Abstract
The development and widespread use of vaccines, defined by the World Health Organization (WHO) as, “biological preparations that improve immunity to a particular disease”, represents one of the most significant strides in medicine. Vaccination was first applied to reduce mortality and morbidity from infectious diseases. The WHO estimates that vaccines prevent 2–3 million human deaths annually, and these numbers would rise by at least 6 million if all children received the recommended vaccination schedule.1 However, the origins of allergen immunotherapy shared the same intellectual paradigm and subsequent innovations in vaccine technology have been applied beyond the prevention of infection, including in the treatment of cancer and allergic diseases. This review will focus on how new, more rational approaches to vaccine development utilize novel biotechnology, target new mechanisms, and shape the immune system response with an emphasis on discoveries that have direct translational relevance to the treatment of allergic diseases.
Keywords: vaccines, immunotherapy, antibodies, sequencing
History of vaccine development
Early advances in vaccinology led to ground-breaking discoveries, from the role of antibodies in the adaptive immune system, to the development of allergen specific immunotherapy (ASIT) for the treatment of allergies. In the 18th century Edward Jenner observed that milkmaids previously infected by cowpox, a zoonotic disease transmitted from cows to humans, were protected from smallpox. This observation led to the first known clinical vaccine trial conducted with cowpox in 1796.2 This same idea, that inoculation with an offending substance may provide future protection against disease, was applied by Noon and Freeman in 1911 to hay fever,3, 4 giving rise to ASIT, which is still widely employed today in the treatment of a variety of allergic diseases.
Attenuated vaccination, discovered by Louis Pasteur when he vaccinated farm animals with attenuated B.antracis in 1881,5 used a slightly modified and less virulent form of the microbe for vaccination. This discovery was followed by a human trial of attenuated rabies virus vaccine in a young boy in 1885.6 The discovery that ‘antitoxins’ in the sera of vaccinated individuals were responsible for protection led to the discovery of antibodies and their role in clinical protection.7 Clinical trials demonstrated that the protective power of immunoglobulins extended from bacterial to viral agents.8
The next generation of vaccines aimed to elicit protection from multiple strains of the infectious agents. Albert Sabin demonstrated protection using 3 attenuated strains of poliovirus,9 while Jonas Salk found that fully inactivated poliovirus from 3 strains could also induce protection.10 Both types of polio vaccines, the oral polio vaccine using 3 attenuated strains approved in 1961 and the trivalent inactivated polio vaccine approved in 1955, are still used today.
The explosion of molecular genetics and recombinant DNA technology led to another revolutionary step in vaccines – that of using specific antigens from infectious agents to administer protective benefit more safely. The previous efforts in vaccinology had required the culturing of infectious agents to produce vaccine candidates, but by using recombinant DNA, subunit vaccines could be manufactured for infectious agents, which were otherwise challenging to culture or highly pathogenic.
Most clinical immunotherapy in use today for treating allergies is technologically where vaccines for infectious agents were decades ago: using whole allergen extracts. The first innovations in allergen immunotherapy have been to apply the concept of subunit vaccines by using the individual dominant allergens that are the known targets of the allergic (IgE) response. Several component vaccines have now been studied in clinical trials for environmental11 as well as peanut12 allergies, and these have been thoroughly reviewed by others.13, 14
Reverse vaccinology
Reverse vaccinology is a new approach to designing vaccines, which combines our rapidly increasing feasibility to sequence whole genomes of microorganisms and apply bioinformatic analyses to those data. Predictive modeling identifies new pathogen targets that are ideally conserved and targets of protective responses. The subsequent expression of candidate targets for screening using human serum from those with effective immunity and for evaluation in murine models can lead to the design of optimal vaccines, particularly to bacterial pathogens.
For example, Neissera meningococcus serotype B causes quickly progressive meningitis with high mortality. Unlike the other strains of meningococcus, its capsular polysaccharide is sialylated like human glycoproteins, interfering with the ability to create a typical conjugate vaccine. Genomic sequencing of meningococcus serotype B led to the identification of 90 new surface antigens, of which about 30% bound to serum antibodies from immune patients.15, 16 Subsequent murine models using selected antigens demonstrated effective protection, and ultimately these antigens contributed to the development of the currently available meningococcus B subunit vaccine.17 This antigenome analysis – the interrogation of the antigenic repertoire of a pathogen using libraries of recombinantly expressed antigens screened with serum antibodies of infected patients and then subsequently evaluated by model organism vaccination experiments – has also been used to identify novel antigens from S. pneumonia.18 Clinical vaccine trials using 3 of these proteins, PhtD, PcpA and Ply, are currently underway.19 A variation on this approach using computational analysis of the binding site of neutralizing antibodies to RSV led to the formulation of a recombinant immunogen, which when combined with an adjuvant, demonstrates protection in animal models.20
As a parallel in allergic diseases, genomic characterization of many major allergens is also underway. The use of reverse vaccinology in the context of allergic disease has already led to novel allergen identification. Der f 24, an ubiquinol-cytochrome c reductase homologue from house dust mite, was found after the Dermatophagoides farine genome was sequenced by high throughput sequencing, followed by identification of predicted genes, expression of selected genes, and validation using immunoblotting, ELISA, and skin testing in allergic donors.21 Further engineered immunotherapy approaches based on novel allergen discoveries may provide a more effective method of inducing more complete protective responses in allergic individuals.
Lessons from HIV vaccinology approaches
Many of the most recent innovations in vaccine immunology have been developed for the treatment and prevention of human immunodeficiency virus type 1 (HIV-1) infection. We therefore focus on this particular pathogen to illustrate new vaccine approaches that target adaptive immunity.
Targeting humoral immunity
As alluded to earlier, antibody-based immunity has been the backbone of most vaccine- mediated protection, and the induction of protective humoral responses has been characterized as the “holy grail” of HIV vaccine research.22 Protective antibodies against HIV have been classified into 2 major types: neutralizing (NAbs) and non-neutralizing antibodies (non-NAbs). Neutralizing antibodies can prevent the infection of target cells by binding to HIV-1 virion envelope glycoproteins (Env). Env-specific non-NAbs can recognize Env expressed on HIV-1- infected cells and contribute antiviral activity through Fc effector functions by inducing antibody-dependent cellular cytotoxicity (ADCC) through Fc-gamma receptor expressing cells such as NK cells.
By comparison, one goal of allergen immunotherapy has been to develop a long-lasting non-IgE antibody response capable of suppressing effective engagement and cross-linking of allergen-specific IgE on effector cells and there may be distinct functional activities of clones within the poly-clonal allergen-specific response.23 By analogy to HIV-specific antibodies, neutralizing antibodies that bind specific epitopes through their Fab, are similar to allergen-specific blocking antibodies,23 while non-neutralizing antibodies are similar to the allergen-specific antibodies that can activate inhibitory receptors such as CD3224 through their Fc region. The relative contributions of these functional antibody attributes to tolerance despite sensitization in the context of allergic diseases are still unknown and may vary between people or specific allergies.
Though administration of allergens as vaccines does not pose an infectious risk, it does pose the risk of triggering an allergic reaction. To mitigate that risk, hypoallergenic vaccines, or those that do not have IgE binding epitopes, have been devised. Many clinical trials using hypoallergenic recombinant protein vaccines are currently underway (Table 1), including for birch pollen allergy (Bet v 1)25 and grass pollen allergy (Phl p 1, Phl p 2, Phl p 5a and b, and Phl p 6).26 It is worth noting that even when IgE-dependent activation is bypassed, clinical studies with these hypoallergenic vaccines have shown that while they do not trigger immediate hypersensitivity reactions, late phase reactions do still occur, possibly related to T-cell mediated effects.27, 28
Table 1:
Clinical trials with novel allergy vaccine products using recombinant and modified allergens.
| Year | Molecule | Description | Clinical trial phase |
|---|---|---|---|
| 1996 | Allervax CAT | Two Fel d 1 peptides | DBPC |
| 2000 | Bet v 1 trimer, fragments | Hypoallergenic Bet v 1 proteins | 2 |
| 2002 | Recombinant grass pollen | Recombinant Phl p 1, Phl p 2, Phl p 5a+b, Phl p 6 | 3 |
| 2002 | Folding variant of Bet v 1 | Hypoallergenic Bet v 1 | 3 |
| 2002 | Recombinant Bet v 1 | Recombinant Bet v 1 | 2 |
| 2006 | Recombinant Bet v 1 (tablets) | Recombinant Bet v 1 (sublingual) | 2 |
| 2009 | E.coli encapsulated recombinant modified Ara h 1, Ara h 2, Ara h 3 | Rectal delivery of vaccine | 1 |
| 2011 | Fcγ1-Fel d 1 fusion protein | Intradermal delivery of fusion protein | Safety |
| 2012 | BM32 | Four hypoallergenic grass allergens | 2 |
| 2012 | ToleroMune Cat | Fel d 1 synthetic peptides | 3 |
| 2012 | AllerT | Bet v 1 peptides | 2 |
| 2013 | FAST-Fish | Mutated parvalbumin | 1/2 |
| 2014 | ToleroMune Grass | Peptides from grass pollen | 2 |
| 2014 | ToleroMune HDM | Peptides from house dust mite | 2 |
| 2014 | ToleroMune Ragweed | Peptides from Amb a 1 | 2 |
| 2015 | ASP4070 | Japanese cedar pollen (Cry j 1, j 2) LAMP based DNA plasmid vaccine | 1 |
| 2016 | ASP0892 | Peanut (Ara h 1, h 2, h 3) LAMP based DNA plasmid vaccine | 1 |
Another strategy to avoid triggering allergic reactions while still preserving T-cell mediated immune protection is immunotherapy using selected peptides derived from allergens. A variety of techniques have been used to define peptides having specific characteristics, such as the induction of blocking antibodies29 or stimulation of T cells30, without the consequence of IgE crosslinking. Several products for treatment of environmental allergies, including cat, dust mite, grass, and ragweed allergy have been studied in clinical trials (Table 1).
The underlying mechanism of efficacy and relative importance of T cells versus antibodies in these engineered recombinant protein and peptide vaccines is still unclear.31 Induction of T cell anergy or deletion of pathogenic Th2 cells may play a dominant role in the clinical efficacy of T cell peptide vaccines. Another mechanism of vaccines lacking IgE epitopes – whether protein or peptide based – may be to increase the pool of allergen-specific T cells that can provide B cell help upon subsequent complete antigenic re-stimulation, to then produce IgE-blocking antibodies. Alternatively in some cases, hypoallergenic vaccines may be able to directly stimulate the induction of allergen-specific blocking antibodies recognizing novel epitopes with the capacity to suppress allergen effector cells through inhibitory receptors such as CD32.24 Elucidation of the underlying immunological mechanisms in engineered vaccine approaches may provide additional insights for improving clinical outcomes after therapy.
Innovative techniques for the study of antigen-specific antibodies
Technological advancements for the study of humoral immunity now allow us to directly study these mechanisms of allergen immunotherapy, both in conventional and engineered allergen immunotherapies. One example of those advances has been the affinity-based labeling of antigen-specific B cells for analysis at the single-cell level, which was pioneered in the context of infectious diseases such as influenza,32,33 HIV,34,35, and dengue,36, and applied more recently in food allergy37. The ability to detect and isolate allergen-specific B cells with high specificity in peanut allergy has provided new insights into the development of allergen-specific repertoires by the use of single-cell sequencing technologies37, 38 and these advances may hold significant promise for the future.
As an example, the isolation of HIV-specific single B cells provided the foundation that has led to the development of a diverse set of new vaccine approaches in that field. Single cell immunoglobulin sequencing of paired heavy and light chains from HIV-specific B cells followed by recombinant antibody production has allowed for further characterization of antigen-specific antibody responses, including antigen binding sites, structural and functional characterization. These tools have also led to the development of therapeutic approaches, including the infusion of neutralizing antibodies for the prevention and treatment of infection, as well as the development of bi-specific antibody therapeutics (see below). Finally, these advances, including characterization of individual antibodies, are significantly complemented by next generation sequencing of the immunoglobulin heavy chain repertoire from circulating or tissue resident B cells. The resulting deep lineage analysis of antibodies over time (e.g., pre/post immunization or infection) has provided a richer understanding of antibody development in disease. Therefore, the development of techniques to study antigen-specific antibodies has led to the discovery of broadly neutralizing antibodies and the subsequent invention of newer treatment modalities in HIV.
Neutralizing antibodies as therapeutics
Identified, isolated, and cloned from single B-cells from HIV-infected individuals, neutralizing antibodies that recognize a broad set of antigens (bNAbs) bind to sites on the viral envelope (Env) that mediate viral binding (Table 2). Passive administration of bNAbs can prevent simian-human immunodeficiency virus infection in non-human primates.39-47 The first human clinical trial HVTN 703 to evaluate the effectiveness of a bNAb VRC01, which was discovered in an elite viral controller and binds CD4bs, to prevent HIV-1 transmission is currently ongoing after demonstrating acceptable human safety.48
Table 2:
HIV-specific antibodies, including both broadly neutralizing (bNAbs) and non-neutralizing antibodies (non-NAbs), in clinical trials.
Early antigen-specific B cell work found that these bNAbs have unusual structural characteristics, including long and hydrophobic heavy chain complementarity determining region 3 (CDR3) loops, a high degree of somatic hypermutation, and autoreactivity – suggesting a strong role for affinity maturation and involvement of immune tolerance mechanisms.49-52 In as many as 50% of infected individuals53, these antibodies arise after 2-4 years of HIV-1 infection54, as the HIV-1 virus mutates within infected individuals, encouraging the development of bNAbs by the immune system.55-57 This continual evolution of host antibodies and HIV proteins, can lead to finely-tuned bNAbs, with long, anionic CDR3 regions that target conserved regions under the mutable Env glycan shield.58 Hence, the unusual structural characteristics of bNAbs can contribute to their functional neutralization ability, given the heavy glycosylation and mutability of Env, which can allow it to escape from immune pressures.
Clinical trials examining the efficacy of passive infusion of monoclonal bNAbs as therapeutic treatment over the past few years has repeatedly demonstrated transient viral suppression at the highest administered bNAb doses, with subsequent resistant HIV-1 viral rebound. Trials have utilized VRC0148, 59, 60 and 3BNC117 targeting CD4bs,61-63 and 10-1037 mAb targeting the high-mannose patch.64 Pre-selection of subjects with susceptible HIV-1 infections did demonstrate a longer period of viral suppression of up to 9.9 weeks with 4 infusions of the monoclonal.62 Despite the observation of bNAb induced viral suppression, the rebound of resistant virus highlights the co-evolution of the host-microbe responses and has prompted the consideration of combination bNAb therapy. Further work on using combination antibody therapy in passive immunization may hold promise.
The correlate of HIV neutralizing antibodies in allergy are allergen-specific blocking antibodies, which also emerge after chronic antigenic stimulation of the adaptive immune system. In allergen immunotherapy, the correlation of blocking antibodies to clinical efficacy of immunotherapy highlights the central importance of these blocking antibodies.65 In vitro, immunotherapy-induced blocking antibodies have been shown to effectively suppress allergen effector cells in allergic rhinitis, asthma, and food allergy. Infusion of allergen-specific IgG antibodies have been protective in animal models of food allergy66 and humanized mice models of asthma.67 Recently, a proof-of-mechanism phase I clinical trial with 2 Fel d 1-specific blocking monoclonal IgG antibodies infused in cat allergic patients successfully decreased nasal provocation scores,68 validating passive immunization with allergen blocking antibodies as a treatment approach for allergic diseases.
Induction of bNAbs: sequential immunization
The very structural characteristics discussed above, such as long hydrophobic heavy chain CDR3 regions and high rates of somatic mutations, thought to enhance the neutralization abilities of bNAbs, also tend to be selected against during clonal evolution and B cell maturation, posing a significant barrier to trying to intentionally induce them as a therapeutic approach. Using systems biology-based ‘antibody-omics’ approaches described further below, the presence of unique B cell lineages that foster the development of bNAbs have been identified and targeted.57
There are also additional limitations due to differences in human and model organism germline V-gene segments, and researchers have developed new murine models that are proving to be critical tools for next generation vaccine design. As an example, the human VH1-02 heavy chain variable segment, which is part of the VRC01 CD4bs bNAb, does not occur in mice, and even the closest primate ortholog does not include all of the amino acids that define a crucial motif for VRC01-like antibodies.68 The use of humanized murine models, such as BLT (human bone marrow-liver-thymus) transplanted model on a Rag2−/−gamma-chain−/−/CD47−/− background, has been limited by their poor germinal center reactions, which are central to antibody development. In addition, wild type mice have short D segments, incapable of producing long H-CDR2 regions, as well as poorly diversified and expressed lambda regions. However, all of these differences can be overcome by the knock-in of specific immunoglobulin heavy and/or light chain arrangements, known as KI models. These models have provided an opportunity to better delineate the mechanism of bNAb development and present a tool for rational vaccine design by sequential immunization.
Using this approach, B cells expressing bNAb precursors are primed by an antigen resulting in memory B cells. Sequential immunization with antigens progressively more similar to the native Env trimer is then used to reactivate antigen-specific memory B cells to undergo further affinity maturation until bNAbs are expressed. Immunization studies with the VRC01gH KI model have been used to develop and test the ability of a specific priming immunogen, eOD-GT8 to elicit memory B cells with VRC01-like antibodies.69, 70 The elicitation of CD4bs-specific antibodies after subsequent immunization with a more native-like gp120 protein, BG505coreGT371 demonstrates the ability of engineered sequential immunogens to shape a human-like B cell repertoire to favor protective antibody production. The ability to optimize the shape of the BCR repertoire represents a new, powerful approach to harnessing the adaptive immune response and has broader implications for interventional treatments for allergic diseases. Sequential immunization with hypoallergenic recombinant allergens to stimulate the development of blocking antibody clones may be a fruitful approach provide an idea for the next generation of recombinant allergen strategies.
The main critique of this strategy is that BCR repertoires in general have been found to be highly individual.72 However, more recently, stereotyped antigen-specific antibodies in the repertoires of unrelated individuals infected by dengue have been identified.73 We have also reported allergen-specific stereotyped antibodies from patients undergoing peanut oral immunotherapy37. Antigen-specific antibodies that are similar in their amino acid sequences imply convergent evolution, or shared pathways, to the development of antigen-specific BCRs in unrelated individuals may be important. Interventions, using either epitopes or unrelated proteins, aimed at fostering the development of protective convergent antibodies that provide long term efficacy after immunotherapy may provide a new therapeutic approach.
Systems approach to antibody-omics
Another new approach to systematically identify the most clinically relevant aspects of the dynamic and polyclonal adaptive immune responses–recognizing the diversity of epitope recognition, somatic antibody variants, Fc domain subclass and glycosylation differences, T cell receptors and HLA alleles–relies on integration of many measures of the humoral response. While our advances in single-cell techniques for BCR analysis and antibody characterization have substantially advanced the field, in vivo, antibodies do not work in isolation but rather as a dynamic swarm of molecules, with Fc and Fab regions that have multiple variations.
Using systems immunology, novel clinically relevant mechanisms of vaccine efficacy in HIV have been identified. The RV144 clinical trial, using a priming step with a canarypox vector ALVAC-HIV (vCP1521) expressing Env, gag and pro followed by protein boosts with AIDSVAX® gp120 and alum adjuvant, resulted in 60.5% vaccine efficacy at 1 year and 31.2% efficacy at 3.5 years.74 Prior work suggested that vaccine induction of neutralizing antibodies may best correlate with vaccine efficacy. Surprisingly, the sera from individuals vaccinated in the RV144 trials did not have neutralizing activity, though higher anti-gp120 antibody levels were correlated with vaccine efficacy. Subsequent work using a systems immunology approach suggested that vaccine protection correlated to FcR mediated ADCC activity, likely due to non-neutralizing antibodies.75 The unexpected but central importance of FcR activity as a marker of clinical efficacy was discovered after an exhaustive humoral characterization, demonstrating the power of a systematic approach to finding novel mechanisms contributing to vaccine efficacy. In the future, a similar systems immunology approach in allergen immunotherapy, measuring the various aspects of antibody responses such as quantity, diversity, affinity, Fc variants, and glycosylation, may ultimately identify unexpected and new mechanisms of clinical efficacy in allergen immunotherapy.
Targeting cellular immunity: DC LAMP vaccines
HIV-specific T cells play an important role in both the acute and chronic phases of HIV infection. In the acute phase, activated CD8+ T cells are necessary for control of viremia, and HIV-specific CD4 T cells play a crucial role in sustaining an effective cytotoxic T cell response.76, 77 The central role of CD4+ T cells in the control of HIV infection has become clear in the last few years. The low viral load in HIV-infected long-term non-progressors was correlated to a higher Gag-specific polyfunctional CD4+ T cell response.78 In non-treated HIV infected individuals, the lower viral load correlated to an expanded HIV-specific CD4+ T cell population that has enhanced cytolytic activity and IFN-gamma production.79
Dendritic cells (DCs) provide a unique bridge between innate and adaptive immunity in their ability to effectively acquire antigens for processing and presentation to T cells. Dendritic cells, in their capacity as professional antigen-presenting cells, can therefore promote a beneficial CD4+ T cell response, if candidate vaccines contain conserved HIV-1 desirable epitopes recognized by CD4+ T cells and target them to DCs using DC-specific endocytic receptors to boost immunogenicity.
Recombinant DNA vaccines, such as those involving injection of plasmids with DNA encoding sequences, are relatively inexpensive and easy to produce. However, these efforts have been hampered by their limited immunogenicity due to low levels of expression.80 One effort to overcome this hurdle is by more effectively targeting the DNA vaccines to the most immunogenic cellular compartment, by targeting vaccine delivery to the endocytic compartment of DCs.
Several strategies to produce DC-targeted vaccines have been considered in the HIV field. Taking advantage of the highly expressed DEC205 endocytic receptor on CD11c+CD8alpha+ DCs in lymph nodes, antigens linked to a DEC205 monoclonal antibody are efficiently internalized and processed by DCs, presented to both CD4 and CD8 T cells, and result in strong cellular and humoral responses.81 This technique has been tried in a murine model of ovalbumin-induced asthma.82 Nanoparticles encapsulating antigens targeted to DCs by DEC205 also elicit strong antigen-specific T cell responses.83
Another method uses lysosomal-associated membrane protein 1 (LAMP-1) to target antigen expression again to the endocytic vesicles of dendritic cells to enhance antigen presentation and immunogenicity.84 This approach has been used in vaccines to promote protective immune responses against viruses,85, 86 but also in allergy.87, 88
In the context of allergic diseases, a LAMP1-based recombinant DNA vaccine has been studied in Phase 1 trials for the treatment of Japanese cedar pollen allergy and peanut allergy. A phase I trial of CryJ2-LAMP plasmid vaccine for Japanese cedar pollen allergy found that the vaccine was well-tolerated with a high rate of conversion of skin testing from positive to negative in allergic subjects.89 The Phase I interventional clinical trial in peanut allergic subjects with ASP0892, which is a LAMP-based recombinant DNA vaccine with Ara h 1, Ara h 2, and Ara h 3, is currently ongoing (NCT02851277).
Arming with cellular and antibody immunity: Using bispecific molecules
In the prevention and treatment of HIV infection, our ability to devise bispecific molecules, engineered to have 2 antigen-binding sites, as another novel therapeutic vaccine approach has been dependent on the technological innovations to isolate, clone, and characterize antigen-specific B cells. For prevention of HIV binding to host cells, bispecific antibodies, such as iMabm3690 and 10E8v2.0/iMab,91 have one arm specific for human CD4 (iMab) blocking the gp120 binding site and the other specific HIV-1 gp 120 (m36 or 10E8v2.0). Viral load reduction and protection from viral challenge after 10E8v2.0/iMab treatment occurred in humanized mouse models.91
To target latently infected cells on viral reactivation, bispecific molecules with a CD3- specific single-chain variable fragment (scFv) linked by a polypeptide to the scFv of an HIV-1 specific antibody such as VRC01 (VRC01-antiCD3), could recruit cytotoxic CD3 T cells. Treatment with VRC01-antiCD3 of in vitro peripheral blood cells from HIV-1 infected subjects reduced the frequency of proviral DNA positive CD4+ T cells.92 Another molecule, a dual-affinity retargeting (DART) molecule with both a CD3-specific arm and HIV-specific arm (CD4-inducible constant regions 1 and 2 and gp41 cluster 1 non-NAbs A32 and 7B2), similarly demonstrated killing of patient-derived HIV-1 infected cells cultured with a latency-reversing drug.93 These results have also been seen with several other DARTs where the HIV-1 specific arm targeted different bNAbs.94 Combinatory treatment with DARTs of different HIV-1 specificities may be possible and especially powerful given their ability to redirect normal resting cytotoxic T cells.
There have been several novel bi-specific molecules under investigation for treatment of allergic diseases, though the development of allergen-specific bi-specific molecules is just emerging. The combination of a recombinant allergen, Phl p 1, and intracellular adhesion molecule-1 (ICAM-1), termed P2/ICAM, has been shown to inhibit in vitro allergen migration through the respiratory layer.95 More recently, a Siglec-engaging tolerance-induced antigenic lipsome (STAL) that target CD22 on the surface of B cells was combined with the major peanut allergen Ara h 2 to target Ara h 2 specific B cells and prevent sensitization in mice.96 The presumed deletion of Ara h 2 specific B cells diminished the allergic response not only to Ara h 2 but also to peanut. This targeting of allergen-specific B cells for the modulation of allergic responses provides a promising new target.
Conclusions and future perspectives
Historically, translation of clinical observations, the scientific tools at our disposal, and our understanding of immunology has guided vaccine development. Our ability to engineer molecules to target immune subsets, use rational design to develop immunogens to shape protective antibody responses, and combine these two strategies to redirect allergic responses illustrate the next generation of vaccine design. Future clinical implementation of these approaches will rely on our nuanced appreciation of the dynamic evolution of immune tolerance in allergic disease. Innovations in vaccine design for infectious disease may continue to hold important implications for the future of therapeutics for allergic diseases as well.
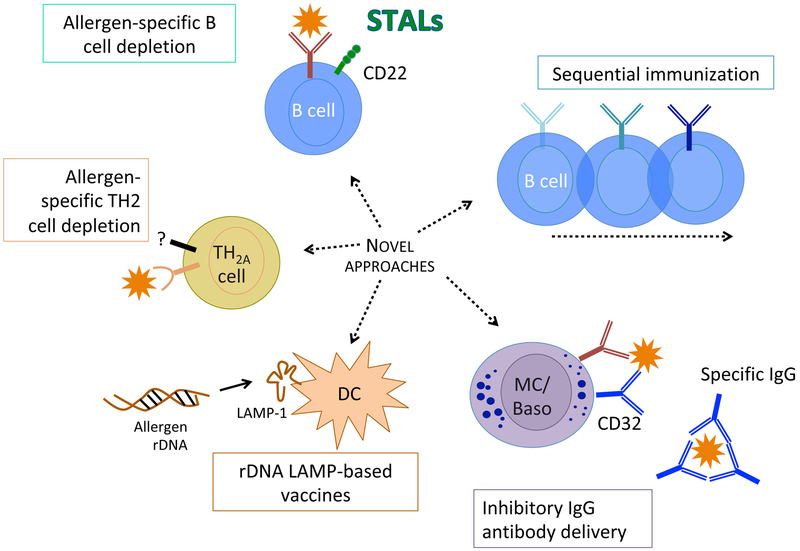
Figure 1: Translational mechanisms for future innovations in immunotherapy for allergies.
Novel approaches for allergen specific immunotherapy include bispecific molecules targeting antigen-specific B cells or allergen-specific Th2 cells, DC-targeting vaccines, sequential immunization strategies to elicit enhanced blocking antibodies, and passive immunization with inhibitory IgG antibodies
Acknowledgments
Funding:
Sarita Patil declares research funding from National Institutes of Health, National Institute of Allergy and Infectious Diseases, American Academy of Asthma, Allergy and Immunology, Food Allergy Research and Education. Clinical trials with Astellas, DBV, Aimmune, and Sanofi. Royalties from UptoDate.
Wayne Shreffler declares research funding from National Institutes of Health, National Institute of Allergy and Infectious Diseases, Food Allergy Science Initiative, Gerber, Sanofi. Clinical trials with Astellas, DBV, Aimmune, and Sanofi. Royalties from UptoDate
Abbreviations:
- ASIT
allergen specific immunotherapy
- HIV-1
human immunodeficiency virus type 1 B cell receptor (BCR)
- NAbs
neutralizing antibodies
- non-Nabs
non-neutralizing antibodies
- Env
HIV-1 virion envelope glycoproteins
- ADCC
antibody-dependent cellular cytotoxicity
- CDR3
immunoglobulin heavy chain complementarity determining region 3 loops
- scFv
single-chain variable fragment
Footnotes
Declaration of Conflict of Interest
This work did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andre FE BR, Bock HL, Clemens J, Datta SK, John TJ, Lee BW, Lolekha S, Peltola H, Ruff TA, Santosham M, Schmitt HJ. Vaccination greatly reduces disease, disability, death and inequity worldwide. Bulletin of the World Health Organization 2008:140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenner E An inquiry into the causes and effects of the variolae vaccinae, a disease discovered in some of the western counties of England, particularly Gloucestershire, and known by the name of the cow pox. London,: Law; 1798. [Google Scholar]
- 3.Noon L PROPHYLACTIC INOCULATION AGAINST HAY FEVER. The Lancet 1911; 177:1572–3. [Google Scholar]
- 4.Freeman J FURTHER OBSERVATIONS ON THE TREATMENT OF HAY FEVER BY HYPODERMIC INOCULATIONS OF POLLEN VACCINE. The Lancet 1911; 178:814–7. [PubMed] [Google Scholar]
- 5.Pasteur L, Chamberland, Roux. Summary report of the experiments conducted at Pouilly-le-Fort, near Melun, on the anthrax vaccination, 1881. Yale J Biol Med 2002; 75:59–62.2588695. [PMC free article] [PubMed] [Google Scholar]
- 6.Pasteur L Méthode pour prévenir la rage après morsure. CRAcadSci 1885; 101:765–72. [Google Scholar]
- 7.Ev Behring. Ueber das Zustandekommen der Diphtherie-Immunität und der Tetanus-Immunitat bei Thieren. 1890. [Google Scholar]
- 8.Hammon WM, Coriell LL, Wehrle PF, Stokes J Jr. Evaluation of Red Cross gamma globulin as a prophylactic agent for poliomyelitis. IV. Final report of results based on clinical diagnoses. J Am Med Assoc 1953; 151:1272–85. [PubMed] [Google Scholar]
- 9.Sabin AB, Hennessen WA, Winsser J. Studies on variants of poliomyelitis virus. I. Experimental segregation and properties of avirulent variants of three immunologic types. J Exp Med 1954; 99:551–76.2136347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salk JE, Krech U, Youngner JS, Bennett BL, Lewis LJ, Bazeley PL. Formaldehyde treatment and safety testing of experimental poliomyelitis vaccines. Am J Public Health Nations Health 1954; 44:563–70.1620937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curin M, Garib V, Valenta R. Single recombinant and purified major allergens and peptides: How they are made and how they change allergy diagnosis and treatment. Ann Allergy Asthma Immunol 2017; 119:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood RA, Sicherer SH, Burks AW, Grishin A, Henning AK, Lindblad R, et al. A phase 1 study of heat/phenol-killed, E. coli-encapsulated, recombinant modified peanut proteins Ara h 1, Ara h 2, and Ara h 3 (EMP-123) for the treatment of peanut allergy. Allergy 2013; 68:803–8.3663889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook QS, Burks AW. Peptide and Recombinant Allergen Vaccines for Food Allergy. Clin Rev Allergy Immunol 2018. [DOI] [PubMed] [Google Scholar]
- 14.Valenta R, Niespodziana K, Focke-Tejkl M, Marth K, Huber H, Neubauer A, et al. Recombinant allergens: what does the future hold? J Allergy Clin Immunol 2011; 127:860–4. [DOI] [PubMed] [Google Scholar]
- 15.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 2000; 287:1816–20. [DOI] [PubMed] [Google Scholar]
- 16.Litt DJ, Savino S, Beddek A, Comanducci M, Sandiford C, Stevens J, et al. Putative vaccine antigens from Neisseria meningitidis recognized by serum antibodies of young children convalescing after meningococcal disease. J Infect Dis 2004; 190:1488–97. [DOI] [PubMed] [Google Scholar]
- 17.Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A 2006; 103:10834–9.2047628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giefing C, Meinke AL, Hanner M, Henics T, Bui MD, Gelbmann D, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med 2008; 205:117–31. 2234372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pichichero ME, Khan MN, Xu Q. Next generation protein based Streptococcus pneumoniae vaccines. Hum Vaccin Immunother 2016; 12:194–205. 4962723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossey I, Gilman MS, Kabeche SC, Sedeyn K, Wrapp D, Kanekiyo M, et al. Potent singledomain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat Commun 2017; 8:14158. 5316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW, Li RQ, et al. The draft genome, transcriptome, and microbiome of Dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J Allergy Clin Immunol 2015; 135:539–48. [DOI] [PubMed] [Google Scholar]
- 22.Ferrari G, Pollara J, Tomaras GD, Haynes BF. Humoral and Innate Antiviral Immunity as Tools to Clear Persistent HIV Infection. J Infect Dis 2017; 215:S152–S9. 5410976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James LK, Shamji MH, Walker SM, Wilson DR, Wachholz PA, Francis JN, et al. Long-term tolerance after allergen immunotherapy is accompanied by selective persistence of blocking antibodies. J Allergy Clin Immunol 2011; 127:509–16. [DOI] [PubMed] [Google Scholar]
- 24.Burton OT, Logsdon SL, Zhou JS, Medina-Tamayo J, Abdel-Gadir A, Noval Rivas M, et al. Oral immunotherapy induces IgG antibodies that act through FcgammaRIIb to suppress IgE-mediated hypersensitivity. J Allergy Clin Immunol 2014; 134:1310–7 e6.4261076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer W, Narkus A, Salapatek AM, Hafner D. Double-blind, placebo-controlled, dose-ranging study of new recombinant hypoallergenic Bet v 1 in an environmental exposure chamber. Allergy 2013; 68:724–31. [DOI] [PubMed] [Google Scholar]
- 26.Niederberger V, Neubauer A, Gevaert P, Zidarn M, Worm M, Aberer W, et al. Safety and efficacy of immunotherapy with the recombinant B-cell epitope-based grass pollen vaccine BM32. J Allergy Clin Immunol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campana R, Mothes N, Rauter I, Vrtala S, Reininger R, Focke-Tejkl M, et al. Non-IgE-mediated chronic allergic skin inflammation revealed with rBet v 1 fragments. J Allergy Clin Immunol 2008; 121:528–30 e1. [DOI] [PubMed] [Google Scholar]
- 28.Purohit A, Niederberger V, Kronqvist M, Horak F, Gronneberg R, Suck R, et al. Clinical effects of immunotherapy with genetically modified recombinant birch pollen Bet v 1 derivatives. Clin Exp Allergy 2008; 38:1514–25. [DOI] [PubMed] [Google Scholar]
- 29.Zieglmayer P, Focke-Tejkl M, Schmutz R, Lemell P, Zieglmayer R, Weber M, et al. Mechanisms, safety and efficacy of a B cell epitope-based vaccine for immunotherapy of grass pollen allergy. EBioMedicine 2016; 11:43–57.5049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worm M, Lee HH, Kleine-Tebbe J, Hafner RP, Laidler P, Healey D, et al. Development and preliminary clinical evaluation of a peptide immunotherapy vaccine for cat allergy. J Allergy Clin Immunol 2011; 127:89–97. [DOI] [PubMed] [Google Scholar]
- 31.O’Hehir RE, Prickett SR, Rolland JM. T Cell Epitope Peptide Therapy for Allergic Diseases. Curr Allergy Asthma Rep 2016; 16:14.4713452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franz B, May KF Jr., Dranoff G, Wucherpfennig K. Ex vivo characterization and isolation of rare memory B cells with antigen tetramers. Blood 2011; 118:348–57.3138687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiche S, Bussmann BM, Dwai Y, Jassoy C. Antibody-mediated binding of fluorescent HIV Gag and influenza nucleoprotein tetramers to blood cells. Immunobiology 2010; 215:223–9. [DOI] [PubMed] [Google Scholar]
- 34.Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, et al. Isolation of a human anti- HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One 2011; 6:e23532.3184076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogunniyi AO, Thomas BA, Politano TJ, Varadarajan N, Landais E, Poignard P, et al. Profiling human antibody responses by integrated single-cell analysis. Vaccine 2014; 32:2866–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox KS, Tang A, Chen Z, Horton MS, Yan H, Wang XM, et al. Rapid isolation of dengue-neutralizing antibodies from single cell-sorted human antigen-specific memory B-cell cultures. MAbs 2016; 8:129–40.4966506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patil SU, Ogunniyi AO, Calatroni A, Tadigotla VR, Ruiter B, Ma A, et al. Peanut oral immunotherapy transiently expands circulating Ara h 2-specific B cells with a homologous repertoire in unrelated subjects. J Allergy Clin Immunol 2015; 136:125–34 e12.4494892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoh RA, Joshi SA, Liu Y, Wang C, Roskin KM, Lee JY, et al. Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J Allergy Clin Immunol 2016; 137:157–67.4699867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sholukh AM, Watkins JD, Vyas HK, Gupta S, Lakhashe SK, Thorat S, et al. Defense-in-depth by mucosally administered anti-HIV dimeric IgA2 and systemic IgG1 mAbs: complete protection of rhesus monkeys from mucosal SHIV challenge. Vaccine 2015; 33:2086–95.4411954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pegu A, Yang ZY, Boyington JC, Wu L, Ko SY, Schmidt SD, et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med 2014; 6:243ra88.4562469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mascola JR, Louder MK, VanCott TC, Sapan CV, Lambert JS, Muenz LR, et al. Potent and synergistic neutralization of human immunodeficiency virus (HIV) type 1 primary isolates by hyperimmune anti-HIV immunoglobulin combined with monoclonal antibodies 2F5 and 2G12. J Virol 1997; 71:7198–206.192059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol 1999; 73:4009–18.104180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofmann-Lehmann R, Vlasak J, Rasmussen RA, Smith BA, Baba TW, Liska V, et al. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J Virol 2001; 75:7470–80.114982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofmann-Lehmann R, Vlasak J, Rasmussen RA, Jiang S, Li PL, Baba TW, et al. Postnatal pre- and postexposure passive immunization strategies: protection of neonatal macaques against oral simian-human immunodeficiency virus challenge. J Med Primatol 2002; 31:109–19. [DOI] [PubMed] [Google Scholar]
- 45.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol 2010; 84:1302–13.2812338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog 2009; 5:e1000433.2674935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, et al. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci U S A 2011; 108:11181–6.3131343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ledgerwood JE, Coates EE, Yamshchikov G, Saunders JG, Holman L, Enama ME, et al. Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin Exp Immunol 2015; 182:289–301.4636891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol 2011; 23:383–90.3139952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haynes BF, Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science 2014; 344:588–9.4162091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 2005; 308:1906–8. [DOI] [PubMed] [Google Scholar]
- 52.Bonsignori M, Alam SM, Liao HX, Verkoczy L, Tomaras GD, Haynes BF, et al. HIV-1 antibodies from infection and vaccination: insights for guiding vaccine design. Trends Microbiol 2012; 20:532–9.3757512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 2014; 28:163–9.4042313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog 2011; 7:e1001251.3020924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moody MA, Gao F, Gurley TC, Amos JD, Kumar A, Hora B, et al. Strain-Specific V3 and CD4 Binding Site Autologous HIV-1 Neutralizing Antibodies Select Neutralization-Resistant Viruses. Cell Host Microbe 2015; 18:354–62.4567706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 2013; 496:469–76.3637846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 2014; 158:481–91.4150607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 2011; 480:336–43.3406929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 2015; 7:319ra206 [DOI] [PubMed] [Google Scholar]
- 60.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, et al. Effect of HIV Antibody VRC01 on Viral Rebound after Treatment Interruption. N Engl J Med 2016; 375:203750.5292134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoofs T, Klein F, Braunschweig M, Kreider EF, Feldmann A, Nogueira L, et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 2016; 352:997–1001.5151174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu CL, Lorenzi JC, et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 2016; 535:556–60.5034582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP, Buckley N, et al. Corrigendum: Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 2016; 535:580. [DOI] [PubMed] [Google Scholar]
- 64.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, et al. Antibody 10–1074 suppresses viremia in HIV-1-infected individuals. Nat Med 2017; 23:185–91.5467219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shamji MH, Ljorring C, Francis JN, Calderon MA, Larche M, Kimber I, et al. Functional rather than immunoreactive levels of IgG4 correlate closely with clinical response to grass pollen immunotherapy. Allergy 2012; 67:217–26. [DOI] [PubMed] [Google Scholar]
- 66.Burton OT, Tamayo JM, Stranks AJ, Koleoglou KJ, Oettgen HC. Allergen-specific IgG antibody signaling through FcgammaRIIb promotes food tolerance. J Allergy Clin Immunol 2018; 141:189–201 e3.5671359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vizzardelli C, Gindl M, Roos S, Mobs C, Nagl B, Zimmann F, et al. Blocking antibodies induced by allergen-specific immunotherapy ameliorate allergic airway disease in a human/mouse chimeric model. Allergy 2017. [DOI] [PubMed] [Google Scholar]
- 68.Crowe JE Jr. Principles of Broad and Potent Antiviral Human Antibodies: Insights for Vaccine Design. Cell Host Microbe 2017; 22:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 2015; 349:156–61.4669217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016; 351:1458–63.4872700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, et al. Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies. Cell 2016; 166:1459–70 e11.5018249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glanville J, Kuo TC, von Budingen HC, Guey L, Berka J, Sundar PD, et al. Naive antibody gene segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci U S A 2011; 108:20066–71.3250199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parameswaran P, Liu Y, Roskin KM, Jackson KK, Dixit VP, Lee JY, et al. Convergent antibody signatures in human dengue. Cell Host Microbe 2013; 13:691–700.4136508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAx to prevent HIV-1 infection in Thailand. N Engl J Med 2009; 361:2209–20. [DOI] [PubMed] [Google Scholar]
- 75.Li SS, Gilbert PB, Tomaras GD, Kijak G, Ferrari G, Thomas R, et al. FCGR2C polymorphisms associate with HIV-1 vaccine protection in RV144 trial. J Clin Invest 2014; 124:3879–90.4151214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turk G, Ghiglione Y, Falivene J, Socias ME, Laufer N, Coloccini RS, et al. Early Gag immunodominance of the HIV-specific T-cell response during acute/early infection is associated with higher CD8+ T-cell antiviral activity and correlates with preservation of the CD4+ T-cell compartment. J Virol 2013; 87:7445–62.3700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalams SA, Buchbinder SP, Rosenberg ES, Billingsley JM, Colbert DS, Jones NG, et al. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J Virol 1999; 73:6715–20.112756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boaz MJ, Waters A, Murad S, Easterbrook PJ, Vyakarnam A. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T cell responses is associated with nonprogression in HIV-1 infection. J Immunol 2002; 169:6376–85 [DOI] [PubMed] [Google Scholar]
- 79.Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, et al. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci Transl Med 2012; 4:123ra25.3918726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Apostolopoulos V, Weiner DB. Development of more efficient and effective DNA vaccines. Expert Rev Vaccines 2009; 8:1133–4. [DOI] [PubMed] [Google Scholar]
- 81.Bonifaz LC, Bonnyay DP, Charalambous A, Darguste DI, Fujii S, Soares H, et al. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med 2004; 199:815–24.2212731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Niezold T, Storcksdieck Genannt Bonsmann M, Maaske A, Temchura V, Heinecke V, Hannaman D, et al. DNA vaccines encoding DEC205-targeted antigens: immunity or tolerance? Immunology 2015; 145:519–33.4515132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saluja SS, Hanlon DJ, Sharp FA, Hong E, Khalil D, Robinson E, et al. Targeting human dendritic cells via DEC-205 using PLGA nanoparticles leads to enhanced cross-presentation of a melanoma-associated antigen. Int J Nanomedicine 2014; 9:5231–46.4235494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lucas CG, Matassoli FL, Pecanha LM, Santillo BT, Oliveira LM, Oshiro TM, et al. Dendritic cells primed with a chimeric plasmid containing HIV-1-gag associated with lysosomal-associated protein-1 (LAMP/gag) is a potential therapeutic vaccine against HIV. FASeB J 2016; 30:2970–84. [DOI] [PubMed] [Google Scholar]
- 85.Arruda LB, Sim D, Chikhlikar PR, Maciel M Jr., Akasaki K, August JT, et al. Dendritic cell- lysosomal-associated membrane protein (LAMP) and LAMP-1-HIV-1 gag chimeras have distinct cellular trafficking pathways and prime T and B cell responses to a diverse repertoire of epitopes. J Immunol 2006; 177:2265–75. [DOI] [PubMed] [Google Scholar]
- 86.Maciel M Jr., Cruz Fda S, Cordeiro MT, da Motta MA, Cassemiro KM, Maia Rde C, et al. A DNA vaccine against yellow fever virus: development and evaluation. PLoS Negl Trop Dis 2015; 9:e0003693.4395287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Su Y, Connolly M, Marketon A, Heiland T. CryJ-LAMP DNA Vaccines for Japanese Red Cedar Allergy Induce Robust Th1-Type Immune Responses in Murine Model. J Immunol Res 2016; 2016:4857869.4867073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Su Y, Romeu-Bonilla E, Anagnostou A, Fitz-Patrick D, Hearl W, Heiland T. Safety and longterm immunological effects of CryJ2-LAMP plasmid vaccine in Japanese red cedar atopic subjects: A phase I study. Hum Vaccin Immunother 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Su Y, Romeu-Bonilla E, Anagnostou A, Fitz-Patrick D, Hearl W, Heiland T. Safety and longterm immunological effects of CryJ2-LAMP plasmid vaccine in Japanese red cedar atopic subjects: A phase I study. Hum Vaccin Immunother 2017; 13:2804–13.5718801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun M, Pace CS, Yao X, Yu F, Padte NN, Huang Y, et al. Rational design and characterization of the novel, broad and potent bispecific HIV-1 neutralizing antibody iMabm36. J Acquir Immune Defic Syndr 2014; 66:473–83.4163016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang Y, Yu J, Lanzi A, Yao X, Andrews CD, Tsai L, et al. Engineered Bispecific Antibodies with Exquisite HIV-1-Neutralizing Activity. Cell 2016; 165:1621–31.4972332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pegu A, Asokan M, Wu L, Wang K, Hataye J, Casazza JP, et al. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun 2015; 6:8447.4633990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sung JA, Pickeral J, Liu L, Stanfield-Oakley SA, Lam CY, Garrido C, et al. Dual-Affinity ReTargeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. J Clin Invest 2015; 125:4077–90.4639974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sloan DD, Lam CY, Irrinki A, Liu L, Tsai A, Pace CS, et al. Targeting HIV Reservoir in Infected CD4 T Cells by Dual-Affinity Re-targeting Molecules (DARTs) that Bind HIV Envelope and Recruit Cytotoxic T Cells. PLoS Pathog 2015; 11:e1005233.4634948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Madritsch C, Eckl-Dorna J, Blatt K, Ellinger I, Kundi M, Niederberger V, et al. Antibody conjugates bispecific for intercellular adhesion molecule 1 and allergen prevent migration of allergens through respiratory epithelial cell layers. J Allergy Clin Immunol 2015; 136:490–3 e11.4530582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Orgel KA, Duan S, Wright BL, Maleki SJ, Wolf JC, Vickery BP, et al. Exploiting CD22 on antigen-specific B cells to prevent allergy to the major peanut allergen Ara h 2. J Allergy Clin Immunol 2017; 139:366–9 e2.5222808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr., Klein F, Gao H, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 2011; 334:1289–93.3232316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011; 333:1633–7.3351836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 2010; 329:811–7.2981354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 2012; 109:E3268–77.3511153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011; 477:466–70.3393110. [DOI] [PMC free article] [PubMed] [Google Scholar]