Abstract
The primary role of bacterial periplasmic binding proteins is sequestration of essential metabolites present at a low concentration in the periplasm and making them available for active transporters that transfer these ligands into the bacterial cell. The periplasmic binding proteins (SiaPs) from the tripartite ATP-independent periplasmic (TRAP) transport system that transports mammalian host–derived sialic acids have been well studied from different pathogenic bacteria, including Haemophilus influenzae, Fusobacterium nucleatum, Pasteurella multocida, and Vibrio cholerae. SiaPs bind the sialic acid N-acetylneuraminic acid (Neu5Ac) with nanomolar affinity by forming electrostatic and hydrogen-bonding interactions. Here, we report the crystal structure of a periplasmic binding protein (SatA) of the ATP-binding cassette (ABC) transport system from the pathogenic bacterium Haemophilus ducreyi. The structure of Hd-SatA in the native form and sialic acid–bound forms (with Neu5Ac and N-glycolylneuraminic acid (Neu5Gc)), determined to 2.2, 1.5, and 2.5 Å resolutions, respectively, revealed a ligand-binding site that is very different from those of the SiaPs of the TRAP transport system. A structural comparison along with thermodynamic studies suggested that similar affinities are achieved in the two classes of proteins through distinct mechanisms, one enthalpically driven and the other entropically driven. In summary, our structural and thermodynamic characterization of Hd-SatA reveals that it binds sialic acids with nanomolar affinity and that this binding is an entropically driven process. This information is important for future structure-based drug design against this pathogen and related bacteria.
Keywords: crystal structure, isothermal titration calorimetry (ITC), ABC transporter, energetics, sugar transport, enthalpy, entropy, nutrient sequestration, sialic acid, virulence, Haemophilus ducreyi, periplasmic binding protein
Introduction
Protein–protein interactions and protein–ligand interactions play a key role in all living organisms, ranging from prokaryotes to eukaryotes (1, 2). These interactions are highly specific and regulate several downstream signaling pathways and cellular metabolic processes (3, 4). Understanding the molecular details of protein–ligand interactions is a key step in the structure-assisted drug discovery process. Electrostatic interactions, hydrogen-bonding interactions, van der Waals interactions, and hydrophobic interactions contribute to the binding affinity of a ligand to a protein. Thermodynamically, both enthalpy (ΔH) and entropy (ΔS) contribute to the binding affinity. The difference in enthalpy and entropy between the bound and unbound forms dictates the spontaneity of the binding reaction. The enthalpy of the system is mainly derived from noncovalent interactions, such as hydrogen bonding, ionic interactions, and van der Waals contacts between polar side chains of the amino acids and ligand. In addition, hydrophobic interactions between ligands and proteins are responsible for the displacement of water, which contributes to either positive or negative net entropy and generates randomness or order within the system. In biological systems, enthalpy–entropy compensation of protein–ligand interactions dictates how spontaneous the reactions will be (5, 6).
Sialic acids are nine-carbon–containing sugars, and the carboxylate group at the C1 position imparts the acidic nature to the molecule. In vertebrates and in associated symbiotic and pathogenic bacteria, these sugars are present as the outermost sugars on glycoproteins and glycolipids of the outermost cell surface (7, 8). Based on structural differences, sialic acids exist in ∼50 different isoforms, and the two most common sugars include N-acetylneuraminic acid (Neu5Ac)3 and N-glycolylneuraminic acid (Neu5Gc). Chemically, Neu5Gc differs from Neu5Ac by a single hydroxyl group at the C5 position (9–11). Most pathogenic bacteria that reside in the mucus-rich regions lack the biosynthetic pathways to make Neu5Ac, so they scavenge and transport host-derived sialic acids for their long-term survival. Sialic acids are transported across the bacterial periplasmic membrane by tripartite ATP-independent periplasmic (TRAP), ABC, or major facilitator superfamily (MFS) transport systems. After transport of sialic acid into the cytoplasm, it is incorporated as the outermost sugar on the lipooligosaccharide (LOS) by CMP-sialic acid synthetase (SiaB) and sialyltransferases (SiaA, Lic3A, Lic3B, or LsgB). Pathogenic bacteria incorporate sialic acid during lipid glycosylation to evade the host immune system by “molecular mimicry” (12). Previously, it was reported that mutating either SiaB or the sialyltransferase gene Lic3A, which encodes for α2,3-sialyltransferase, exhibited less resistance to the killing effects of human serum (13, 14). In addition to lipid glycosylation, part of the internalized sialic acid is often converted into fructose 6-phosphate by a pathway of enzymes that include NanA, NanK, NanE, NagA, and NagB that enables the bacteria to use them as an energy source (15).
Gram-negative bacteria, such as Haemophilus influenzae, Pasteurella multocida, Fusobacterium nucleatum, and Vibrio cholera, utilize the TRAP transport system for sialic acid transport (16, 17). The sialic acid–binding proteins (SiaPs) of the TRAP system have been structurally and functionally characterized (18–20). These proteins contain two domains, and sialic acid binds between these two domains in a conserved binding pocket. In SiaPs, ligand binding generates conformational changes in the hinge region, which cause the movement of two domains to close on the substrate. This mechanism is called either the Venus flytrap mechanism or Pac-Man model (21, 22). Thermodynamic studies have shown that the binding of sialic acids to SiaPs is an enthalpically favored process.
However, Haemophilus ducreyi, another Gram-negative bacteria, has been reported to use the ABC transport system for sialic acid transport, utilizing ATP hydrolysis to import sugars across the periplasmic membrane (23, 24). H. ducreyi is a pathogenic Gram-negative coccobacillus belonging to the pasteurellaceae family. It causes a sexually transmitted genital disease called chancroid (25). This disease is a major public health concern in developing and low socio-economic level countries due to an increased risk of co-transmission with HIV (26, 27). The pathogenicity of H. ducreyi in humans is caused by several virulence factors, including the hemolytic toxin (28, 29) and cytolethal-distending toxin (30–32). In addition, the LOS of H. ducreyi acts as a putative virulence factor that aids in bacterial adherence to human skin fibroblasts and mucosal epithelial cells (33, 34). LOS terminates with N-acetyllactosamine and sialyl-N-acetyllactosamine, which is heavily sialylated (35).
In H. ducreyi, the ABC transport system that transports sialic acid is structurally encoded by the four-gene operon SatABCD. In this operon, SatA encodes a periplasmic binding protein that binds to sialic acid in the periplasm. SatBCD encodes the ABC-type integral membrane transporter that transports the delivered sugar using the energy derived from ATP hydrolysis (36). Similar to the maltose-binding protein system, it is expected that SatA bound to Neu5Ac/Neu5Gc anchors onto the transmembrane domain of the ABC transporter. This signals ATP binding, the sialic acid is then released from SatA, and the sugar is transported from the periplasm to the cytoplasm by SatBCD (37–39).
ABC transporters import and efflux a variety of different substrates across the lipid bilayer, including ions, peptides, proteins, and sugars. Although several periplasmic binding proteins of ABC transporters that transport a wide range of other substrates have been described previously, the structural and molecular mechanism of sialic acid binding to SatA in ABC transporters is not known. The sequence similarity between SiaP from TRAP transporters and SatA from ABC transporters is ∼20–25%; thus, it is important to structurally and functionally characterize SatA from ABC transporters.
Here, we report the structures of the sialic acid–binding protein of H. ducreyi, which uses the ABC transporter system (Hd-SatA), in unliganded and ligand-bound forms. In addition, we have thermodynamically characterized Hd-SatA binding to its ligands Neu5Ac and Neu5Gc. Site-directed mutagenesis of residues that bind to the sugars followed by measurement of binding affinities shed light on the contribution of these residues to the binding phenomenon.
Results
Hd-SatA unliganded, Neu5Ac-bound, and Neu5Gc-bound structures
Hd-SatA was crystallized in three different forms: unliganded, Neu5Ac-bound, and Neu5Gc-bound. Hd-SatA has low sequence identity (20–25%) with other periplasmic sugar-binding proteins, and there are no homologous structures available in the Protein Data Bank. We first determined the crystal structure of selenomethionine-derivatized Hd-SatA bound to Neu5Gc to 2.4 Å resolution by the multiple-wavelength anomalous dispersion method (Fig. 1). These crystals belong to the space group C121, and the model was refined to an R-factor of 0.17 and free R-factor of 0.25. The crystal structure shows two molecules in the asymmetric unit, and the electron density clearly shows the presence of Neu5Gc in the binding pocket. The crystals of the Hd-SatA–Neu5Ac complex diffracted to a higher resolution of 1.49 Å, and these crystals belong to the P212121 space group. This crystal structure was determined by molecular replacement using the Hd-SatA–Neu5Gc structure as the search model. It was refined to an R-factor of 0.17 and a free R-factor of 0.197. In the Hd-SatA–Neu5Ac structure, the electron density map clearly shows the presence of Neu5Ac in the binding pocket (Fig. 2, inset). The crystals of the Hd-SatA unliganded form diffracted to 2.1 Å, and the structure was determined by molecular replacement using the Hd-SatA–Neu5Ac structure as the search model. These crystals belong to the space group P1211 and were refined to an R-factor of 0.20 and a free R-factor of 0.26. The crystal structures of unliganded Hd-SatA and Hd-SatA–Neu5Ac forms have one molecule in the asymmetric unit.
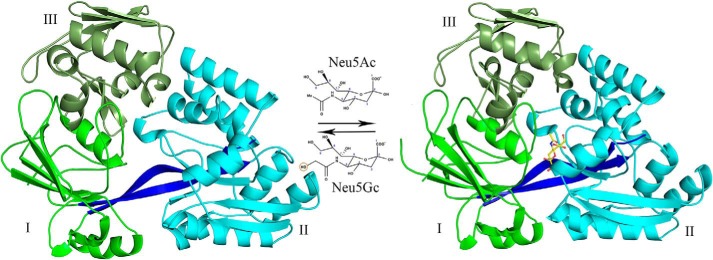
Figure 1.
Structure of Hd-SatA in complex with Neu5Ac. A cartoon representation shows the crystal structures of Hd-SatA in unliganded form (left) and ligand-bound conformations (right). Neu5Ac is represented by a ball-and-stick model. Structural representations of Neu5Ac and Neu5Gc are also shown. The domains are marked as I, II, and III. The program PyMOL (Schrödinger) (54) was used to create the figure.
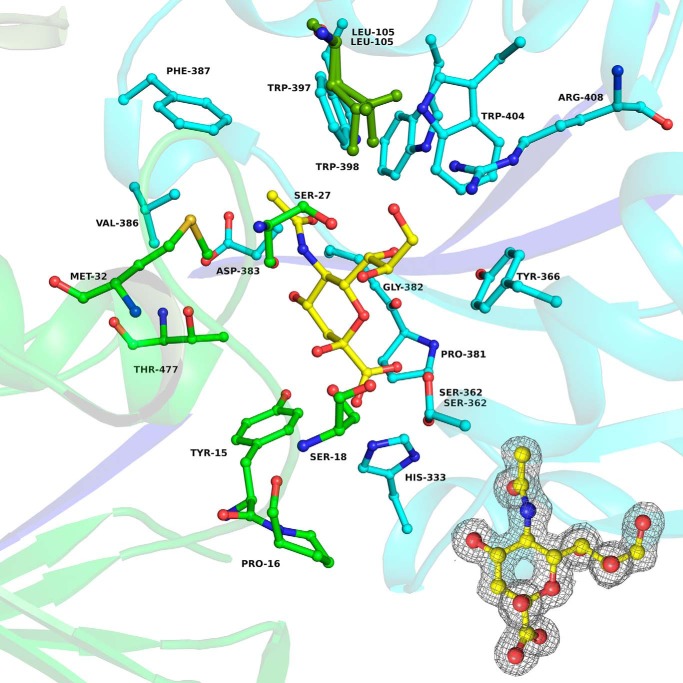
Figure 2.
Binding pocket of Hd-SatA with Neu5Ac. A ball-and-stick diagram shows Neu5Ac and its interacting residues in Hd-SatA. Inset, 2Fo − Fc map at 1σ level showing the electron density map of Neu5Ac in the binding pocket of Hd-SatA. The program PyMOL (Schrödinger) (54) was used to create the figure.
The details of the crystallography data and refinement statistics are presented in Table 1. The coordinates and structure factors for Hd-SatA unliganded, Hd-SatA–Neu5Ac and Hd-SatA–Neu5Gc have been deposited in the Protein Data Bank (PDB) with accession numbers 5ZA4, 5Z99, and 5YYB, respectively.
Table 1.
Crystallography data and refinement statistics
Statistics for the highest-resolution shell are shown in parentheses.
| Parameters | Unliganded | With Neu5Ac | With Neu5Gca |
|---|---|---|---|
| Wavelength | 1.03 | 0.9793 | 0.9546 |
| Resolution range (Å) | 29.43–2.19 (2.27–2.19) | 19.52 − 1.49 (1.52 − 1.49) | 28.99 − 2.48 (2.59 -2.48) |
| Space group | P 1 21 1 | P 21 21 21 | C 1 2 1 |
| Unit cell | |||
| a (Å) | 43.9 | 73.4 | 134.4 |
| b (Å) | 79.3 | 89.6 | 70.1 |
| c (Å) | 68.4 | 90.8 | 113.1 |
| α, β, γ (degrees) | 90, 91.2, 90 | 90, 90, 90 | 90, 114.2, 90 |
| Total reflections | 90,314 (7117) | 604,746 (23,207) | 124,762 (11,816) |
| Unique reflections | 24,030 (1961) | 93,977 (4237) | 33,076 (3289) |
| Multiplicity | 3.8 (3.6) | 6.4 (5.5) | 3.8 (3.6) |
| Completeness (%) | 99.5 (94.9) | 96.8 (88.6) | 97.2 (85.6) |
| Mean I/σ(I) | 7.00 (3.00) | 19 (4.9) | 7.2 (2.3) |
| Wilson B-factor | 31.7 | 11.2 | 36.6 |
| Rmerge | 0.178 (1.175) | 0.058 (0.538) | 0.116 (0.549) |
| Rmeas | 0.209 (1.382) | 0.063 (0.591) | 0.135 (0.641) |
| Rpim | 0.108 (0.72) | 0.024 (0.237) | 0.069 (0.329) |
| CC1/2 | 0.985 (0.37) | 0.999 (0.807) | 0.993 (0.502) |
| Reflections used in refinement | 24,004 (2281) | 93,931 (9021) | 33,071 (2840) |
| Reflections used for Rfree | 1191 (102) | 4864 (459) | 1647 (118) |
| Rwork | 0.20 (0.26) | 0.17 (0.26) | 0.17 (0.26) |
| Rfree | 0.26 (0.29) | 0.19 (0.28) | 0.25 (0.38) |
| Number of non-hydrogen atoms | 3974 | 4555 | 7869 |
| Macromolecules | 3847 | 3972 | 7667 |
| Ligands | 21 | 44 | |
| Solvent | 127 | 562 | 202 |
| Protein residues | 488 | 485 | 964 |
| Root mean square deviation | |||
| Bonds (Å) | 0.008 | 0.005 | 0.01 |
| Angles (degrees) | 0.96 | 0.8 | 1.02 |
| Ramachandran (%) | |||
| Favored | 94.86 | 96.27 | 93.96 |
| Allowed | 4.94 | 3.73 | 5.73 |
| Outliers | 0 | 0 | 0 |
| B-factor | |||
| Average | 28.91 | 18.23 | 40.86 |
| Macromolecules | 28.83 | 16.75 | 40.29 |
| Ligands | 9.97 | 32.53 | |
| Solvent | 27.26 | 29.01 | 38.44 |
| PDB code | 5ZA4 | 5Z99 | 5YYB |
a This data set is also the data set from the peak wavelength of selenomethionine protein used to determine the structure.
The overall structure contains three α/β domains, highlighted in Fig. 1. The structure contains an N-terminal domain that is larger than the C-terminal domain due to the presence of an extended region in the N terminus and can be divided into two domains, I and III. The N-terminal domain contains amino acids from the N terminus and C terminus. Domain I contains residues 1–38, 160–239, and 461–487 (from the C terminus), which form six β strands that are sandwiched between three α helices. Domain III is composed of residues 39–159, which form six β strands and two α helices. Domain II contains the C terminus residues 244–456, which form six β strands enclosed by seven α helices. The N- and C-terminal domains are connected by two β strands, which are composed of residues 240–243 and 457–460, and these two strands may serve as hinge regions. The crystal structure clearly shows that the ligand is buried deep inside the binding pocket formed between domains I and II, and there are no apparent interactions of the ligand with domain III. Upon ligand binding, both domains I and II come together to trap the ligand, akin to the Venus fly trap mechanism, which is similar to the pattern that is observed in other sugar-binding proteins.
The Dali server search results showed that many structures are structurally similar to Hd-SatA, but they have less than 25% sequence identity. Some of the structures with the highest matching Z scores are dipeptide-binding protein DppA from Escherichia coli (with a Z score of 40.9) (PDB code 1DPP) (40), periplasmic oligopeptide-binding protein AppA from Bacillus subtilis (PDB code 1XOC) (41), antibiotic agrocinopine A transport protein in Agrobacterium tumefaciens (PDB code 4ZE9) (42), and periplasmic lipoprotein HbpA in Haemophilus parasuis (PDB code 3M8U) (43).
The sialic acid–binding pocket of Hd-SatA
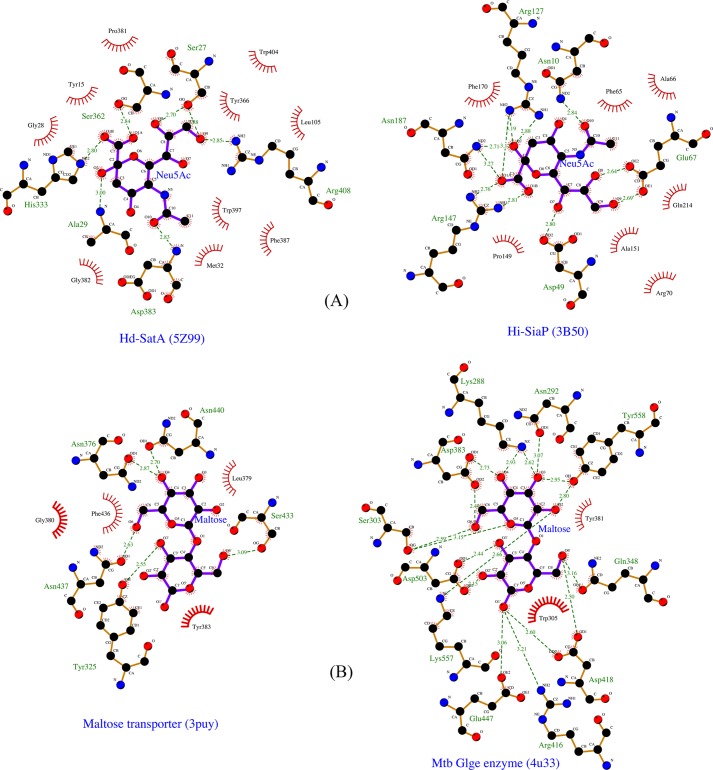
The binding of Neu5Ac in the binding pocket of Hd-SatA is depicted in Fig. 2. There are several hydrogen-bonding interactions observed between different amino acids in the binding pocket and the Neu5Ac molecule. In SiaPs of TRAP transporters, the major interactions are salt bridges between an Arg and the carboxylate group (C1 position) of sialic acid (18, 19). In Hd-SatA, there are no amino acids in the binding pocket that make salt bridges with the ligand. There is only one Arg in the binding pocket, at position 408, and it forms a hydrogen bond with the hydroxyl group at the C9 position. His333 and Ser362 are two polar residues closest to the carboxylate group at C1, and they form hydrogen bonds with the carboxylate group. Ser18 and Ser362 form a hydrogen bond with the hydroxyl group at C8, and Ser27 forms a hydrogen bond with hydroxyl groups at positions C8 and C9. Pro16 has a water-mediated hydrogen-bonding interaction with O2. Tyr15, Thr477, and Asp383 also display water-mediated hydrogen-bonding interactions with the hydroxyl group at position C4. The binding pocket of Hd-SatA–Neu5Gc is similar to Hd-SatA–Neu5Ac, and there are no structural changes between these two sugar-bound forms.
In summary, SiaPs from TRAP transporters have more charged residues in their binding pocket, and their interactions with sialic acids are mostly mediated by hydrogen bonds, ionic interactions, and salt bridges. However, Hd-SatA has few charged residues and more polar residues in the binding pocket, and its interaction with Neu5Ac is mainly mediated by hydrogen bonds and hydrophobic interactions.
Hinge region analysis
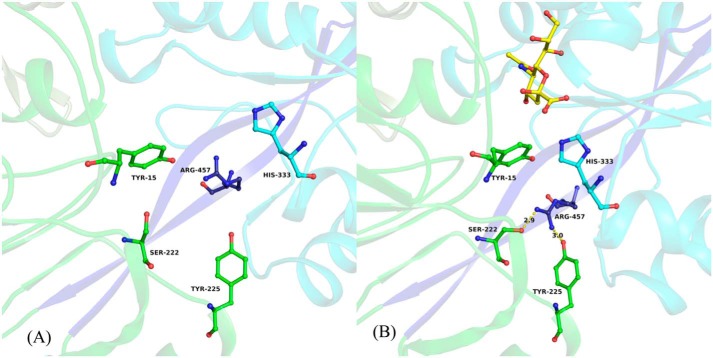
The superposition of open and closed conformations of Hd-SatA resulted in a root mean square deviation of 3.496 for 3641 Cα atoms. The relative movement of domains in the open and closed conformations was analyzed using DYNDOM. DYNDOM classified the structure as a two-domain protein, where domain I is considered a fixed domain and is composed of residues 1–240 and 460–487. Domain II is considered a mobile domain and is composed of residues 244–456. Domain II moves at a rotation of 31° relative to domain 1 and contributes a small component of translation by 0.5 Å. The two hinge regions, which are responsible for the domain movement, are formed by residues 241–243 and 457–459 (Fig. 3). Comparative analyses of φ and ψ angles of unliganded Hd-SatA and Hd-SatA–Neu5Ac bound structures show that there are large changes in the amino acids in the two hinge regions. Main-chain torsion angles between Phe241 and Tyr242, between His456 and Arg457, and between Arg457 and Lys458 are primarily responsible for the hinge movement. In both cases, the largest differences are in the φ angle. Further structural analysis reveals that there are significant conformational changes of amino acids in the binding pocket upon ligand binding. In the open conformation, Arg457 from hinge two is oriented toward the binding pocket, and His333 and Tyr15 from domains I and II are far away from the binding pocket. Upon ligand binding, the Arg457 side chain moves away from the binding pocket to form hydrogen bonds with Ser222 and Tyr225, and amino acids His333 and Tyr15 move closer to the ligand to form hydrogen bonds with the Neu5Ac. Thus, there are a series of conformational changes upon ligand binding that lead to the opening and closing of the two domains of the protein (Fig. 3).
Figure 3.
Close-up view of hinge region and amino acids from the binding pocket of Hd-SatA. Neu5Ac and its interacting residues in the binding pocket of Hd-SatA are shown in a ball-and-stick model. A close-up view clearly shows the conformational changes in the amino acids before (A) and after (B) Neu5Ac binding. Neu5Ac is shown in B. The two β strands that form the hinge are shown in dark blue color. The figure was created using PyMOL (Schrödinger) (54).
Thermodynamics of Neu5Ac and Neu5Gc binding to Hd-SatA and its mutants
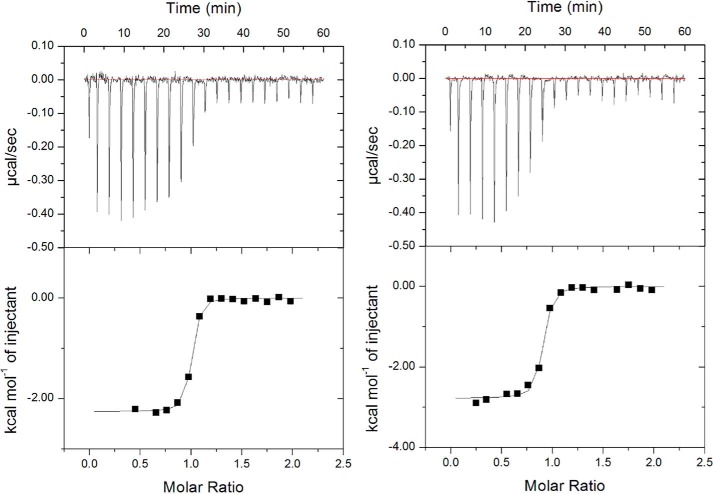
Isothermal titration calorimetry (ITC) was used to measure the thermodynamics of the interactions between Hd-SatA and its ligands Neu5Ac and Neu5Gc. Fitting the data using single-site binding mode analysis indicates that Hd-SatA binds to Neu5Ac and Neu5Gc in a 1:1 stoichiometric ratio with nanomolar binding affinity. Further binding studies showed that the entropy contribution has a larger effect on the overall free energy of the system compared with enthalpy (Kd = 133 ± 22 nm, ΔH = −2.21 ± 0.023 kcal/mol and −TΔS = −7.172 kcal/mol). These results are similar to Hd-SatA binding to Neu5Gc (Kd = 277 ± 58 nm, ΔH = −2.986 ± 0.056 kcal/mol and −TΔS = −5.96 kcal/mol) (Fig. 4). Therefore, the interaction between Hd-SatA and sialic acid is an entropically driven process. Based on the determined structures and thermodynamic properties of SiaPs from TRAP transporters, we anticipated that the enthalpic contribution to the free energy of binding in Hd-SatA can be increased by substituting a few polar residues to charged residues in the binding pocket. To test this, we made a total of 10 substitutions in the binding pocket of Hd-SatA, S27N, S27K, S362K, S362R, S362A, H333K, H333R, H333A, R408A, and R408K, and their thermodynamic properties were probed using ITC. Ser27, which forms hydrogen bonds with the hydroxyl groups at positions C8 and C9, was modified to Lys and Asn. ITC binding studies showed that S27K had no effect on the interaction, but S27N bound endothermically to Neu5Ac (Fig. S1) and exothermically to Neu5Gc. Ser362 and His333, which form hydrogen bonds with the C1 carboxylate of Neu5Ac, were mutated to S362K, S362R, S362A, H333K, H333R, and H333A, respectively. Ser362 and His333 were individually mutated to Arg and Lys to investigate whether an ATP transporter can be made to have similar binding properties as periplasmic binding proteins in TRAP transporters via salt bridge formation (18). However, these substitutions had binding properties similar to the WT protein, although there was no measurable binding affinity with the H333K substitution. The R408A and R408K substitutions did not show any measurable binding to Neu5Ac. The binding affinities, including the enthalpic and entropic contributions of sialic acids binding to Hd-SatA WT and its mutants, are presented in Table 2. The thermodynamic studies show that there were few minor changes in the thermodynamic properties of the protein due to site-directed mutagenesis.
Figure 4.
Thermodynamic studies of Hd-SatA. Isothermal calorimetric titration shows the binding of Hd-SatA to Neu5Ac (A) and Neu5Gc (B).
Table 2.
Binding affinities and thermodynamic parameters of native protein and proteins with site-specific amino acid substitutions
ITC with a Microcal instrument was used for thermodynamic measurements, and data were analyzed using SEDPHAT (for Hd-SatA WT) and Origin software (for Hd-SatA mutants). NB, no binding.
| Site no. | Hd-SatA protein | Ligand | Kd | ΔH | −TΔS |
|---|---|---|---|---|---|
| kcal/mol | kcal/mol | ||||
| 1 | WT | Neu5Ac | 133 ± 22 nm | −2.21 ± 0.023 | −7.172 |
| Neu5Gc | 277 ± 58 nm | −2.986 ± 0.056 | −5.96 | ||
| 2 | S27N | Neu5Ac | 5.52 ± 1.18 μm | 2.458 ± 0.116 | −9.625 |
| Neu5Gc | 57.8 ± 19.3 μm | −0.635 ± 0.11 | −5.155 | ||
| 3 | S27K | Neu5Ac | 24 ± 3.04 μm | −2.725 ± 0.126 | −3.576 |
| 4 | S362R | Neu5Ac | 4.83 ± 0.44 μm | −1.107 ± 0.017 | −6.138 |
| 5 | S362A | Neu5Ac | 418 ± 179 nm | −0.47 ± 0.0133 | −8.224 |
| 6 | S362K | Neu5Ac | 5.37 ± 0.22 μm | −1.63 ± 0.03 | −5.542 |
| 7 | H333R | Neu5Ac | 115 ± 52 nm | −2.850 ± 0.055 | −6.615 |
| 8 | H333A | Neu5Ac | 31 ± 10 nm | −5.724 ± 0.052 | −4.529 |
| 9 | H333K | Neu5Ac | NB | NB | NB |
| 10 | R408A | Neu5Ac | NB | NB | NB |
| 11 | R408K | Neu5Ac | NB | NB | NB |
Database search to identify proteins binding to the same ligand with different binding modes
In a PDB database search, we found two proteins, a maltose transporter (PDB code 3PUY) and a bacterial maltosyltransferase (PDB code 4U33), that bound to the same ligand (maltose) with different modes of binding. Previously, it was reported that maltose binding to an MBP-maltose transporter is an entropically driven process (44). The MBP-maltose transporter has a large hydrophobic binding pocket; hence, the displacement of water molecules might be the reason for the entropically favored binding process. However, unlike the data presented here, no experimental evidence is present to establish whether the thermodynamics of binding is different in the maltose-binding protein of bacterial maltosyltransferase.
Discussion
Pathogenic bacteria have evolved different mechanisms to evade the host immune system, including coating their outer membranes with host nine-carbon sugars, known as sialic acids, that are scavenged from the hosts using TRAP/ABC/MFS transporter systems. Pathogenic bacteria display these sugars as the outermost moiety to mimic eukaryotic outer membrane composition, which aids in eluding the host immune response. Although several studies have been reported in the last few decades related to sialic acid scavenging proteins and sialic acid incorporation, structural information at the atomic level on sialic acid binding modes as well as the amino acid environment of the different transport systems are just beginning to be revealed. Here, we have structurally and functionally characterized the sialic acid–binding protein from H. ducreyi that belongs to the ABC transport system. The results from structural and thermodynamic studies support the conclusion that Hd-SatA binds to sialic acids with nanomolar affinity, and this process is entropically driven.
Hd-SatA is a three-domain protein, where sialic acid binds between domains I and II. Domain III is a small part of the N-terminal domain, which does not interact with the ligand. Initially, Fukami-Kobayashi et al. (45) classified substrate-binding proteins based on their β sheet topology as Class I and Class II proteins. After this initial classification, many more proteins were structurally and functionally characterized with a wide range of ligands. Nearly a decade later, Berntsson et al. (46) further classified these substrate-binding proteins into a total of six groups (Cluster A to Cluster F) based on their structural similarity. In this classification, SiaPs from TRAP transporters were categorized in Cluster E. All other clusters have proteins that belong to ABC transport systems. SatA from H. ducreyi can be categorized into Cluster C because it has an extra N-terminal domain. Cluster C has other proteins that belong to Class II ABC transporters, where they bind to a number of different ligands, including di- and oligopeptides, nickel, and arginine. Based on previous structural reports, it was hypothesized that proteins like AppA from B. subtilis and OppA from Lactococcus lactis have an extra domain to accommodate large ligands, such as oligopeptides (41, 47); however, in the case of Hd-SatA, the reason and necessity for the extra domain is still unknown.
SiaPs from H. influenzae, P. multocida, V. cholera, and F. nucleatum, which utilize the TRAP transport system, have been structurally and functionally characterized (18–20). The common features of sialic acid–binding proteins from ABC and TRAP transport systems are as follows: (i) the ligand binds between two globular domains, and (ii) the amino acids in the hinge regions are responsible for opening and closing the two domains upon ligand binding. But the salient features of SiaPs from TRAP transporters are as follows: (i) two arginine residues (Arg127 and Arg147; numbers correspond to H. influenzae SiaP) in the binding pocket form salt bridges with the C1 carboxylate group of the sialic acid, and (ii) Glu and His from the hinge region form and break a series of hydrogen bonds upon ligand binding (18). These features show that the binding pocket has charged residues and can form salt bridges and hydrogen bonds with the negatively charged ligand. In the case of Hd-SatA, there are only two charged residues in the binding pocket, Arg408 and Asp383. The other residues in the binding pocket are either polar or hydrophobic (Tyr15, Ser27, Gly28, Ala29, Leu105, Pro381, His333, Asp383, and Trp397). This is an important difference between SiaP and SatA, where one ligand (sialic acid) is binding to two different proteins (SiaP and SatA) in two different amino acid environments. Results from thermodynamic studies confirmed that the binding of sialic acid in Hd-SatA is a more entropically favored process, whereas binding is a more enthalpically favored process in the SiaP of the TRAP transport system (Table S1). The structures provide a molecular explanation for the observed thermodynamic properties.
We calculated the binding pocket volumes in the sialic acid–binding proteins of the TRAP and ABC transport systems using the CASTp (Computed Atlas of Surface Topography of proteins) server (48). The binding pocket volumes of H. influenzae SiaP in the open and closed conformations are 834.5 and 191.9 Å3. This shows that upon ligand binding, there is a modest change in the volume of 643 Å3. In Hd-SatA, the binding pocket volumes of the open and closed conformations are 1490.4 and 242 Å3. Thus, upon ligand binding, there is a large volume change of 1200 Å3 (Fig. S2). This large volume change upon ligand binding may displace a lot of water molecules, which may account for the increased entropic contribution in ABC transporters. This analysis shows that the binding mechanism of sialic acid–binding proteins is determined by the amino acid composition in the binding pocket and the cavity size in the protein.
These preliminary structural and protein homology studies suggest that the binding of the sialic acid by heterologous peptides is the result of convergent evolution; the sialic acid–binding proteins of the ABC transport system and TRAP transport system evolved independently to accomplish similar sialic acid binding functions. However, this hypothesis requires further testing.
This interesting result persuaded us to search the PDB database to explore whether there are other proteins that bind to the same ligand with a different motif of the binding pocket. Our database search shows that the maltose transporter from E. coli and bacterial maltosyltransferase from Mycobacterium tuberculosis both bind maltose but utilize two distinct binding pockets. The binding pocket of the maltose transporter (PDB code 3PUY) is mainly composed of polar and hydrophobic residues, such as Tyr, Phe, Gly, Leu, Asn, Phe, and Ser. This binding pocket amino acid composition is similar to the binding pocket of Hd-SatA. It was previously reported that binding of maltose to the MBP-maltose transporter is an entropically driven process. This supports the concept that, in both the MBP-maltose transporter and Hd-SatA, the hydrophobic binding pocket is responsible for entropic binding to their respective ligands. In the bacterial maltosyltransferase, the binding pocket has many charged residues, including Arg, Lys, Asp, and Glu, which is similar to TRAP transporter SiaPs. Based on the previous structural and thermodynamic studies of SiaPs from the TRAP system, we hypothesize that binding of maltose to bacterial maltosyltransferase might be an enthalpically driven process (Fig. 5).
Figure 5.
Ligplot diagrams showing the ligands and its interacting residues different proteins. A, Ligplot representation of the binding pocket of Hd-SatA and H. influenzae SiaP in the presence of their ligand Neu5Ac. B, Ligplot representation of maltose transporter and bacterial maltosyltransferase in the presence of their ligand maltose. Ligands are represented in purple color. Carbon, nitrogen, and oxygen are represented in black, blue, and red, respectively. The residues with red spikes that are oriented toward the ligand represent hydrophobic interactions.
In summary, pathogenic bacteria have evolved to bind sialic acids with similar binding affinities, using different binding topologies. The topological and chemical properties of the binding pocket lead to different energetic processes that govern binding. This study of SatA of H. ducreyi from the ABC transport system and analysis of previously reported SiaPs from the TRAP transport system together provides important and divergent structural details about sialic acid–binding proteins in two different transport systems. This information is important for future structure-based drug design against these pathogenic bacteria.
Experimental procedures
Cloning and mutagenesis of Hd-SatA WT
The DNA encoding Neu5Ac-binding protein (SatA) from H. ducreyi in PET101D-TOPO was amplified by PCR and subcloned into the BamHI and XhoI sites of a pET21a (Novagen) vector as a fusion with a C-terminal His tag (NCBI accession number WP_010945473). The gene sequence was verified by DNA sequencing and designated Hd1669 (FSS-End) pET21a. Site-specific substitutions S362R, S362A, S362K, S27K, S27N, H333R, H333K, H333A, R408A, and R408K were generated in Hd-SatA using site-directed mutagenesis with corresponding mutagenic primers. These substitutions were confirmed by sequencing.
Protein expression and purification
The protein expression of the recombinant plasmids was carried out in E. coli BL21(DE3) star cells. The cells were transformed with the plasmid and grown overnight at 37 °C in lysogeny broth medium containing 100 μg/ml ampicillin. The cells were reinoculated with 1% overnight culture in fresh lysogeny broth medium containing ampicillin. Then these cells were cultured at 37 °C until the absorbance reached 0.6, and the cultures were induced with 100 μm isopropyl β-d-thiogalactopyranoside. The cells were incubated at 25 °C for 4 h. Later, the cells were harvested at 13,000 rpm for 30 min. For protein purification, each 1-liter pellet was resuspended in 25 ml of resuspension buffer containing 20 mm HEPES, 150 mm NaCl, 5 mm imidazole (pH 8.0), which was further supplemented with a protease inhibitor mixture tablet lacking EDTA (Roche Applied Science). The resuspended pellet was treated with lysozyme and DNase and incubated on ice for 30 min following lysis using an Emulsiflex C3 from Avestin at 15,000 p.s.i. The lysate was centrifuged at 13,000 rpm for 30 min and further purified on a Talon affinity column (Bio-Rad) using Profinia (Bio-Rad). The column was first equilibrated with resuspension buffer, and the lysate was loaded onto the column. Following a wash with 10 column volumes of wash buffer (20 mm HEPES, 500 mm NaCl, 5 mm imidazole, pH 8.0), the protein was eluted with four column volumes of elution buffer containing 20 mm HEPES, 150 mm NaCl, and 500 mm imidazole, pH 8.0. The eluted protein was dialyzed overnight in 20 mm HEPES and 10 mm NaCl (pH 8.0). The protein was further purified by anion-exchange chromatography (column Hi Trap Q FF, GE Healthcare). The column was first equilibrated with Buffer A (20 mm HEPES, 10 mm NaCl, pH 8.0) and loaded with the dialyzed protein. The protein was eluted with a gradient of Buffer A to the same buffer containing 1 m NaCl. The protein eluted at a salt concentration of 150 mm. The protein fractions were pooled and purified using size-exclusion chromatography (Sepharose 200 column, GE Healthcare) using 20 mm Tris-Cl, 100 mm NaCl (pH 8.0). The samples corresponding to the protein peak were pooled, and concentration was measured using the Bradford assay (Bio-Rad). To express selenomethionine-derivatized protein of Hd-SatA, a protocol similar to that of Doublié et al. (49) was used. Further, the protein was purified using the same method as the native protein.
Crystallization, structure determination, and refinement
Hd-SatA bound with Neu5Gc
To obtain crystals of the protein–ligand complex, selenomethionine-derivatized Hd-SatA protein at a concentration of 50 mg/ml was mixed with Neu5Gc at a molar ratio of 1:10. The hanging-drop vapor diffusion method was used for crystallization trials. Crystals were obtained in a buffer containing 20 mm MES (pH 6.2), 60 mm NaCl with 30% PEG 1500 as a precipitant. The Hd-SatA–Neu5Gc crystals were mounted in loops, and X-ray data were collected at three different wavelengths (Molecular Biology Consortium, Advanced Light Source, using a NOIR1 detector). For phasing, data were collected from selenomethionine-incorporated crystals of Neu5Gc-bound Hd-SatA at selenium peak wavelength (0.9778 Å). Further selenomethionine-substituted protein structure refinement was carried out using the automated SOLVE-RESOLVE pipeline (50). Further refinements were done using Phenix, and model building was carried out using COOT (51). The coordinates and structure factors for the Hd-SatA–Neu5Gc complex have been deposited in the PDB with accession number 5YYB.
Hd-SatA bound with Neu5Ac
Hd-SatA protein at a concentration of 50 mg/ml was mixed with Neu5Ac at a molar ratio of 1:10. The crystals were obtained in buffer containing 20 mm MES, pH 6.2, 60 mm NaCl with 30% PEG 1500 as a precipitant by the hanging-drop diffusion method. The Hd-SatA–Neu5Ac complex crystals were mounted in loops, and X-ray data were collected at the IMCA-CAT beamline using the MAR-CCD detector at the Advanced Photon Source (Argonne, IL). The data were indexed, integrated, and scaled using HKL2000, and further molecular replacement was carried out using the Hd-SatA–Neu5Gc structure as a search model. Manual model building was performed using COOT, and further refinements were performed using Phenix. The coordinates and structure factors for the Hd-SatA–Neu5Ac complex have been deposited in the PDB with accession number 5Z99.
Hd-SatA without ligand
The crystals of Hd-SatA were grown by the hanging-drop vapor diffusion method by mixing 1 μl of protein at 40 mg/ml concentration with the 1 μl of the reservoir. Crystals were obtained from the reservoir solution containing 0.2 m sodium iodide and 20% (w/v) PEG 3350. These crystals were mounted in loops, and X-ray data were collected from the IMCA-CAT beamline using the MAR-CCD detector at the Advanced Photon Source (Argonne, IL). Molecular replacement was performed using Neu5Ac-bound Hd-SatA as a search model. Manual model building was performed using COOT, and further refinements were performed using Phenix. The coordinates and structure factors for Hd-SatA have been deposited in the PDB with accession number 5ZA4.
Thermodynamic studies of Hd-SatA
Thermodynamic studies were performed using isothermal calorimetry (MicroCal ITC microcalorimeter, Malvern, GE Healthcare). All titrations were carried out at 25 °C, and the protein samples were in a buffer containing 20 mm HEPES and 10 mm NaCl (pH 8.0). Protein concentrations were measured using the Bradford assay (Bio-Rad). The concentrations of Hd-SatA WT and proteins with site specific amino acid substitutions were varied in different titrations. Hd-SatA WT ITC titrations with Neu5Ac and Neu5Gc was carried out from three independent preparations and technical triplicates for each preparation. All ITC data from proteins with mutations in Hd-SatA were technical replicates. Each experiment consisted of 20 injections, with the first injection 1 μl and the rest of the injections 2 μl with 180 s between the titrations. The nonspecific heats of dilution liberated during the titration of the sugars and protein were calculated by averaging the heat liberated during the last 3–5 injections after saturation. The raw heat released during each injection was calculated by deleting these heats of dilution.
Global fit analysis from technical replicates was done for Hd-SatA WT using SEDPHAT (52). The analysis was repeated with three different protein preparations, and all global fit analysis led to similar values. All of the mutant protein data were analyzed using the ORIGIN ITC software package (Microcal) using a single-site binding model, and errors are fit to curves. In the ORIGIN software, nonlinear least squares analysis was used to calculate the values of stoichiometry, affinity, change in enthalpy (ΔH), and change in entropy (ΔS).
Database search to identify proteins that bind to the same ligand with different binding modes
To identify proteins where the same ligand bound to completely different amino acids, the complete database of 121,700 PDB structures was downloaded. Of these structures, 98,011 had no ligand bound to them, so these structures were eliminated. From the remaining structures, ligands were extracted, and a directory was created for each ligand. Approximately 23,700 structures were sorted into groups based on their ligand specificity. As we were interested in finding two proteins with dissimilar binding sites, only the ligands to which more than one protein bound were retained. Finally, this whole process resulted in the sorting of 6015 ligands. The majority of current methods use binding pocket geometry rather than the physicochemical properties or the microenvironment of the binding site; therefore, they may report the similar binding sites as dissimilar ligand-binding sites. For example, a replacement from Arg to Lys may be considered as a dissimilarity. Therefore, we chose the pocket feature algorithm, which considers the similarity of the binding site as a function of shared microenvironments rather than the geometric orientations of the amino acids in the binding site. “Microenvironment” refers to residues around a given center within a radius value of 7.5 Å. This feature calculates the microenvironment in protein A and B; further, the Tanimoto coefficient was calculated based on similar properties (53). A score of 7 or greater signifies similar binding pockets, whereas scores between 0 and 1 signify dissimilar binding sites. We selected a score of 0 to identify completely dissimilar sites. All ligands with a molecular mass less than 110 Da were deleted, including the proteins that bound to these molecules. Among these, ligands bound to sites other than the active site were also ignored. The resulting structures were then visually examined.
Accession numbers
Structure factors and coordinates of Hd-SatA unliganded, Hd-SatA–Neu5Ac, and Hd-SatA–Neu5Gc structures have been deposited in Protein Data Bank with accession codes 5ZA4, 5Z99, and 5YYB, respectively.
Author contributions
T. G. S., J. C. M., G. A. H., S. P. M., and R. S. designed the research and performed all experiments. M. A. A and R. S. M. conceived the project and helped design experiments and interpret the data. T. G. S., S. P. M., and J. C. M. prepared constructs and did expression studies of the protein. Selenomethionine protein purifications, crystallization, and data collection were carried out by G. A. H. Data processing was done by T. G. S. and S. R. ITC experiments were done by T. G. S. and S. P. M. Modeling studies were done by S. S. Results were discussed with all of the authors. T. G. S. and R. S. wrote the manuscript with contributions from coauthors.
Supplementary Material
Acknowledgments
We thank the ESRF Access Program of RCB (supported by Department of Biotechnology Grant BT/INF/22/SP22660/2017). We thank the Advance Photon Source (IMCA-CAT beamline 17-ID) and Advanced Light Source (beamline 4.2.2, MBC) for providing the beam time for this project. The NCBS X-ray Facility was supported by Grant BT/PR5081/INF/156/2012 from the DBT.
This work was supported by Department of Biotechnology (DBT)-Indo Swedish Grant BT/IN/SWEDEN/06/SR/2017, DBT-B-life Grant BT/PR12422/MED/31/287/214, and a grant from the Department of Science and Technology (DST) (to S. R.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (codes 5ZA4, 5Z99, and 5YYB) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Neu5Ac
- N-acetylneuraminic acid
- Neu5Gc
- N-glycolylneuraminic acid
- SatA
- sialic acid transport A gene
- Hd-SatA
- H. ducreyi sialic acid transport A gene
- SiaP
- sialic acid–binding protein
- TRAP
- tripartite ATP-independent periplasmic
- ITC
- isothermal calorimetry
- LOS
- lipooligosaccharide
- PDB
- Protein Data Bank
- MBP
- maltose-binding protein.
References
- 1. De Las Rivas J., and Fontanillo C. (2010) Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 6, e1000807 10.1371/journal.pcbi.1000807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuzmanov U., and Emili A. (2013) Protein-protein interaction networks: probing disease mechanisms using model systems. Genome Med. 5, 37 10.1186/gm441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawson T., and Nash P. (2000) Protein-protein interactions define specificity in signal transduction. Genes Dev. 14, 1027–1047 [PubMed] [Google Scholar]
- 4. Sevimoglu T., and Arga K. Y. (2014) The role of protein interaction networks in systems biomedicine. Comput. Struct. Biotechnol. J. 11, 22–27 10.1016/j.csbj.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saini R. D. (2017) Thermodynamics of protein-ligand interactions and their analysis. J. Proteins Proteomics 8, 205–217 [Google Scholar]
- 6. Du X., Li Y., Xia Y. L., Ai S. M., Liang J., Sang P., Ji X. L., and Liu S. Q. (2016) Insights into protein–ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 17, E144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schauer R., Srinivasan G. V., Wipfler D., Kniep B., and Schwartz-Albiez R. (2011) O-Acetylated sialic acids and their role in immune defense. Adv. Exp. Med. Biol. 705, 525–548 10.1007/978-1-4419-7877-6_28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vimr E. R., Kalivoda K. A., Deszo E. L., and Steenbergen S. M. (2004) Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68, 132–153 10.1128/MMBR.68.1.132-153.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayakawa T., Satta Y., Gagneux P., Varki A., and Takahata N. (2001) Alu-mediated inactivation of the human CMP-N-acetylneuraminic acid hydroxylase gene. Proc. Natl. Acad. Sci. U.S.A. 98, 11399–11404 10.1073/pnas.191268198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Varki A. (2010) Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc. Natl. Acad. Sci. U.S.A. 107, 8939–8946 10.1073/pnas.0914634107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varki A., and Schauer R. (2009) Sialic acids. in Essentials of Glycobiology, 2nd Ed (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E., eds) pp. 1–17, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 12. Varki A. (2008) Sialic acids in human health and disease. Trends Mol. Med. 14, 351–360 10.1016/j.molmed.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hood D. W., Cox A. D., Gilbert M., Makepeace K., Walsh S., Deadman M. E., Cody A., Martin A., Månsson M., Schweda E. K., Brisson J. R., Richards J. C., Moxon E. R., and Wakarchuk W. W. (2001) Identification of a lipopolysaccharide α-2,3-sialyltransferase from Haemophilus influenzae. Mol. Microbiol. 39, 341–350 10.1046/j.1365-2958.2001.02204.x [DOI] [PubMed] [Google Scholar]
- 14. Hood D. W., Makepeace K., Deadman M. E., Rest R. F., Thibault P., Martin A., Richards J. C., and Moxon E. R. (1999) Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33, 679–692 10.1046/j.1365-2958.1999.01509.x [DOI] [PubMed] [Google Scholar]
- 15. Apicella M. A. (2012) Nontypeable Haemophilus influenzae: the role of N-acetyl-5-neuraminic acid in biology. Front. Cell. Infect. Microbiol. 2, 19 10.3389/fcimb.2012.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Severi E., Hood D. W., and Thomas G. H. (2007) Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822 10.1099/mic.0.2007/009480-0 [DOI] [PubMed] [Google Scholar]
- 17. Almagro-Moreno S., and Boyd E. F. (2009) Sialic acid catabolism confers a competitive advantage to pathogenic vibrio cholerae in the mouse intestine. Infect. Immun. 77, 3807–3816 10.1128/IAI.00279-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gangi Setty T., Cho C., Govindappa S., Apicella M. A., and Ramaswamy S. (2014) Bacterial periplasmic sialic acid-binding proteins exhibit a conserved binding site. Acta Crystallogr. D Biol. Crystallogr. 70, 1801–1811 10.1107/S139900471400830X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnston J. W., Coussens N. P., Allen S., Houtman J. C. D., Turner K. H., Zaleski A., Ramaswamy S., Gibson B. W., and Apicella M. A. (2008) Characterization of the N-acetyl-5-neuraminic acid-binding site of the extracytoplasmic solute receptor (SiaP) of nontypeable Haemophilus influenzae strain 2019. J. Biol. Chem. 283, 855–865 10.1074/jbc.M706603200 [DOI] [PubMed] [Google Scholar]
- 20. Müller A., Severi E., Mulligan C., Watts A. G., Kelly D. J., Wilson K. S., Wilkinson A. J., and Thomas G. H. (2006) Conservation of structure and mechanism in primary and secondary transporters exemplified by SiaP, a sialic acid binding virulence factor from Haemophilus influenzae. J. Biol. Chem. 281, 22212–22222 10.1074/jbc.M603463200 [DOI] [PubMed] [Google Scholar]
- 21. Felder C. B., Graul R. C., Lee A. Y., Merkle H. P., Sadee W., and Chemistry P. (1999) The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS Pharmsci. 1, E2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oh B. H., Pandit J., Kang C. H., Nikaido K., Gokcen S., Ames G. F., and Kim S. H. (1993) Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without. J. Biol. Chem. 268, 11348–11355 [PubMed] [Google Scholar]
- 23. Thomas G. H. (2016) Sialic acid acquisition in bacteria: one substrate, many transporters. Biochem. Soc. Trans. 44, 760–765 10.1042/BST20160056 [DOI] [PubMed] [Google Scholar]
- 24. Haines-Menges B. L., Whitaker W. B., Lubin J. B., and Boyd E. F. (2015) Host sialic acids: a delicacy for the pathogen with discerning taste. Microbiol. Spectr. 3, 10.1128/microbiolspec.MBP-0005-2014 10.1128/microbiolspec.MBP-0005-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spinola S. M., Bauer M. E., and Munson R. S. (2002) Immunopathogenesis of Haemophilus ducreyi infection (Chancroid). Infect. Immunity 70, 1667–1676 10.1128/IAI.70.4.1667-1676.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cohen M. S. (1998) Sexually transmitted diseases enhance HIV transmission: no longer a hypothesis. Lancet 351, Suppl. 3, 5–7; Correction (1998) Lancet352, 2026 10.1016/S0140-6736(98)90002-2 [DOI] [PubMed] [Google Scholar]
- 27. Inamadar A. C., and Palit A. (2002) Chancroid: an update. Indian J. Dermatol. Venereol. Leprol. 68, 5–9 [PubMed] [Google Scholar]
- 28. Alfa M. J., DeGagne P., and Totten P. A. (1996) Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect. Immun. 64, 2349–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wood G. E., Dutro S. M., and Totten P. A. (1999) Target cell range of Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun. 67, 3740–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cortes-Bratti X., Chaves-Olarte E., Lagergård T., and Thelestam M. (1999) The cytolethal distending toxin from the chancroid bacterium Haemophilus ducreyi induces cell-cycle arrest in the G2 phase. J. Clin. Invest. 103, 107–115 10.1172/JCI3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ceelen L. M., Decostere A., Ducatelle R., and Haesebrouck F. (2006) Cytolethal distending toxin generates cell death by inducing a bottleneck in the cell cycle. Microbiol. Res. 161, 109–120 10.1016/j.micres.2005.04.002 [DOI] [PubMed] [Google Scholar]
- 32. Cope L. D., Lumbley S., Latimer J. L., Klesney-Tait J., Stevens M. K., Johnson L. S., Purven M., Munson R. S. Jr., Lagergard T., Radolf J. D., and Hansen E. J. (1997) A diffusible cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. U.S.A. 94, 4056–4061 10.1073/pnas.94.8.4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alfa M. J., and DeGagne P. (1997) Attachment of Haemophilus ducreyi to human foreskin fibroblasts involves LOS and fibronectin. Microb. Pathog. 22, 39–46 10.1006/mpat.1996.0089 [DOI] [PubMed] [Google Scholar]
- 34. Gibson B. W., Campagnari A. A., Melaugh W., Phillips N. J., Apicella M. A., Grass S., Wang J., Palmer K. L., and Munson R. S. (1997) Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J. Bacteriol. 179, 5062–5071 10.1128/jb.179.16.5062-5071.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Melaugh W., Campagnari A. A., and Gibson B. W. (1996) The lipooligosaccharides of Haemophilus ducreyi are highly sialylated. J. Bacteriol. 178, 564–570 10.1128/jb.178.2.564-570.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Post D. M. B., Mungur R., Gibson B. W., and Munson R. S. Jr. (2005) Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect. Immun. 73, 6727–6735 10.1128/IAI.73.10.6727-6735.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hollenstein K., Frei D. C., and Locher K. P. (2007) Structure of an ABC transporter in complex with its binding protein. Nature 446, 213–216 10.1038/nature05626 [DOI] [PubMed] [Google Scholar]
- 38. Oldham M. L., Davidson A. L., and Chen J. (2008) Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18, 726–733 10.1016/j.sbi.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davidson A. L., Dassa E., Orelle C., and Chen J. (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364, table of contents 10.1128/MMBR.00031-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dunten P., and Mowbray S. L. (1995) Crystal structure of the dipeptide binding protein from Escherichia coli involved in active transport and chemotaxis. Protein Sci. 4, 2327–2334 10.1002/pro.5560041110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Levdikov V. M., Blagova E. V., Brannigan J. A., Wright L., Vagin A. A., and Wilkinson A. J. (2005) The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J. Mol. Biol. 345, 879–892 10.1016/j.jmb.2004.10.089 [DOI] [PubMed] [Google Scholar]
- 42. El Sahili A., Li S. Z., Lang J., Virus C., Planamente S., Ahmar M., Guimaraes B. G., Aumont-Nicaise M., Vigouroux A., Soulère L., Reader J., Queneau Y., Faure D., and Moréra S. (2015) A pyranose-2-phosphate motif is responsible for both antibiotic import and quorum-sensing regulation in Agrobacterium tumefaciens. PLOS Pathog. 11, e1005071 10.1371/journal.ppat.1005071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vergauwen B., Elegheert J., Dansercoer A., Devreese B., and Savvides S. N. (2010) Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA. Proc. Natl. Acad. Sci. U.S.A. 107, 13270–13275 10.1073/pnas.1005198107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomson J., Liu Y., Sturtevant J. M., and Quiocho F. A. (1998) A thermodynamic study of the binding of linear and cyclic oligosaccharides to the maltodextrin-binding protein of Escherichia coli. Biophys. Chem. 70, 101–108 10.1016/S0301-4622(97)00113-0 [DOI] [PubMed] [Google Scholar]
- 45. Fukami-Kobayashi K., Tateno Y., and Nishikawa K. (1999) Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290 10.1006/jmbi.1998.2454 [DOI] [PubMed] [Google Scholar]
- 46. Berntsson R. P., Smits S. H. J., Schmitt L., Slotboom D., and Poolman B. (2010) A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 [DOI] [PubMed] [Google Scholar]
- 47. Maqbool A., Horler R. S. P., Muller A., Wilkinson A. J., Wilson K. S., and Thomas G. H. (2015) The substrate-binding protein in bacterial ABC transporters: dissecting roles in the evolution of substrate specificity. Biochem. Soc. Trans. 43, 1011–1017 10.1042/BST20150135 [DOI] [PubMed] [Google Scholar]
- 48. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., and Liang J. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 10.1093/nar/gkl282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Doublié S., Kapp U., Aberg A., Brown K., Strub K., and Cusack S. (1996) Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 384, 219–221 10.1016/0014-5793(96)00316-X [DOI] [PubMed] [Google Scholar]
- 50. Terwilliger T. (2004) SOLVE and RESOLVE: automated structure solution, density modification, and model building research papers. J. Synchrotron Radiat. 11, 49–52 10.1107/S0909049503023938 [DOI] [PubMed] [Google Scholar]
- 51. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Houtman J. C. D., Brown P. H., Bowden B., Yamaguchi H., Appella E., Samelson L. E., and Schuck P. (2007) Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 10.1110/ps.062558507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu T., and Altman R. B. (2011) Using multiple microenvironments to find similar ligand-binding sites: application to kinase inhibitor binding. PLoS Comput. Biol. 7, e1002326 10.1371/journal.pcbi.1002326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. DeLano W. L. (2018) The PyMol Molecular Graphics System, version 2.1, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.