells were traced blind (barring some image artifacts that differentiated labeling method) and all analyses of traces were performed electronically through Neurolucida Explorer, void of manual biases. Sholl Analysis was characterized by concentric rings, with the first placed at the soma of the cell, then at 50 µm intervals beyond the soma. Parameter values returned by Neurolucida Explorer for each trace were averaged across raters and across cells within each genotype group and parameter measure. Specifically, the parameters quantified by Branch Structure Analysis were apical and basal dendritic total length, number of branches, average length, and tortuosity, or curvature, of the dendrite.
The parameters quantified by Sholl analysis were apical and basal dendritic length and branch intersections. Sholl Analysis values were collected at 10 μm intervals and averaged across 50 μm intervals away from the neuron soma. For total arbor analysis, the animal’s apical and basal total lengths were multiplied by their respective tortuosity to get their full length, the final product being “apical total arbor path” and “basal total arbor path”.
Statistical Approach
Each animal had three cells collected from hippocampal brain slices (n = 93), with each cell traced by three independent raters. A power analysis was performed to determine appropriate sample size—with a type I error rate of 0.05, a power of 0.80, three groups (ABc, BAc, WT) and the number of dendritic parameters, an n of 9 per genotype group was the requisite sample size (G*Power 3.0, Faul et al., 2009). Then, each cell was averaged across the three rater traces in Microsoft Excel to minimize interrater differences and to ensure that each observation was independently assessed. Finally, the three cells were averaged within-subject to produce a single measure for each dendritic parameter per animal, which was compared across gender, labeling method, driver line, or genotype by repeated-measures ANOVA in SPSS (IBM SPSS Statistics, Armonk, NY). Multivariate ANOVA was used to compare apical and basal total arbor path. This parametric test was performed instead of an alternative, nonparametric test as the following criteria were met: 1) independent observations, as performed by within-subject averaging, 2) the presence of a normal distribution, unless than sample size is > 25, which the sample size of animals in this study is 31, and 3) sphericity. Where Mauchly’s test indicated that the assumption of sphericity was not met, the Greenhouse-Geisser correction was applied. When a main or interaction effect was significant, Multivariate ANOVA was used to compute pairwise differences. Bonferroni’s correction was used for all statistical tests, such that Bonferroni corrected p-values were used for all independent variable comparisons. As such, all post hoc and pairwise comparisons use the Bonferroni correction. For all tests, p < 0.05 was considered statistically significant. All figures display group means, and error bars are +/− standard error of the means. Reporting of statistics is located in the Appendices, in Tables A1 through J1.
Equipment and Settings
Golgi-Cox labeled brain sections were imaged on an Olympus AX70 Brightfield microscope while eGFP labeled brain slices were imaged with a Nikon Eclipse 80i confocal microscope. Each image has a resolution of 96 dpi and a 24-bit depth. Both Golgi-Cox impregnated and eGFP labeled sections were mounted on glass slides using a super resolution antifade mounting medium (H-1000, Vector Labs) and covered with thin glass coverslips (12–548-5P, Fisher Scientific). Tissue was processed and images were collected at room temperature. Neurolucida software (Version 11.01.1, MBF Bioscience) was used to collect and analyze bright field images. EZ-C1 software (Version 3.91, Build 891, Nikon) was used to collect and analyze confocal images. Confocal images were volume rendered. All images were saved as .tiff files, then traced in Neurolucida. No manipulations were performed on the images during tracing. The cells and their respective traces presented in Fig. 1A and 1B were placed side-by-side in Neurolucida software, then saved as a .jpg. These images each have pixel dimensions of 640 × 480.
Data Availability
The datasets that were generated and analyzed for the current study are available from the corresponding author upon request.
Results
Since only two or fewer raters registered apical branch order values at the 8th branch order or 51st annulus, analysis stopped at the 7th branch order or 50th annulus, respectively. Similarly, only two or fewer raters measured basal branch order values at the 5th branch order or 20th annulus. Therefore, data analyses for basal dendrites proceeded to the 4th branch order or 19th annulus.
Comparisons Across Age and Gender Demonstrated Method, but not Genotype, Differences
There was a main effect of method on age, with significant differences present between Thy1_eGFP mice and animals used for Golgi-Cox labeling [F(1,29) = 5.498, p = 0.026], with animals in the Golgi-Cox group being significantly older (eGFP: 43.3 ± 4.8 days; Golgi-Cox: 61.1 ± 5.9 days). Despite this, there was no main effect of genotype on age for the eGFP [F(2,13) = 1.127, p = 0.354] or the Golgi-Cox analyses [F(2,12) = 1.245, p = 0.323]. There was no genotype by age interaction, either.
For eGFP labeled neurons, there were no main effects of gender found in any parameter in any analysis (Table A1). For Golgi-Cox labeled neurons, branch order analysis only showed effects of gender in average apical length and apical tortuosity (Table A1). Here, females displayed greater average dendritic length (post hoc: p = 0.039) and tortuosity compared to males (post hoc: p = 0.011) (Table A1). In Sholl analysis, Golgi-Cox labeled cells revealed an interaction between gender and annulus in basal length [RMANOVA, F(1.629,21.183) = 11.047, p = 0.001], and basal branch intersections [F(1.620,21.062) = 11.284, p = 0.001]. Here, females had lower basal length and branch intersections at 50 and 100µm, equal basal length and intersections at 150µm, and higher basal length and intersections at 200µm compared to males. Since gender effects were minimal, and only found in Golgi-Cox labeled neurons, data were collapsed across gender in all measures.
No Major Differences Present Across WTs and Driver Lines
In eGFP labeled neurons, branch analyses of both apical and basal dendrites revealed no differences between WTs in any measures except for basal tortuosity, where dendritic branches in neurons from WT-ABc mice were more tortuous than in WT-BAc animals (p = 0.043) (Table B1). No measures showed significance with Golgi-Cox labeling. For Sholl analyses, neither method revealed parameter differences between WTs (Table B1). Total arbor path measures also demonstrated no differences between WT groups in eGFP and Golgi-Cox labeled cells. Since these two WT lines differed in only one of 14 total measures, the WT-ABc and WT-BAc mice were collapsed into one group henceforth called WTs. Basal tortuosity of eGFP labeled neurons were analyzed independently, with WTs not collapsed (Fig. 2H).
Figure 2:
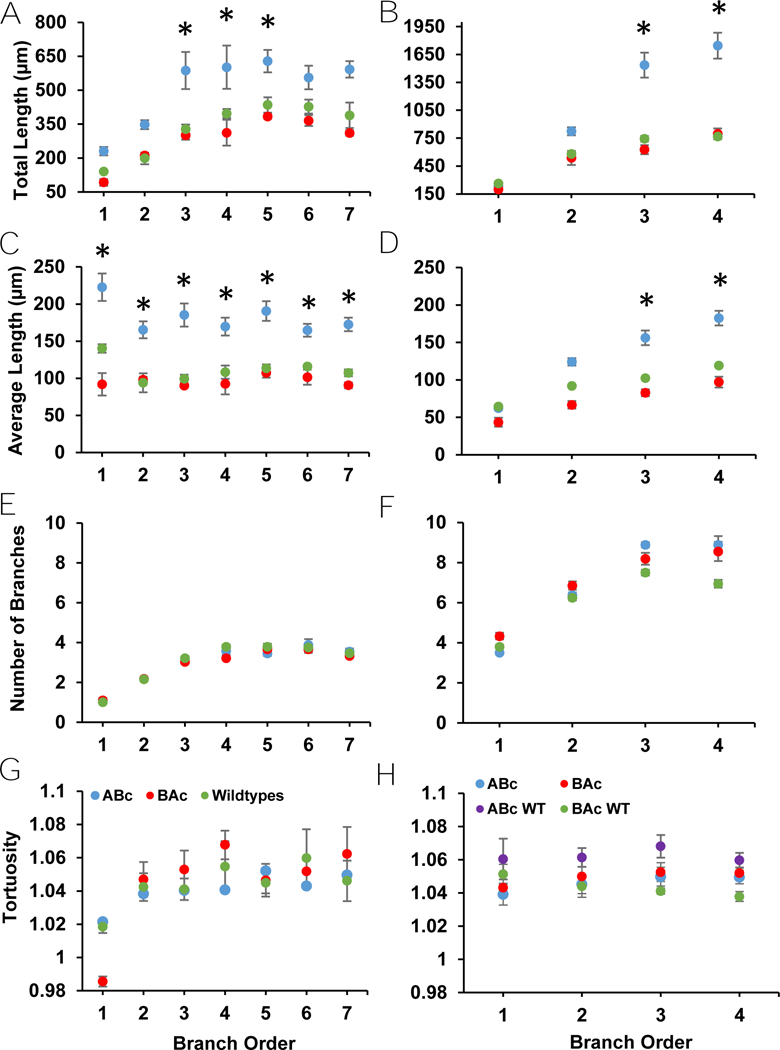
Apical and basal dendritic parameters from branch analyses of eGFP labeled neurons. A) Total apical length showed a trend for an increase in cells from ABc relative to WT and BAc animals. B) There was a trend for an increase in total basal length in neurons from ABc relative to WT and BAc mice. C) Average apical length showed a trend for an increase in neurons from ABc relative to WT and BAc mice. D) Average basal length showed a trend for an increase in neurons from ABc relative to WT and BAc mice. E) Apical number of branches in eGFP labeled neurons did not differ across genotypes. F) Basal number of branches did not differ across genotypes. G) Apical tortuosity did not differ across genotypes. H) Basal tortuosity, which has WTs separated for this analysis, showed no significant differences.
Two separate tTA driver lines were used to express the GluN2 chimeras: line-84 from the University of Oregon and C57BL/6-Tg(Camk2a-tTA) from Jackson Labs. There were no main effects of driver line on any parameter in any analysis in eGFP, or Golgi-Cox labeled neurons (Table C1). The singular exception was the measure of apical length in Sholl analysis of Golgi-Cox labeled neurons (Table C1), with the Jackson Mayford tTA line displaying longer apical dendrites. As such, data from both driver lines were collapsed except for the Sholl analysis apical length in Golgi-Cox labeled neurons, where driver lines were analyzed separately (Fig. 5A).
Figure 5:
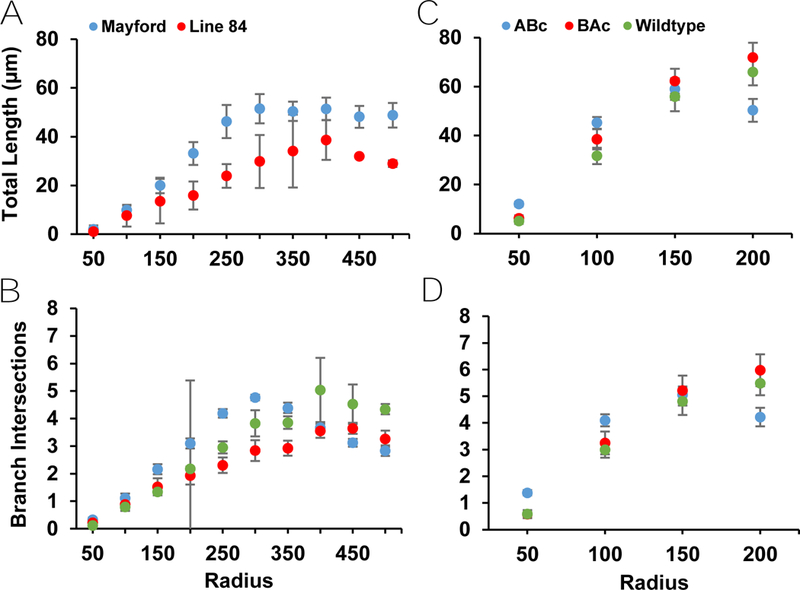
Apical and basal dendritic measures from Sholl analysis of Golgi-Cox labeled neurons. Driver lines had to be separated for the Sholl analysis of apical total length. A) Line-84 shows significantly less apical length than Mayford tTA, with no genotype differences shown in either line. B) Additionally, the Sholl analysis demonstrated no differences between genotypes in basal total length. C) This analysis also revealed no differences between genotypes in apical branch intersections or D) basal branch intersections.
Significant Methodological Differences Found Across Dendritic Parameters
Main effects of method presented primarily in basal dendritic parameters (total arbor, number of branches, and total length); the only apical measure that differed was total arbor path (Table D1). eGFP labeled neurons demonstrated higher basal total length and number of branches, as well as more extensive apical and basal arbor paths, than Golgi-Cox labeled neurons (Table D1). Interactions between method and branch order presented in apical average length (p = 0.022) and basal number of branches (p = 0.002) (Table D1). Apical average length was increased in primary and secondary branches of eGFP labeled cells compared to Golgi-Cox labeled cells. At higher branch orders, apical average length did not differ between the two methods. eGFP cells demonstrated a higher number of branches than Golgi-Cox cells selectively at higher branch orders (Table D1). These results suggest more complete dendritic labeling by eGFP expression compared to Golgi-Cox impregnation.
Further, there was a main effect of method present in all Sholl analysis parameters (Table D1). Basal parameters in the Sholl analysis demonstrated a significant interaction between method and annulus (basal length: p < 0.001; basal branch intersections: p < 0.001) (Table D1). Compared to Golgi-Cox, eGFP labeled cells were longer (apical length, p = 0.025; basal length, p < 0.001) and had a higher number of branches (apical branch intersections, p < 0.048; basal branch intersections, p < 0.001), most especially at annuli further from the soma (Table D1). This again supports more complete labeling by eGFP compared to Golgi-Cox. Due to these significant differences, effects of genotype on dendritic parameters were analyzed separately for each labeling method.
Genotype Differences for eGFP Labeled Neurons Found Primarily in Apical Dendrite
Branch Analyses
There was no effect of genotype for total apical dendritic length, apical and basal number of branches, basal average length, and apical and basal tortuosity (Table E1) (Fig. 2). However, total basal dendritic length and tortuosity and apical average length all demonstrated significant main effects of genotype in the branch analysis (Table E1) (Fig. 2). There was no genotype by branch order interaction in any of the branch analysis measures, with the sole exception of total basal dendritic length (Table E1).
ABc mice demonstrated a trend for longer total basal dendritic length compared to WT mice (p = 0.068), though not to BAc mice (p = 0.135) (Fig. 2B). Additionally, ABc animals showed a trend for longer average apical length compared to WT (p = 0.064) and BAc animals (p = 0.082) (Table E1) (Fig. 2A). BAc and WT animals did not differ from each other in either total basal dendritic length or average apical length (Fig. 2). The interaction present between branch order and genotype in total basal dendritic length revealed a greater divergence between genotypes at later branch orders compared to earlier branch orders (Table E1). Here, ABc mice demonstrated similar primary and secondary lengths, but much longer tertiary and quaternary lengths, than their BAc and WT counterparts (Fig. 2B). It is likely that this divergence occluded significant post hoc comparisons, while still yielding a significant main effect of genotype in total basal length.
All branch analysis measures except for average apical length and basal tortuosity demonstrated a main effect of branch order (Table E1) (Fig. 2). Total apical dendritic length and total basal dendritic length both increased as branch order increased (Table G1) (Fig. 2). Similarly, apical and basal number of branches both increased as branch order increased (Table G1) (Fig. 2). Basal average length, but not apical average length, increased with increasing branch order (Table G1) (Fig. 2). Apical tortuosity, but not basal tortuosity, also increased as branch order increased (Table G1) (Fig. 2). Increases in dendritic length and number of branches with increasing branch order are similar to reports from prior studies of CA1 pyramidal neurons (Vida, 2010; Bannister & Larkman, 1995) and were fully expected.
Sholl Analyses
No main effect of genotype, nor an interaction between genotype and annulus, presented on basal length or basal number of branches (Table E1) (Fig. 3). Main effects of genotype on apical dendritic length and apical branch intersections, along with an interaction between genotype and annulus for intersections, were seen (Table E1) (Fig. 3). ABc mice demonstrated a trend for decreased apical dendritic length compared to WT mice (p = 0.054), but not to BAc mice (p = 0.105) (Fig. 3A). Apical dendritic length did not differ between BAc and WT animals (Fig. 3A). Similarly, the number of apical branch intersections was reduced in ABc mice relative to WT (p = 0.014) and BAc mice (p = 0.025) (Fig. 3C). The number of apical branch intersections did not differ between cells from BAc and WT mice (Fig. 3C). The interaction between annulus and apical branch intersections was characterized by a linear increase in apical branch intersections across annuli in ABc animals, while BAc and WT animals demonstrated an initial linear increase at proximal annuli, which then plateaued at more distal annuli (Fig. 3C).
Figure 3:
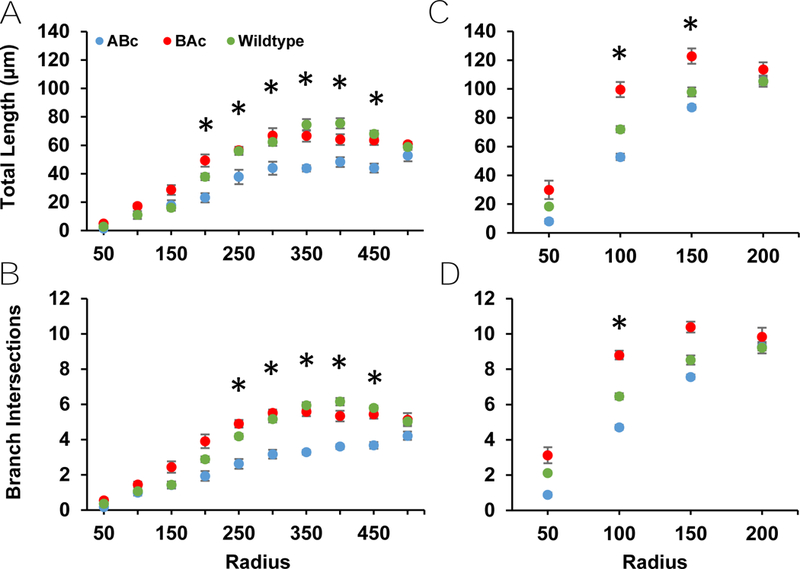
Apical and basal dendritic parameters from Sholl analysis of eGFP labeled neurons. A) ABc cells demonstrate a lower total apical length compared to BAc and WT cells. B) A similar effect is shown in total basal length, with ABc cells showing decreased length. C) There was a statistically significant decrease in apical branch intersections among ABc cells compared to BAc and WT cells. D) This effect persisted in basal branch intersections, though the difference here was not statistically significant.
All Sholl analysis measures demonstrated a main effect of annulus (Table E1) (Fig. 3). Apical and basal length both increased as annulus increased (Table H1) (Fig. 3). Apical and basal branch intersections also increased as annulus level increased (see Table H1) (Fig. 3).
No Genotype Differences for Golgi-Cox Labeled Neurons Found in Any Dendritic Parameter
Branch Analyses
There was no main effect of genotype for any branch analysis measure in Golgi-Cox stained cells (Table F1) (Fig. 4). All measures except for basal tortuosity demonstrated a main effect of branch order (Table F1) (Fig. 4H). Total apical and basal length both increased as branch order increased (Table I1) (Fig. 4). Apical and basal number of branches increased variably as branch order increased (Table I1) (Fig. 4). Apical average length showed a main effect of branch order, however only one comparison (1 vs. 5, p = 0.007) was significant (Fig. 4). Basal average length showed a main effect of branch order but post hoc tests revealed no differences (Fig. 4D). Like apical average length, apical tortuosity demonstrated a main effect of branch order, with one comparison significant (1 vs. 5, p = 0.007) (Fig. 4G). No genotype by branch order interaction effects were seen in any branch analysis measure (Table F1).
Figure 4:
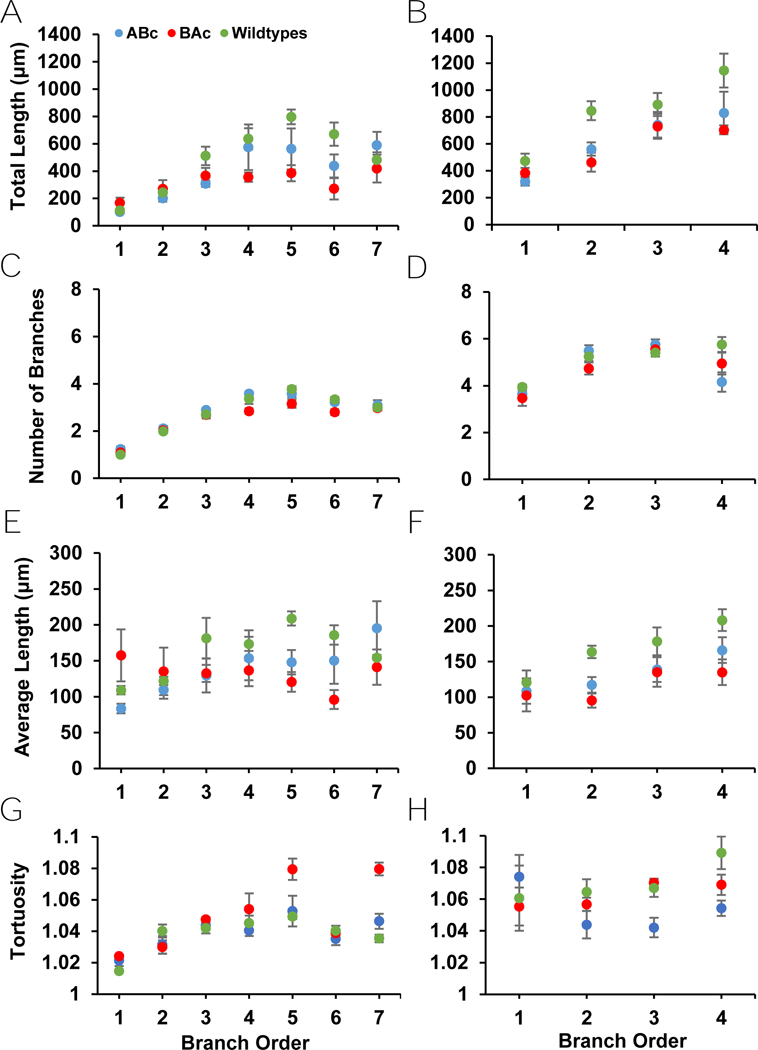
Apical and basal dendritic measures from branch analyses of Golgi-Cox labeled neurons. A) No differences between genotypes were present in total apical length or B) total basal length. C) Additionally, Golgi-Cox labeled neurons showed no difference in genotypes in either apical average length or D) basal average length. E) Similarly, these neurons showed no differences between genotypes in apical number of branches or F) basal number of branches. G) There were also no differences present between genotypes for apical tortuosity or H) basal tortuosity.
Sholl Analyses
There was no main effect of genotype for any of the measures in Golgi-Cox labeled neurons (Table F1) (Fig. 5). All measures demonstrated a main effect of annulus (Table F1) (Fig. 5). In apical length, there was a significant difference found between driver lines—as such, Mayford and Line-84 mice were analyzed separately for Sholl analysis (Fig. 5A). The Line-84 driver line showed insufficient power, so annulus main effects were only analyzed in the Mayford driver line. Mayford mice demonstrated an increase in apical length (p < 0.001) and in basal length (p < 0.001) as annulus increased (Table J1). Apical and basal branch intersections increased as annulus level increased (Table J1). No genotype by annulus interaction effect was present for any Sholl measure.
ABc Mice Showed Increased Total Arbor Path for eGFP- and Golgi-Cox Labeled Neurons
Total arbor path eGFP:
Total arbor path was calculated by summing the products of each branch order’s total length and tortuosity. There was a significant main effect of genotype for apical total arbor [F(2,13) = 6.109, p = 0.013] and basal total arbor [F(2,12) = 4.015, p = 0.044] (Fig. 6A). ABc mice demonstrated a higher apical arbor path than WT mice (post hoc: p = 0.017) and a trend for high apical arbor path compared to BAc mice (p = 0.066) (Fig. 6A). There was no difference in apical arbor between BAc and WT cells (Fig. 6A). Basal total arbor path showed the same trends: an increase in ABc mice basal arbor path compared to WT mice (post hoc: p = 0.017) with a trend in the same measure for BAc mice (post hoc: p = 0.066) (Fig. 6). There was no difference between BAc and WT basal arbor paths (Fig. 6A).
Figure 6:
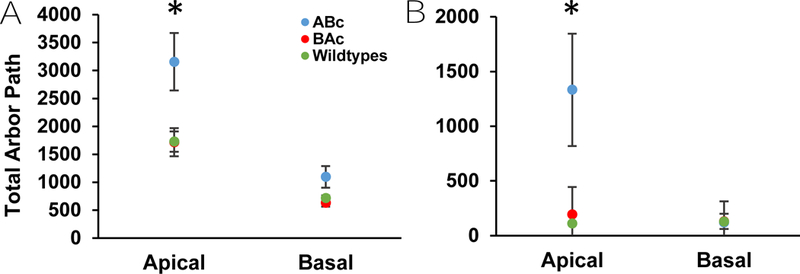
Analysis of total arbor path in apical and basal dendrites in Thy-1 eGFP and Golgi-Cox labeled cells. A) Thy-1 eGFP labeled neurons show a significant increase in total apical and basal arbor path for ABc cells compared to BAc and WT cells. B) Golgi-Cox labeled neurons also show a significant increase in total apical arbor path for ABc cells over BAc and WT cells, though no such effect was shown for the total basal arbor path.
Total arbor path Golgi-Cox:
There was a significant main effect of genotype on apical total arbor for Golgi-Cox labeled neurons [F(2,12) = 24.724, p < 0.001] (Fig. 6B). ABc neurons demonstrated an increased apical arbor path compared to BAc neurons (post hoc: p < 0.001) and WT neurons (post hoc: p < 0.001) (Fig. 6B). BAc and WT mice did not differ (Fig. 6B). In contrast, there was no main effect of genotype present for basal total arbor path [F(2,12) = 0.058, p = 0.944] (Fig. 6B).
Discussion
Overall, when observed with eGFP labeling, CA1 pyramidal neurons in ABc mice showed measures of more extensive dendritic arbors compared to those from BAc and WT animals. For branch order analyses, increased values were observed for apical average length and basal total length in ABc compared to BAc and WT neurons. For Sholl analyses, ABc cells showed decreased apical length and apical branch intersections near to the soma when compared to BAc and WT cells. This discrepancy in apical length measures between branch order and Sholl analyses is most likely due to a total increased length at higher branch orders, which may have been omitted in the Sholl analysis due to the restriction in total linear extent from the soma imposed by the subjective raters (apical: 500 μm; basal: 200 μm). In agreement with the branch order analyses, ABc animals demonstrated a higher apical total arbor path and basal total arbor path than WTs. Since these experiments were performed in pubertal mice, which have a largely GluN2A native background, the predominant molecular change in ABc mice (compared to WTs) is increased expression of the GluN2B C-terminus. This suggests that the GluN2B C-terminus, the intracellular signaling component of the NMDAR, is primarily responsible for influencing dendritic growth.
Genotype effects on some but not all measures of neuron morphology suggest that subfamilies of dendritic parameters may have distinct molecular factors that define them. ABc expression altered apical average length, apical Sholl length and branch intersections, basal total length, and total arbor path for both apical and basal dendrites. Apical total length, apical and basal tortuosity, apical and basal number of branches, basal average length, basal branch intersections and basal Sholl length were not affected by genotype. Applying the same logic as in the opening paragraph of the discussion, parameters affected by ABc expression are likely more directly impacted by the GluN2B C-terminus, because the N-terminus and TMDs of ABc match the WT background. This is further supported by the lack of effect in the BAc line where the molecular alteration is replacement of the N-terminus and TMDs of the native GluN2A subunits with those of the transgenic GluN2B subunits, which would directly alter calcium conductance and not intracellular signaling. Other morphological parameters may be governed by different NMDAR properties or are not altered by GluN2 subunit content. For instance, the chimeric changes did not impact the number of branches in either the apical or basal dendrites, suggesting that this measure is not impacted by changes in NMDAR signaling. Previous reports of NMDAR involvement in the regulation of dendritic morphology involved stress (Blanke & VanDongen, 2009) or developing animals (Maki, Aman, Amico-Ruvio, Kussius & Popescu, 2012), supporting the idea that some aspects of dendrite morphology may be more stable and less susceptible to perturbation in healthy adult animals.
Transgene expression was not assessed on a cell-by-cell basis. As such, variability in transgene expression levels between mouse lines could influence results. However, it has been shown previously that, when targeting the hemagglutinin epitope of the chimeric GluN2 subunits, immunohistochemical labeling of transgenic subunit levels in area CA1 were not different between ABc and BAc mice (Sanders et al., 2018). As such, the genotype effects on morphology reported here are likely not due to variations in transgene expression. Incorporation of additional histological techniques to more directly relate transgene expression to dendritic morphology was avoided due to incompatibility with Golgi-Cox staining and introduction of a confound when comparing between labeling methods. Thus, it remains unclear if the NMDAR containing chimeric GluN2 subunits are added to or replace native NMDARs. This will be clarified in future Western blot experiments by quantifying GluN1 to test for addition and with a pull-down strategy to subtract HA-labeled GluN2 subunits from the total GluN2 pool to test for replacement. Also, single-cell PCR may be applied in future studies to determine the variability in transgene expression levels across neurons within subjects.
The observation that dendritic morphology is sensitive to the intracellular signaling properties of GluN2 subunits provides a foundation for additional studies of downstream molecular signaling. Relative to NMDARs containing GluN2A, NMDARs that incorporate GluN2B associate more readily with calmodulin-dependent protein kinase II (CaMKII), the obligatory kinase for the induction of activity-dependent synaptic potentiation (Shipton & Paulsen, 2014). Additionally, NMDARs with GluN2B subunits influence synaptic efficacy and spine shape through selective interactions with RasGRF-1 (Li, Tian, Hartley & Feig, 2006) and α-actinin (Wyszynski et al., 1997), respectively. Finally, GluN2B subunits lack CDK5 and PKA phosphorylation sites and have fewer PKC phosphorylation sites that are present in GluN2A subunits and regulate channel dynamics (Sanz-Clemente, Nicoll, & Roche, 2013; Chen & Roche, 2007). Thus, activation of one or more of these GluN2B CTD signaling streams could underlie the increased dendritic growth observed in the ABc line. More generally, activation of GluN2B CTD signaling streams in WT mice may be largely responsible for dendritic remodeling and synaptic connectivity, and hence large-scale network connectivity.
Dendritic morphology has been shown to impact various electrophysiological measures in both computational and experimental models (Duijnhouwer, Remme, van Ooyen, & van Pelt, 2001; Ferrante, Migliore, & Ascoli, 2013; Krichmar, Nasuto, Scorcioni, Washington, & Ascoli, 2002). Type of firing, firing frequency, impedance mismatch, spike initiation, and more are all affected by dendritic morphology (Ferrante et al., 2013; Krichmar et al., 2002). For example, cells in area CA3 with more expansive dendritic trees are prone to higher spiking frequencies and lower burst frequencies than cells with smaller dendritic trees (Ferrante et al., 2013; Krichmar et al., 2002). Therefore, higher spiking frequencies might be expected in CA1 pyramidal neurons in ABc mice having more expansive dendritic arbors compared to BAc and WT mice. Additionally, CA1 cells with longer apical dendrites are more sensitive to locationspecific synapse integration (Ferrante et al., 2013; Krichmar et al., 2002), and membrane area of CA1 cells impacts spike threshold (Ferrante et al., 2013; Krichmar et al., 2002). Further, in cultured hippocampal neurons, the spiking rate tends to increase as the number of dendritic terminations increases (Krichmar et al., 2002; Krichmar, Velasquez, & Ascoli, 2006). Thus, CA1 pyramidal neurons in ABc mice would be expected to exhibit alterations in synapse integration and spike threshold, and spike rate. Electrophysiological changes such as these likely impact neuronal plasticity, though investigation of the direct impacts of dendritic morphology on functional plasticity is lacking (Ferrante et al., 2013; Krichmar et al., 2002). Electrophysiological experiments in hippocampal slices taken from ABc and BAc mice may help to clarify how dendritic morphology influences activity-dependent synaptic plasticity.
Analyses of dendritic morphology parameters resulting from traces of eGFP and Golgi-Cox labeled CA1 pyramidal neurons revealed a more complete registry of both apical and basal dendrites by eGFP labeling, though differences were especially prominent in basal measures. While branch analysis noted method differences solely in basal measures, Sholl analysis demonstrated a main effect of method in every measure, suggesting Sholl analysis may be more sensitive to labeling method. eGFP and Golgi-Cox labeled neurons diverged substantially in their significance values for genotype-level differences, with eGFP labeled neurons always having pvalues closer to or lesser than 0.05 when compared to Golgi-Cox neurons. This suggests that eGFP expression may reveal genotype differences more readily than Golgi-Cox staining. There were fewer branch intersections and shorter branch lengths per annulus, and a much lower total arbor path, found in Golgi-Cox compared to eGFP neurons, further suggesting incomplete labeling. These results could be due to the heavy tissue processing required for Golgi staining or the background noise inherent in Golgi staining, which reduces the resolution of finer processes (Fig. 1B). Additionally, these results could be due to lack of blindness to labeling method during the neuron selection and tracing processes. However, it is unlikely that the differences in dendritic parameters across labeling methods is a result of a lack of blind analysis because the results from the branch order analyses matched the Sholl analyses, which were performed independently and subsequent to imaging and tracing. Combined with the comparative simplicity of tissue processing, these findings support Thy-1_eGFP labeling as a potential replacement for Golgi-Cox labeling when performing morphological assessment of excitatory neurons in the mouse forebrain.
While the collapse of the categorical variables of age, gender, WT status, and driver line designation were largely appropriate for most measures, some did show significant differences. In the case of basal tortuosity, WTs appeared to differ (Table B1); however, given that this is the only measure where the WTs differed, and its p-value being close to 0.05, this difference is likely a type I error.
Regarding age, Golgi-Cox assigned animals were slightly but significantly older than Thy-1_eGFP animals; this age difference was primarily found in the BAc and WT animals, with ABc animals not differing in age between the methodologies. While this specific age difference could be masking potential BAc effects, it is unlikely, given that BAc animals and WT animals did not differ from one another in any dendritic measure in either methodology. Further, when comparing between animals at 30 and 60 or 70 days of age, there is no difference in relevant dendritic parameters for CA1 pyramidal cells (Jakubowska-Dogru et al., 2017; Brunette et al., 2010). In Jakubowska-Dogru et al., mean dendritic field area, apical and basal dendritic length, apical and basal number of branches, and apical and basal mean highest branch order did not differ between control animals from P30 to P60 (2017). In Brunette et al., maximal apical length, branch width, total Sholl ring crossings, and peak branching distance did not differ in control animals from P30 to P70 (2010). Neither of these papers found age differences in any measured dendritic parameter, suggesting that postnatal age did not impact the results reported in this study.
In the case of gender, females displayed greater apical average dendritic length and tortuosity compared to males. Though this effect was only seen in Golgi-Cox labeled neurons, there is some precedence for the finding in that prior Golgi-Cox labeling of CA3 pyramidal neurons showed greater proximal dendritic branching in females and greater distal branching in males (Juraska, Fitch, & Washburne, 1989). This was specifically seen in the apical dendrite, while no sex differences were seen in the basal dendrites. In contrast, a study in Golgi-Cox labeled CA1 pyramidal neurons showed a higher number of dendritic branch intersections in males along all areas of the dendritic tree; apical and basal dendrites both showed these differences proximally and distally (Markham, Mckian, Stroup, & Juraska, 2005). Combined, these findings support differential effects of gender on CA3 and CA1 pyramidal neurons. However, due to the lack of gender differences observed with eGFP labeling in the current study, it is possible that these sex-specific findings were products of the methodology. As such, Golgi-Cox staining may be more susceptible to sex influences on neuronal labeling.
It is known that the NMDAR GluN2 subunit composition is altered over the third postnatal week of development: NMDARs composed primarily of GluN2B switch to a predominantly GluN2A composition (Dumas, 2005). As such, understanding the impacts of the chimeric GluN2 subunits in development may illuminate the mechanisms behind this switch, and the subsequent changes that occur. Transgene expression of the subunit begins at postnatal day 14 (P14), leaving a window of influence for the subunit on neuronal morphology. Given prior behavioral experiments (Sanders et al., 2013), it is possible that the genotype differences shown in this study originate before the hippocampus matures. However, since the experiment reported here was performed in adult animals, it is not currently possible to determine whether the results are a consequence of altering the NMDAR switch, or if these changes are displayed prior to the end of the third postnatal week. As such, future research should examine the impact of the chimeric GluN2 subunit switch on the development of dendritic morphology, and the separate impact of GluN2A and GluN2B on certain parameters of morphology.
By expressing chimeric GluN2 subunits in transgenic mice, this study was able to differentiate effects of NMDAR-dependent calcium conductance dynamics and direct intracellular signaling on dendritic morphology in hippocampal pyramidal neurons. The growth effects of overexpression of the GluN2B CTD support the notion that NMDARs with GluN2B subunits potently regulate dendritic morphology through activation of postsynaptic signaling streams that do not rely on calcium entry through the channel pore. This new perspective paves the way for a greater understanding of the molecular underpinnings of normal and pathological dendritic plasticity and also provides a framework for the development of novel treatments for medical conditions associated with aberrant dendritic growth, including stress-related (Hardingham & Bading, 2010; Vyas, Mitra, Rao & Chattarji, 2002; Christian, Miracle, Wellman, & Nakazawa, 2011; McEwen, 2005) and neurodevelopmental disorders (Dang et al., 2017; Martinez-Cerdeno, 2017; Jacobs, Cheng, & Doering, 2016).
eGFP expression reveals a greater dendritic arbor extent than Golgi-Cox staining
Gender differences in morphology were found solely in Golgi-Cox stained neurons
ABc mice have a longer total arbor path, average apical length and total basal length
BAc mice and wildtype mice did not differ in neuron morphology
NMDAR-dependent intracellular signaling regulates dendritic morphology
Grant Support/Acknowledgments:
This work was supported by the Thomas and Kate Miller Jeffress Memorial Trust Fund, the Department of Defense (ONR#N00014-10-1-0198), and the National Institutes of Health (1R15AG045820).
Appendices
Appendix A
Table A1:
Comparison of morphology measures across gender for each method
| eGFP | Golgi-Cox | |
|---|---|---|
| Apical total length | F(1,14) = 1.879, p = 0.192 | F(1,13) = 2.254, p = 0.157 |
| Apical number of branches | F(1,14) = 0.015, p = 0.903 | F(1,13) = 0.004, p = 0.951 |
| Apical average length | F(1,14) = 0.993, p = 0.336 | F(1,13) = 5.903, p = 0.030* |
| Apical tortuosity | F(1,14) = 0.005, p = 0.943 | F(1,13) = 6.301, p = 0.026* |
| Apical total arbor path | F(1,14) = 0.594, p = 0.454 | F(1,13) = 2.897, p = 0.113 |
| Apical length (Sholl) | F(1,14) = 0.275, p = 0.608 | F(1,13) = 2.968, p = 0.109 |
| Apical branch intersections (Sholl) | F(1,14) = 0.106, p = 0.750 | F(1,13) = 0.645, p = 0.437 |
| Basal total length | F(1,14) = 0.552, p = 0.470 | F(1,13) = 2.152, p = 0.166 |
| Basal number of branches | F(1,14) = 0.020, p = 0.889 | F(1,13) = 0.636, p = 0.440 |
| Basal average length | F(1,14) = 0.894, p = 0.360 | F(1,13) = 2.619, p = 0.130 |
| Basal tortuosity | F(1,14) = 0.496, p = 0.493 | F(1,13) = 2.339, p = 0.150 |
| Basal total arbor path | F(1,14) = 0.483, p = 0.498 | F(1,13) = 3.224, p = 0.096 |
| Basal length (Sholl) | F(1,14) = 0.878, p = 0.365 | F(1,13) = 0.001, p = 0.978 |
| Basal branch intersections (Sholl) | F(1,14) = 0.851, p = 0.372 | F(1,13) = 0.007, p = 0.932 |
Appendix B
Table B1:
Comparisons of morphology measures across WTs for each method
| eGFP | Golgi-Cox | |
|---|---|---|
| Apical total length | F(3,12) = 1.990, p = 1.000 | F(3,11) = 0.761, p = 1.000 |
| Apical number of branches | F(3,12) = 0.135, p = 1.000 | F(3,11) = 0.525, p =1.000 |
| Apical average length | F(3,12) = 2.775, p = 1.000 | F(3,11) = 0.326, p = 1.000 |
| Apical tortuosity | F(3,12) = 0.125, p = 1.000 | F(3,11) = 0.744, p = 1.000 |
| Apical total arbor path | F(3,12) = 3.901, p = 1.000 | F(3,11) = 15.110, p = 1.000 |
| Apical length (Sholl) | F(3,12) = 3.272, p = 1.000 | F(3,11) = 0.512, p = 1.000 |
| Apical branch intersections (Sholl) | F(3,12) = 4.720, p = 1.000 | F(3,11) = 0.830, p = 1.000 |
| Basal total length | F(3,12) = 2.464, p = 1.000 | F(3,11) = 0.706, p = 1.000 |
| Basal number of branches | F(3,12) = 1.072, p = 1.000 | F(3,11) = 0.242, p = 1.000 |
| Basal average length | F(3,12) = 2.272, p = 1.000 | F(3,11) = 0.713, p = 1.000 |
| Basal tortuosity | F(3,12) = 4.335, p = 0.043 | F(3,11) = 1.463, p = 0.475 |
| Basal total arbor path | F(3,12) = 2.644, p = 1.000 | F(3,11) = 0.076, p = 1.000 |
| Basal length (Sholl) | F(3,12) = 2.066, p = 1.000 | F(3,11) = 0.159, p = 1.000 |
| Basal branch intersections (Sholl) | F(3,12) = 1.977, p = 1.000 | F(3,11) = 0.151, p = 1.000 |
Appendix C
Table C1:
Comparisons of morphology measures across driver-line for each method
| eGFP | Golgi-Cox | |
|---|---|---|
| Apical total length | F(1,14) = 1.500, p = 0.241 | F(1,13) = 0.991, p = 0.338 |
| Apical number of branches | F(1,14) = 0.276, p = 0.607 | F(1,13) = 0.005, p = 0.947 |
| Apical average length | F(1,14) = 2.453, p = 0.140 | F(1,13) = 0.670, p = 0.428 |
| Apical tortuosity | F(1,14) = 0.059, p = 0.812 | F(1,13) = 0.132, p = 0.722 |
| Apical total arbor path | F(1,14) = 2.086, p = 0.171 | F(1,13) = 0.951, p = 0.347 |
| Apical length (Sholl) | F(1,14) = 3.183, p = 0.096 | F(1,13) = 6.473, p = 0.024* |
| Apical branch intersections (Sholl) | F(1,14) = 2.997, p = 0.105 | F(1,13) = 0.006, p = 0.941 |
| Basal total length | F(1,14) = 2.203, P = 0.160 | F(1,13) = 0.084, P = 0.776 |
| Basal number of branches | F(1,14) = 1.595, p = 0.227 | F(1,13) = 0.626, p = 0.443 |
| Basal average length | F(1,14) = 1.142, p = 0.303 | F(1,13) = 0.041, p = 0.842 |
| Basal tortuosity | F(1,14) = 1.529, p = 0.237 | F(1,13) = 0.047, p = 0.832 |
| Basal total arbor path | F(1,14) = 2.814, p = 0.116 | F(1,13) = 0.837, p = 0.377 |
| Basal length (Sholl) | F(1,14) = 0.015, p = 0.906 | F(1,13) = 0.442, p = 0.518 |
| Basal branch intersections (Sholl) | F(1,14) = 0.036, p = 0.853 | F(1,13) = 0.334, p = 0.573 |
Appendix D
Table D1:
Comparisons of morphology measures across method
| Method Main Effects (eGFP vs. Golgi-Cox) | |
|---|---|
| Apical total length | F(1,29) = 0.377, p = 0.544 |
| Apical number of branches | F(1,29) = 3.259, p = 0.081 |
| Apical average length | F(1,29) = 0.641, p = 0.430 |
| Apical tortuosity | F(1,29) = 0.024, p = 0.879 |
| Apical total arbor path | F(1,29) = 29.326, p < 0.001* |
| Apical length (Sholl) | F(1,29) = 5.592, p < 0.025* |
| Apical branch intersections (Sholl) | F(1,29) = 4.245, p < 0.048* |
| Basal total length | F(1,29) = 0.158, p = 0.694 |
| Basal number of branches | F(1,29) = 18.458, p < 0.001* |
| Basal average length | F(1,29) = 4.454, p = 0.044* |
| Basal tortuosity | F(1,29) = 2.013, p = 0.167 |
| Basal total arbor path | F(1,29) = 70.096, p < 0.001* |
| Basal length (Sholl) | F(1,29) = 37.528, p < 0.001* |
| Basal branch intersections (Sholl) | F(1,29) = 40.150, p < 0.001* |
Appendix E
Table E1:
Comparisons of morphology across genotype and branch order/annulus in eGFP
| Genotype | Branch order / Annulus | Interaction effect | |
|---|---|---|---|
| Apical total length | F(2,13) = 3.216, p = 0.073 |
F(6,78) = 10.919, p < 0.001* |
F(12,78) = 0.380, p = 0.967 |
| Apical number of branches | F(2,13) = 0.092, p = 0.912 |
F(2.132,27.721) = 32.525, p < 0.001* |
F(4.265,27.721) = 0.208, p = 0.939 |
| Apical average length |
F(2,13) = 4.394, p = 0.035* |
F(3.209,41.713) = 0.424, p = 0.869 | F(6.417,41.713) = 0.424, p = 0.869 |
| Apical tortuosity | F(2,13) = 0.187, p = 0.832 |
F(6,78) = 10.312, p < 0.001* | F(12,78) = 1.663, p = 0.092 |
| Apical length (Sholl) |
F(2,13) = 4.378, p = 0.035* |
F(2.650,34.448) = 42.238, p < 0.001* |
F(5.300,34.448) = 1.544, p = 0.199 |
| Apical branch intersections (Sholl) |
F(2,13) = 7.175, p = 0.008* |
F(2.528,32.866) = 61.115, p < 0.001* |
F(5.056,32.866) = 2.632, p = 0.041* |
| Basal total length |
F(2,13) = 3.963, p = 0.045* |
F(1.428,18.570) = 20.172, p < 0.001* |
F(2.857,18.570) = 3.671, p = 0.033* |
| Basal number of branches | F(2,13) = 1.178, p = 0.339 |
F(1.608,20.899) = 46.669, p < 0.001* |
F(3.215,20.899) = 1.509, p = 0.240 |
| Basal average length | F(2,13) = 3.694, p = 0.054 |
F(3,39) = 20.524, p < 0.001* | F(6,39) = 2.066, p = 0.080 |
| Basal tortuosity | F(2,13) = 0.736, p = 0.498 |
F(3,36) = 0.155, p = 0.926 | F(3,36) = 0.523, p = 0.848 |
| Basal length (Sholl) | F(2,13) = 3.094, p = 0.080 |
F(2.060,26.777) = 151.170, p < 0.001* |
F(4.120, 26.777) = 1.860, p = 0.145 |
| Basal branch intersections (Sholl) | F(2,13) = 3.084, p = 0.080 |
F(2.010,26.124) = 141.717, p < 0.001* |
F(4.019,26.124) = 1.989, p = 0.125 |
Appendix F
Table F1:
Comparisons of morphology across genotype and order/annulus in Golgi-Cox
| Genotype | Branch order / Annulus | Interaction effect | |
|---|---|---|---|
| Apical total length | F(2,12) = 1.223, p = 0.329 |
F(3.584,43.004) = 11.961, p < 0.001* |
F(7.167,43.004) = 1.434, p = 0.216 |
| Apical number of branches | F(2,12) = 0.584, p = 0.573 |
F(2.368,28.410) = 25.618, p < 0.001* |
F(4.735,28.410) = 0.301, p = 0.901 |
| Apical average length | F(2,12) = 0.480, p = 0.630 |
F(6,72) = 2.325, p = 0.042* |
F(12,72) = 1.727, p = 0.079 |
| Apical tortuosity | F(2,12) = 1.005, p = 0.395 |
F(2.796,33.556) = 6.210, p = 0.002* |
F(5.593,33.556) = 1.587, p = 0.185 |
| Apical length (Sholl) | Line-84: F(1,1) = 0.350, p = 0.660 Mayford: F(2,9) = 0.915, p = 0.435 |
F(2.272,20.449) = 22.585, p < 0.001* |
F(4.544,20.449) = 1.681, p = 0.188 |
| Apical branch intersections (Sholl) | F(2,12) = 0.623, p = 0.553 |
F(2.356,28.276) = 22.306, p < 0.001* |
F(4.713,28.276) = 1.613, p = 0.192 |
| Basal total length | F(2,12) = 1.091, p= 0.367 |
F(1.392,16.703) = 5.235, p = 0.026* |
F(2.784,16.703) = 0.323, p = 0.795 |
| Basal number of branches | F(2,12) = 0.144, p = 0.867 |
F(1.332,15.983) = 5.212, p = 0.028* |
F(2.664,15.983) = 0.611, p = 0.757 |
| Basal average length | F(2,12) = 1.150, p = 0.349 |
F(1.965,23.580) = 3.625, p = 0.043* |
F(3.930,23.580) = 0.532, p = 0.711 |
| Basal tortuosity | F(2,12) = 0.264, p= 0.773 |
F(2.004,24.045) = 0.943, p = 0.404 |
F(4.007,24.045) = 1.155, p = 0.352 |
| Basal length (Sholl) | F(2,12) = 0.245, p= 0.786 |
F(1.396,16.753) = 42.560, p < 0.001* |
F(2.792,16.753) = 1.276, p = 0.314 |
| Basal branch intersections (Sholl) | F(2,12) = 0.246, p= 0.786 |
F(2.768,52.168) = 41.329, p < 0.001* |
F(2.768,52.168) = 1.406, p = 0.276 |
Appendix G
Table G1:
eGFP Branch Order Main Effects
| Post Hoc Comparisons Analysis | |
|---|---|
| Apical total length | 1 vs. 3, p = 0.025; 1 vs. 4, p = 0.011; 1 vs. 5, p = 0.001; 1 vs. 6, p < 0.001; 1 vs. 7, p = 0.004; 2 vs. 5 and 2 vs. 6, p = 0.008; all others nonsignificant |
| Basal total length | 1 vs. 2 and 1 vs. 3, p < 0.001; 1 vs. 4, p = 0.002; 2 vs. 3, p = 0.020, 2 vs. 4, p = 0.080; all others nonsignificant |
| Apical number of branches | 1 vs. 2, 3, 4, 5, 6, and 7, p < 0.001; 2 vs. 3 and 2 vs. 4, p < 0.001; 2 vs. 5, p = 0.001; 2 vs. 6, p = 0.008; 2 vs. 7, p = 0.006; all others nonsignificant |
| Basal number of branches | 1 vs. 2, 3, and 4, p < 0.001; 2 vs. 3, p < 0.001; 2 vs. 4, p = 0.075; all others nonsignificant |
| Apical average length | No significant post hoc comparisons |
| Basal average length | 1 vs. 2, p = 0.009; 1 vs. 3, p < 0.001; 1 vs. 4, p = 0.001; 2 vs. 4, p = 0.025; all others nonsignificant |
| Apical tortuosity | 1 vs. 2 and 1 vs. 7, p = 0.001; 1 vs. 3, p = 0.008; 1 vs. 4, p = 0.007; 1 vs. 5, p = 0.003; 1 vs. 6, p = 0.002; all others nonsignificant |
| Basal tortuosity | No significant post hoc comparisons |
Appendix H
Table H1:
eGFP Annulus Level Main Effects
| Post Hoc Comparisons Analysis | |
|---|---|
| Apical length | 1 vs. 2, 3, 5, 6, and 7, p < 0.001; 2 vs. 6 and 2 vs. 7, p < 0.001; 3 vs. 6 and 3 vs. 7, p < 0.001; 1 vs. 4 and 2 vs. 5, p = 0.001; 2 vs. 3, p = 0.029; 2 vs. 4, p = 0.015; 3 vs. 5, p = 0.002; 4 vs. 6, p = 0.016; all others nonsignificant |
| Basal length | 1 vs. 2, 3, and 4, p < 0.001; 2 vs. 3 and 2 vs. 4, p < 0.001; all others nonsignificant |
| Apical branch intersections | 1 vs. 2, 3, 4, 5, 6, and 7, p < 0.001; 2 vs. 5, 6, and 7, p < 0.001; 3 vs. 6 and 3 vs. 7, p < 0.001; 4 vs. 6, p < 0.001; 2 vs. 3, p = 0.010; 2 vs. 4 and 5 vs. 6, p = 0.008; 3 vs. 4, p = 0.036; 3 vs. 5 and 4 vs. 5, p = 0.001; 4 vs. 7, p = 0.003; 4 vs. 8, p = 0.007; 4 vs. 9, p = 0.022; all others nonsignificant |
| Basal branch intersections | 1 vs. 2, 3, and 4, p < 0.001; 2 vs. 3 and 2 vs. 4, p < 0.001; all others nonsignificant |
Appendix I
Table I1:
Golgi-Cox Branch Order Main Effects
| Post Hoc Comparisons Analysis | |
|---|---|
| Apical total length | 1 vs. 3, p = 0.005; 1 vs. 4, p = 0.004; 1 vs. 5, p = 0.003; 1 vs. 6, p = 0.013; 1 vs. 7, p = 0.014; 2 vs. 4, p = 0.024; 2 vs. 5, p = 0.007; 2 vs. 6, p = 0.044; 2 vs. 7, p = 0.009; all others nonsignificant |
| Basal total length | No significant post hoc comparisons |
| Apical number of branches | 1 vs. 2, 3, 4, 5, and 6, p < 0.001; 1 vs.7, p = 0.001; 2 vs. 3, 4, and 5, p = 0.001; 2 vs. 6, p = 0.029; 3 vs. 5, p = 0.022; all others nonsignificant |
| Basal number of branches | 1 vs. 2, p = 0.001; 1 vs. 3, p = 0.038; all others nonsignificant |
| Apical average length | 1 vs. 5, p = 0.007; all others nonsignificant |
| Basal average length | No significant post hoc comparisons |
| Apical tortuosity | 1 vs. 5, p = 0.007; all others nonsignificant |
| Basal tortuosity | No significant post hoc comparisons |
Appendix J
Table J1:
Golgi-Cox Annulus Level Main Effects
| Post Hoc Comparisons Analysis | |
|---|---|
| Apical length | 1 vs. 2, p = 0.055; 1 vs. 3 and 2 vs. 3, p = 0.006; 1 vs. 4, p = 0.013; 1 vs. 5, p = 0.020; 1 vs. 6, p = 0.001; 1 vs. 7 and 2 vs. 7, p < 0.001; 2 vs. 4, p = 0.028; 2 vs. 5, p = 0.038, 2 vs. 6, p = 0.002; 3 vs. 6, p = 0.015; 3 vs. 7, p = 0.005; all others nonsignificant |
| Basal length | 1 vs. 2, 3, and 4, p < 0.001; 2 vs. 3, p = 0.006; all others nonsignificant |
| Apical branch intersections | 1 vs. 2, p = 0.004; 1 vs. 2, 3, 4, 5, 6 and 7, p < 0.001; 2 vs. 3, 6 and 7, p < 0.001; 3 vs. 6, p = 0.003; 2 vs. 4, p = 0.002; 2 vs. 5 and 3 vs. 7, p = 0.001; 3 vs. 5 and 6 vs. 4, p = 0.035; 4 vs. 7, p = 0.025; all others nonsignificant |
| Basal branch intersections | 1 vs. 2, 3, and 4, p < 0.001; 2 vs. 3, p = 0.008; all others nonsignificant |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests Statement:
The authors declare no competing interests.
References
- Aow J, Dore K & Malinow R Conformational signaling required for synaptic plasticity by the NMDA receptor complex. Proc. Natl. Acad. Sci. U. S. A 112, 14711–14716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister NJ & Larkman AU Dendritic morphology of CA1 pyramidal neurones from the rat hippocampus: I. Branching patterns. J. Comp. Neurol 360, 150–160 (1995). [DOI] [PubMed] [Google Scholar]
- Berberich S, Jensen V, Hvalby Ø, Seeburg PH & Köhr G The role of NMDAR subtypes and charge transfer during hippocampal LTP induction. Neuropharmacology 52, 77–86 (2007). [DOI] [PubMed] [Google Scholar]
- Birnbaum JH, Bali J, Rajendran L, Nitsch RM & Tackenberg C Calcium fluxindependent NMDA receptor activity is required for Aβ oligomer-induced synaptic loss. Cell Death Dis 6, e1791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanke ML & VanDongen AMJ Activation Mechanisms of the NMDA Receptor. in Biology of the NMDA Receptor (ed. Van Dongen AM) (CRC Press/Taylor & Francis, 2009). [PubMed] [Google Scholar]
- Brunette KE, Tran PV, Wobken JD, Carlson ES & Georgieff MK Gestational and neonatal iron deficiency alters apical dendrite structure of CA1 pyramidal neurons in adult rat hippocampus. Developmental Neuroscience 32, 238–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B-S & Roche KW Regulation of NMDA Receptors by Phosphorylation. Neuropharmacology 53, 362–368 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian KM, Miracle AD, Wellman CL & Nakazawa K Chronic stress-induced hippocampal dendritic retraction requires CA3 NMDA receptors. Neuroscience 174, 26–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C NMDA receptor as a newly identified member of the metabotropic glutamate receptor family: clinical implications for neurodegenerative diseases. Mol. Cells 36, 99–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang T et al. Autism-associated Dyrk1a truncation mutants impair neuronal dendritic and spine growth and interfere with postnatal cortical development. Mol. Psychiatry (2017). 10.1038/mp.2016.253 [DOI] [PMC free article] [PubMed]
- Dingledine R, Borges K, Bowie D & Traynelis SF The glutamate receptor ion channels. Pharmacol. Rev 51, 7–61 (1999). [PubMed] [Google Scholar]
- Dore K et al. Unconventional NMDA Receptor Signaling. J. Neurosci. Off. J. Soc. Neurosci 37, 10800–10807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore K, Aow J & Malinow R Agonist binding to the NMDA receptor drives movement of its cytoplasmic domain without ion flow. Proc. Natl. Acad. Sci. U. S. A 112, 14705–14710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore K, Aow J & Malinow R The Emergence of NMDA Receptor Metabotropic Function: Insights from Imaging. Front. Synaptic Neurosci 8, 20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijnhouwer J, Remme MWH, van Ooyen A, & van Pelt J (2001). Influence of dendritic topology on firing patterns in model neurons. Neurocomputing, 38–40, 183–189. [DOI] [PubMed] [Google Scholar]
- Dumas TC Developmental regulation of cognitive abilities: modified composition of a molecular switch turns on associative learning. Prog. Neurobiol 76, 189–211 (2005). [DOI] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Buchner A, & Lang A-G Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behavior Research Methods, 41, 11491160 (2009). [DOI] [PubMed] [Google Scholar]
- Ferrante M, Migliore M, & Ascoli GA (2013). Functional impact of dendritic branch point morphology. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 33(5), 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JS et al. GluN2B-Containing NMDA Receptors Regulate AMPA Receptor Traffic through Anchoring of the Synaptic Proteasome. J. Neurosci. Off. J. Soc. Neurosci 35, 8462–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE & Bading H Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat. Rev. Neurosci 11, 682–696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle F, Dehmel M, Leemhuis J, Fischer C & Meyer D Role of GluN2A and GluN2B subunits in the formation of filopodia and secondary dendrites in cultured hippocampal neurons. Naunyn. Schmiedebergs Arch. Pharmacol 385, 171–180 (2012). [DOI] [PubMed] [Google Scholar]
- Jacobs S, Cheng C & Doering LC Hippocampal neuronal subtypes develop abnormal dendritic arbors in the presence of Fragile X astrocytes. Neuroscience 324, 202–217 (2016). [DOI] [PubMed] [Google Scholar]
- Jakubowska-Dogru E, Elibol B, Dursun I, & Yuruker S Effects of prenatal binge-like ethanol exposure and maternal stress on postnatal morphological development of hippocampal neurons in rats. International Journal of Developmental Neuroscience 61, 4050 (2017). [DOI] [PubMed] [Google Scholar]
- Jan Y-N & Jan LY Dendrites. Genes Dev 15, 2627–2641 (2001). [DOI] [PubMed] [Google Scholar]
- Juraska JM, Fitch JM & Washburne DL The dendritic morphology of pyramidal neurons in the rat hippocampal CA3 area. II. Effects of gender and the environment. Brain Res 479, 115–119 (1989). [DOI] [PubMed] [Google Scholar]
- Kannangara TS et al. Deletion of the NMDA Receptor GluN2A Subunit Significantly Decreases Dendritic Growth in Maturing Dentate Granule Neurons. PLOS ONE 9, e103155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konur S & Ghosh A Calcium Signaling and the Control of Dendritic Development. Neuron 46, 401–405 (2005). [DOI] [PubMed] [Google Scholar]
- Krichmar JL, Nasuto SJ, Scorcioni R, Washington SD, & Ascoli GA (2002). Effects of dendritic morphology on CA3 pyramidal cell electrophysiology: a simulation study. Brain Research, 941(1), 11–28. [DOI] [PubMed] [Google Scholar]
- Krichmar JL, Velasquez D, & Ascoli GA (2006). Effects of β-Catenin on Dendritic Morphology and Simulated Firing Patterns in Cultured Hippocampal Neurons. The Biological Bulletin, 211(1), 31–43. [DOI] [PubMed] [Google Scholar]
- Li L-J et al. Glycine Potentiates AMPA Receptor Function through Metabotropic Activation of GluN2A-Containing NMDA Receptors. Front. Mol. Neurosci 9, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian X, Hartley DM & Feig LA Distinct Roles for Ras-Guanine NucleotideReleasing Factor 1 (Ras-GRF1) and Ras-GRF2 in the Induction of Long-Term Potentiation and Long-Term Depression. J. Neurosci 26, 1721–1729 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C & Malenka RC NMDA Receptor-Dependent Long-Term Potentiation and LongTerm Depression (LTP/LTD). Cold Spring Harb. Perspect. Biol 4, a005710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki BA, Aman TK, Amico-Ruvio SA, Kussius CL & Popescu GK C-terminal Domains of N-Methyl-d-aspartic Acid Receptor Modulate Unitary Channel Conductance and Gating. J. Biol. Chem 287, 36071–36080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC & Bear MF LTP and LTD: an embarrassment of riches. Neuron 44, 5–21 (2004). [DOI] [PubMed] [Google Scholar]
- Markham JA, Mckian KP, Stroup TS & Juraska JM Sexually dimorphic aging of dendritic morphology in CA1 of hippocampus. Hippocampus 15, 97–103 (2005). [DOI] [PubMed] [Google Scholar]
- Martínez-Cerdeño V Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev. Neurobiol 77, 393–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Wang J, Kandel ER & O’Dell TJ CaMKII regulates the frequency-response function of hippocampal synapses for the production of both LTD and LTP. Cell 81, 891–904 (1995). [DOI] [PubMed] [Google Scholar]
- McEwen BS Glucocorticoids, depression, and mood disorders: structural remodeling in the brain. Metabolism 54, 20–23 (2005). [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B & Seeburg PH Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12, 529–540 (1994). [DOI] [PubMed] [Google Scholar]
- Nabavi S et al. Metabotropic NMDA receptor function is required for NMDA receptordependent long-term depression. Proc. Natl. Acad. Sci. U. S. A 110, 4027–4032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachernegg S, Strutz-Seebohm N & Hollmann M GluN3 subunit-containing NMDA receptors: not just one-trick ponies. Trends Neurosci 35, 240–249 (2012). [DOI] [PubMed] [Google Scholar]
- Sanders EM et al. Developmental Modification of Synaptic NMDAR Composition and Maturation of Glutamatergic Synapses: Matching Postsynaptic Slots with Receptor Pegs. Biol. Bull 224, 1–13 (2013). [DOI] [PubMed] [Google Scholar]
- Sanders EM et al. (2018) Separate functional properties of NMDARs regulate distinct aspects of spatial cognition. Learning and Memory 25: 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Clemente A, Nicoll RA & Roche KW Diversity in NMDA receptor composition: many regulators, many consequences. Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry 19, 62–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipton OA & Paulsen O GluN2A and GluN2B subunit-containing NMDA receptors in hippocampal plasticity. Philos. Trans. R. Soc. Lond. B. Biol. Sci 369, 20130163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein IS, Gray JA & Zito K Non-Ionotropic NMDA Receptor Signaling Drives Activity-Induced Dendritic Spine Shrinkage. J. Neurosci. Off. J. Soc. Neurosci 35, 12303–12308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida I Morphology of Hippocampal Neurons. in Hippocampal Microcircuits 27–67 (Springer, New York, NY, 2010). [Google Scholar]
- Vissel B, Krupp JJ, Heinemann SF & Westbrook GL A use-dependent tyrosine dephosphorylation of NMDA receptors is independent of ion flux. Nat. Neurosci 4, 587–596 (2001). [DOI] [PubMed] [Google Scholar]
- Vyas A, Mitra R, Rao BSS & Chattarji S Chronic Stress Induces Contrasting Patterns of Dendritic Remodeling in Hippocampal and Amygdaloid Neurons. J. Neurosci 22, 6810–6818 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M et al. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature 385, 439–442 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets that were generated and analyzed for the current study are available from the corresponding author upon request.


