Abstract
The linear ubiquitin chain assembly complex (LUBAC) regulates NF-κB activation by modifying proteins with linear (M1-linked) ubiquitination chains. Although LUBAC also regulates the apoptosis pathway, the precise mechanism by which LUBAC regulates apoptosis remains not fully defined. Here, we report that LUBAC-mediated M1-linked ubiquitination of cellular FLICE-like inhibitory protein (cFLIP), an anti-apoptotic molecule, contributes to tumor necrosis factor (TNF) α-induced apoptosis. We found that deficiency of RNF31, the catalytic subunit of the LUBAC complex, promoted cFLIP degradation in a proteasome-dependent manner. Moreover, we observed RNF31 directly interact with cFLIP, and LUBAC further conjugated M1-linked ubiquitination chains at Lys-351 and Lys-353 of cFLIP to stabilize cFLIP, thereby protecting cells from TNFα-induced apoptosis. Together, our study identifies a new substrate of LUBAC and reveals a new molecular mechanism through which LUBAC regulates TNFα-induced apoptosis via M1-linked ubiquitination.
Keywords: apoptosis, ubiquitylation (ubiquitination), tumor necrosis factor (TNF), caspase, cell death, cFLIP, linear ubiquitination, LUBAC, RNF31
Introduction
Tumor necrosis factor (TNF)3α is a cytokine that plays roles in various cellular processes, such as proliferation, differentiation, and death. It has been reported that it mainly activates two different signaling pathways: nuclear factor (NF)-κB activation and cell death (1, 2). Once TNFα binds to its receptor, signaling molecules such as the TNF receptor 1-associated DEATH domain, receptor-interacting serine/threonine-protein kinase 1 (RIPK1), and TNF receptor-associated factors (TRAFs) are recruited and form the receptor-signaling complex. This complex induces the activation of the IκB kinase (IKK) complex, which further induces the degradation of inhibitor of κB-α (IκB-α) and translocation of NF-κB transcriptional factors, sequentially (3). At the same time, RIPK1/FAS-associated death domain (FADD)-caspase 8 complex is assembled to activate apoptosis (4, 5). When caspase 8 activity is inhibited, RIPK1 can form a complex with RIPK3 to trigger necroptosis (6–8). Because the cell-death process is critical for homeostasis, it is tightly regulated by various inhibitory mechanisms. For example, cellular FLICE-like inhibitory protein (cFLIP) interacts and forms a heterodimer with caspase 8, inhibiting activation of caspase 8 and apoptosis signaling (9). In addition, B-cell lymphoma 2 (BCL2) family proteins and inhibitor of apoptosis proteins (cIAPs) directly and indirectly suppress caspases activation (10–12).
Ubiquitination is a key posttranslational modification (PTM) in TNFα-induced signaling. For example, Lys-48–linked ubiquitination regulates degradation of IκBα (13) and FADD (14), and Lys-63–linked ubiquitination of NEMO, RIPK1, and TRAFs triggers the functional activation of these molecules (15), which further recruit downstream molecules. Recently, researchers discovered M1-linked ubiquitination to be a novel type of ubiquitination involved in the TNFα signaling pathway, especially NF-κB signaling (16). To date, linear ubiquitin chain assembly complex (LUBAC) was identified as the only ligase complex for M1-linked ubiquitination. LUBAC has three components: RNF31 (also named as HOIP), HOIL-1(also named as HOIL-1L or RBCK1), and Sharpin. In particular, RNF31 has a major role in activation of NF-κB signaling by conjugating M1-linked ubiquitination chains with NEMO and RIPK1, whereas HOIL-1 and Sharpin are involved in the functional activation of RNF31 (17). Genetic studies demonstrated that defects in LUBAC components attenuate TNFα-induced NF-κB activation and gene expression (18–20). In addition to TNFα, LUBAC regulates interleukin-1, CD40-, lymphotoxin β-, Toll-like receptor-, T cell receptor-, and nucleotide-binding oligomerization domain containing 2 (NOD2)-mediated cellular events through regulating the NF-κB pathway (21–23). Recent studies reported M1-linked ubiquitination to have an NF-κB-independent function as well (24, 25). Additionally, biochemical and mouse developmental data have suggested that LUBAC has a role in apoptosis signaling. For example, Sharpin-deficient mouse embryonic fibroblasts were highly susceptible to TNFα-induced cell death (20), and cell death-dependent skin inflammation was observed in Sharpin-deficient mice (26, 27). RNF31 or HOIL-1-knockout (KO) mice are embryonic lethal due to massive cell death (28–30). However, the exact molecular mechanism of M1-linked ubiquitination in TNFα-mediated cell death remains largely unknown. Because the deregulation of cell survival and death is the main cause of many diseases such as cancer and autoimmune disease, the clear understanding of cell death regulation is critical for the therapeutic strategies to control these diseases.
In the present study, we determined the molecular mechanism of RNF31 in the regulation of apoptosis pathway. We identified cFLIP as a new substrate of LUBAC, and found that LUBAC conjugates M1-linked ubiquitination to cFLIP, and stabilizes it to protect cells from TNFα-induced apoptosis. These findings provide molecular insight into the regulatory mechanism of apoptosis by M1-linked ubiquitination, and modulating this mechanism may be a novel therapeutic approach for diseases oriented from deregulation of cell death.
Results
Deficiency of RNF31 promotes cFLIP degradation in TNFα-induced apoptosis
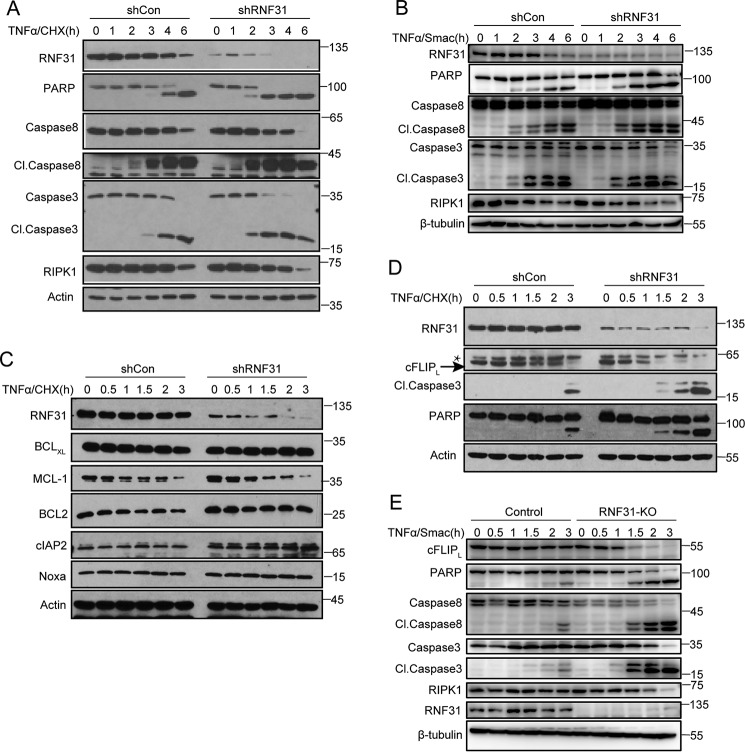
Previous studies have shown that loss of LUBAC components could promote TNFα-induced apoptosis (18, 28, 29). But it is unclear whether LUBAC directly regulates apoptosis. First, we generated stable HeLa cells in which RNF31 was silenced using a short hairpin RNA (shRNA) system and compared them with control HeLa cells. After TNFα plus cycloheximide (CHX) stimulation, RNF31-silenced cells were highly sensitive to apoptosis, which were indicated by cleavage of caspase 3/8 and PARP (Fig. 1A and Fig. S1A). Smac is a mitochondrial protein that can be released into the cytosol to promote caspase activation in the cytochrome c/Apaf-1/caspase 9 pathway (31). TNFα plus Smac mimetics stimulation also triggered more apoptosis in RNF31-silenced cells (Fig. 1B and Fig. S1B). To elucidate the molecular mechanism by which RNF31 regulates the apoptosis pathway, we examined the levels of anti-apoptotic molecules in TNFα-treated HeLa cells. RNF31 silencing did not alter the level of most of anti-apoptotic molecules, including BCL2 family proteins (BCLxL, BCL2, and NOXA), MCL-1 and cIAP2 (Fig. 1C), but we found that the level of cFLIP decreased markedly in RNF31-silenced HeLa cells upon treatment with TNFα and CHX (Fig. 1D and Fig. S1C). To confirm this result, we generated RNF31 knockout cells by CRISPR-Cas9 technique. After TNFα and Smac mimetics stimulation, cFLIP also degraded faster in RNF31 knockout cells (Fig. 1E and Fig. S1D). Above all, deficiency of RNF31 could promote TNFα-induced apoptosis and accelerate the degradation of cFLIP.
Figure 1.
cFLIP degrades faster in RNF31-deficient cells. A, Western blot analysis of the indicated proteins in control and RNF31-silenced HeLa cells after treatment with TNFα and CHX (10 ng/ml and 10 μg/ml, respectively) for the indicated times. B, Western blot analysis of the indicated proteins in control and RNF31-silenced HeLa cells after treatment with TNFα/Smac mimetics (20 ng/ml and 5 μm, respectively) for the indicated times. C and D, Western blot analysis of lysates of control and RNF31-silenced HeLa cells stimulated with TNFα and CHX (10 ng/ml and 10 μg/ml, respectively) for the indicated times. *, nonspecific band. E, Western blot analysis of lysates of control and RNF31-deleted Jurkat cells stimulated with TNFα/Smac mimetics (20 ng/ml and 5 μm, respectively) for the indicated periods.
Decrease of cFLIP in RNF31-deficient cells results from proteasome-dependent manner
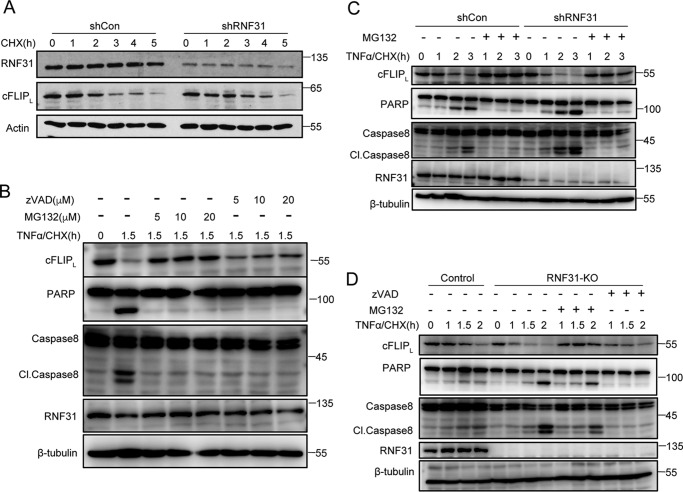
To determine whether the rapid decrease of cFLIP in RNF31-deficient cells depended on activation of TNFα signaling, we treated control and RNF31-silenced HeLa cells with CHX only. We observed a similar pattern of decreased cFLIP in control and RNF31-silenced cells upon CHX treatment (Fig. 2A), indicating that loss of RNF31 did not alter the basal turnover rate for cFLIP. Previous studies reported that treatment with the proteasome inhibitor bortezomib reduced the severity of skin problems in Sharpin-deficient mice (32), suggesting that deficiency of LUBAC could result in degradation of some molecules. cFLIP could be ubiquitinated and led to proteasome-dependent degradation (33, 34). Furthermore, cFLIP also was identified as a substrate of caspase 8 and can be cleaved by activated caspase 8 during apoptosis (9). So, we pre-treated HeLa cells with caspase inhibitor Z-VAD-FMK or the proteasome inhibitor MG132 to determine whether the decreased cFLIP resulted from caspase activity or occurred in a degradation-dependent manner. We found that pre-treatment with Z-VAD-FMK fully inhibited cleavage of PARP and caspase 8 but only slightly restored the level of cFLIP (Fig. 2B and Fig. S2A). However, pre-treatment with MG132 fully blocked the decrease of cFLIP, cleavage of PARP and caspase 8(Fig. 2B and Fig. S2A), suggesting that the TNFα/CHX-induced decrease of cFLIP mainly results from the proteasome-dependent degradation.
Figure 2.
cFLIP degrades in RNF31-deficient cells through proteasome-dependent process. A, Western blot analysis of lysates of CHX-treated (20 μg/ml) control and RNF31 silenced HeLa cells. B, Western blot analysis of HeLa cells pretreated with different dose of Z-VAD-FMK or MG132 and then stimulated with TNFα and CHX (10 ng/ml and 10 μg/ml, respectively) for the indicated periods. C, Western blot analysis of control and RNF31-silenced HeLa cells pretreated with MG132 (10 μm) for 1.5 h and then stimulated with TNFα and CHX (10 ng/ml and 10 μg/ml, respectively) for the indicated periods. D, Western blot analysis of control and RNF31-deleted Jurkat cells pretreated with Z-VAD-FMK (10 μm) or MG132 (10 μm) and then stimulated with TNFα and CHX (10 ng/ml and 10 μg/ml, respectively) for the indicated periods.
To confirm the role of proteasome-mediated degradation in the increased sensitivity of RNF31-silenced cells to apoptosis, we pre-treated control and RNF31-silenced HeLa cells with MG132 and then induced apoptosis with stimulation of TNFα plus CHX. Pre-treatment with MG132 completely blocked the decrease of cFLIP in RNF31-silenced cells (Fig. 2C and Fig. S2B), and the apoptosis sensitization in RNF31-silencing cells was also fully blocked (Fig. 2C), indicating that the sensitization of RNF31-silencing cells to apoptosis is mainly mediated by a proteasome-dependent degradation pathway. Moreover, in RNF31 knockout cells, degradation of cFLIP is fully suppressed by MG132 treatment, but not by Z-VAD-FMK treatment (Fig. 2D and Fig. S2C). Taken together, these data suggested that deficiency of RNF31 promotes cFLIP degradation in a proteasome-dependent manner.
RNF31 interacts with cFLIP
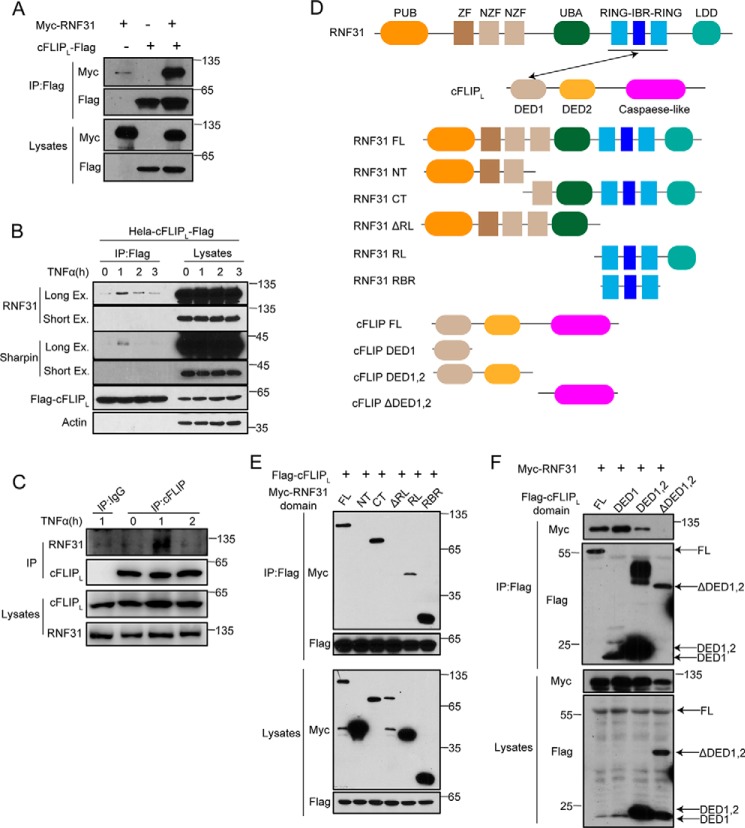
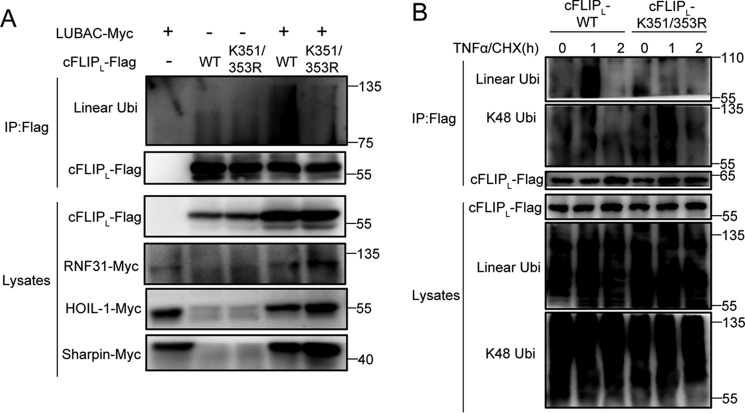
Next, we examined whether RNF31 have a direct interaction with cFLIP. We expressed Myc-tagged RNF31 together with FLAG-tagged cFLIP in HEK293T cells, and found that RNF31 were co-precipitated with cFLIP (Fig. 3A and Fig. S2D). In addition, stimulation of HeLa cells stably expressing FLAG-tagged cFLIP with TNFα led to inducible interaction between cFLIP and LUBAC components, including RNF31 and Sharpin (Fig. 3B). Similarly, TNFα induces endogenous interaction between RNF31 and cFLIP (Fig. 3C). RNF31 and cFLIP are composed of multiple domains (Fig. 3D). Therefore, we determined which domains in cFLIP and RNF31 are responsible for their interaction with each other. The domain mapping by expressing full-length cFLIP together with different RNF31 domains indicated that the RING-IBR-RING (RBR) domain of RNF31 is essential for its binding with cFLIP (Fig. 3E). Furthermore, a co-immunoprecipitation experiment with full-length RNF31 and constructs encoding different cFLIP domains demonstrated that death effector domain 1 (DED1) of cFLIP is critical for the interaction between RNF31 and cFLIP (Fig. 3F). Taken together, these data suggested that the RBR domain of RNF31 and DED1 domain of cFLIP are responsible for their interaction.
Figure 3.
RNF31 interacts with cFLIP. A, Western blot analysis of immunoprecipitates from 293T cells transiently transfected with the indicated plasmids using anti-FLAG beads. B, Western blot analysis of immunoprecipitates from HeLa cells stably expressing FLAG-cFLIP after stimulation with TNFα (20 ng/ml) for the indicated times using anti-FLAG beads. C, Western blot analysis of immunoprecipitates from Jurkat cells after stimulation with TNFα (20 ng/ml) for the indicated times using cFLIP antibody-conjugated beads, and IgG as control. D, schematic of domains of RNF31 and cFLIP and their truncated mutants. E and F, Western blot analysis of immunoprecipitates using anti-FLAG beads and total lysates of 293T cells transfected with the plasmids encoding the indicated domains. FL, full-length; NT, N-terminal fragment; CT, C-terminal fragment; ΔRL, RBR-LDL domain-deleted fragment; RL, RBR-LDL domain; RBR, RBR domain; DED1, DED1 domain; DED1,2, DED1 and DED2 domain; ΔDED1,2, DED1 and DED2 domain-deleted fragment.
RNF31 regulates cFLIP stability through M1-linked ubiquitination
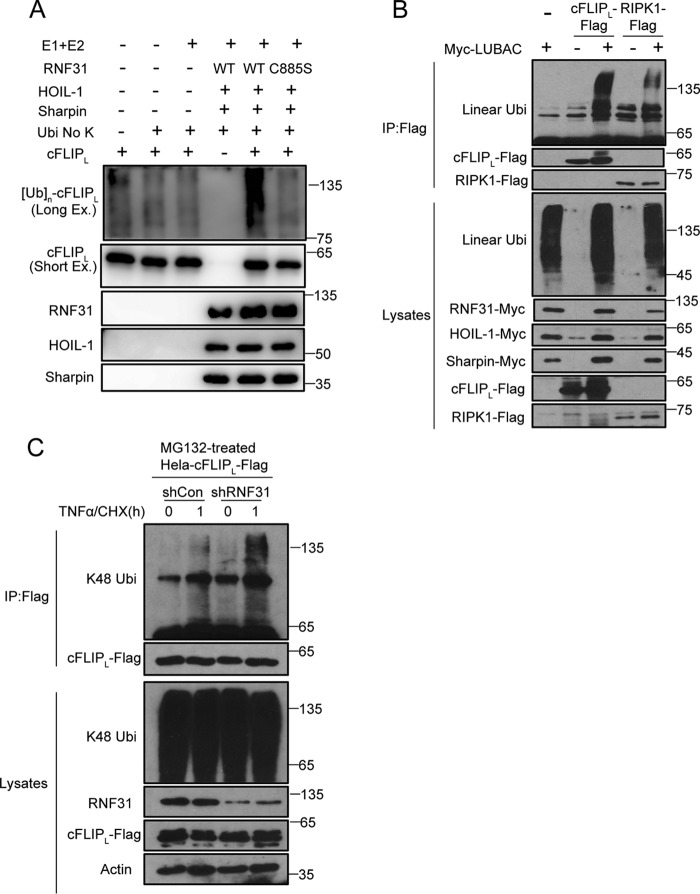
RNF31 directly interact with cFLIP, indicating that cFLIP may be a substrate of LUBAC components. To test our hypothesis, we performed an in vitro ubiquitination assay to determine whether cFLIP is a substrate of LUBAC. We incubated recombinant cFLIP with or without recombinant LUBAC, E1, E2, and lysine KO ubiquitin (all lysine residues are mutated to arginine), and then detected M1-linked ubiquitination of cFLIP. The results showed LUBAC conjugates M1-linked ubiquitination chains to cFLIP (Fig. 4A). However, catalytically dead mutant C885S of RNF31 failed to induce M1-linked ubiquitination chains to cFLIP (Fig. 4A), suggesting that the catalytic activity of RNF31 is critical for the M1-linked ubiquitination of cFLIP. Besides, expression of LUBAC promoted M1-linked ubiquitination of cFLIP as well as the known substrate RIPK1 in HEK293T cells (Fig. 4B). Because cFLIP degrades rapidly in RNF31-deficient cells through a proteasome-dependent manner, we hypothesized that M1-linked ubiquitination stabilizes cFLIP by competing with Lys-48–linked ubiquitination, thereby suppressing proteasome-mediated degradation of cFLIP. We detected the Lys-48–linked ubiquitination of cFLIP in control and RNF31-silenced HeLa cells introduced with FLAG-cFLIP, and observed that silencing of RNF31 promoted Lys-48–linked ubiquitination of cFLIP at the endogenous level upon treatment with TNFα and CHX (Fig. 4C and Fig. S2E). These results were consistent with the results of MG132 treatment, and suggested that M1-linked ubiquitination of cFLIP can compete with Lys-48 ubiquitination to stabilize cFLIP.
Figure 4.
M1-linked ubiquitination of cFLIP inhibits Lys-48-linked ubiquitination. A, LUBAC-induced in vitro ubiquitination of cFLIP. The recombinant FLAG-cFLIP was incubated as indicated and probed with anti-cFLIP antibody. E1 is UBE1, and E2 is UbcH5c. Long Ex, long exposure time; Short Ex, short exposure time. B, Western blot analysis of immunoprecipitates (IP) from 293T cells transiently transfected with the indicated constructs using anti-FLAG beads. C, Western blot analysis of immunoprecipitates from lysates of control and RNF31-silenced HeLa cells stably expressing FLAG-cFLIP after pretreatment with MG132 followed by treatment with TNFα and CHX (20 ng/ml and 10 μg/ml, respectively) for 1 h using anti-FLAG beads.
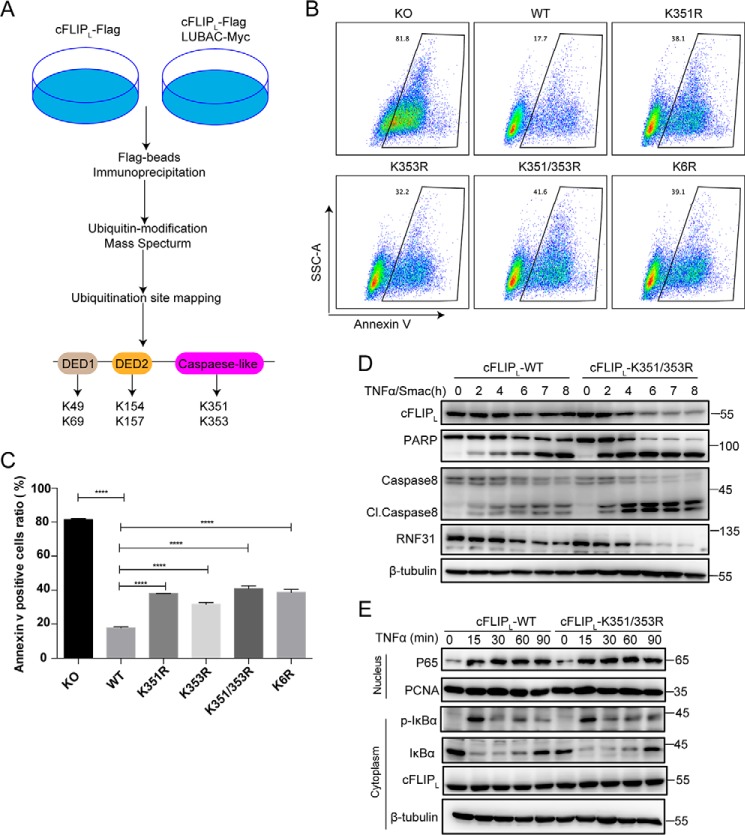
Lys-351 and Lys-353 in cFLIP are the functional sites for M1-linked ubiquitination
To determine which residues in cFLIP are the sites for RNF31-mediated ubiquitination and contribute to the regulation of apoptosis, we performed LC and MS to find the sites of M1-linked ubiquitination on cFLIP by LUBAC (Fig. 5A). We co-expressed FLAG-tagged cFLIP with LUBAC complex in HEK293T cells, and expression of cFLIP alone as control. Using FLAG antibody-conjugated beads to pull down the cFLIP protein, and then detected ubiquitination sites on cFLIP by mass spectrometry. Finally, six candidate ubiquitination sites that were dependent on the expression of LUBAC were detected (Lys-49, Lys-69, Lys-154, Lys-157, Lys-351, and Lys-353) (Fig. 5A).
Figure 5.
K351R and K353R mutations in cFLIP promotes apoptosis and cFLIP degradation. A, schematic approach to identify M1-linked ubiquitination sites on cFLIP. B, the representative pictures of FACS results. cFLIP KO Jurkat cells were reconstituted with WT and ubiquitination site mutants of cFLIP. After being stimulated with TNFα/Smac mimetics (20 ng/ml and 5 μm, respectively) for 5 h, apoptotic cells were analyzed by the annexin V staining and FACS. K6R refers to K49R/K69R/K154R/K157R/K351R/K353R. C, the statistical results of B, and statistical significance was determined using two-tailed unpaired t test (n = 3, ****, p < 0.0001). D, Western blot analysis of the indicated proteins in cFLIP-WT and cFLIP-K351R/K353R mutant Jurkat cells after treatment with TNFα/Smac mimetics (20 ng/ml and 5 μm, respectively) for the indicated times. E, Western blot analysis of the cytoplasmic and nuclear proteins in cFLIP-WT and cFLIP-K351R/K353R mutant Jurkat cells after treatment with TNFα (20 ng/ml) for the indicated times.
Because the above data suggested that M1-linked ubiquitination of cFLIP can protect cells from TNFα-induced apoptosis, we hypothesized that mutations of these M1-linked ubiquitination sites on cFLIP should promote TNFα-induced apoptosis. We replaced those identified lysine residues with arginine (K49R, K69R, K154R, K157R, K351R, K353R) in different combinations, and expressed these cFLIP mutants in cFLIP KO Jurkat cells generated by the CRISPR-Cas9 method (Fig. S3, A–C). We then monitored the sensitivity of these cells to TNFα/CHX-induced apoptosis. The results showed that cFLIP KO cells reconstituted with cFLIP-K351R/K353R double mutations were more sensitive to TNFα/CHX-induced apoptosis than cells reconstituted with other cFLIP mutants, but comparable with cells expressing the cFLIP mutant with all six lysine residues converted to arginine (simplify to K6R) (Fig. S3D), indicating that Lys-351 and Lys-353 are the main M1-linked ubiquitination sites contributing to the protection of cells from TNFα-induced apoptosis. To further determine which M1-linked ubiquitination site in cFLIP is functionally important for preventing TNFα-induced apoptosis, we generated cFLIP mutants with K351R or K353R alone and expressed these mutants in cFLIP KO cells. We found that cells expressing K351R or K353R enhance both the TNFα/Smac mimetics and TNFα/CHX-induced apoptosis compared with WT cFLIP-reconstituted cells (Fig. 5, B and C, and Fig. S3, E and F). These data indicated that Lys-351 and Lys-353 may play a redundant role in protecting cells from TNFα-induced apoptosis. Consistent with the annexin V staining, we found that more and earlier cleavage of PARP and caspase 8 in cFLIP KO cells reconstituted with K351R/K353R double mutant (Fig. 5D), and mutated cFLIP also degraded faster than WT cFLIP (Fig. 5D). Finally, we monitored the NF-κB activation in K351R/K353R double-mutated cells. The results showed that K351R/K353R mutation of cFLIP did not affect TNFα-induced NF-κB activation, indicated by phosphorylation of IκBα, degradation of IκBα, and translocated p65 in the nucleus (Fig. 5E). These results suggested that linear ubiquitination of cFLIP protected the cell from TNFα-induced apoptosis through a NF-κB-independent manner.
To further confirm whether Lys-351 and Lys-353 are the main M1-linked ubiquitination sites of cFLIP, we performed the ubiquitin assay in HEK293T cells. The results also showed that mutation of K351R/K353R significantly blocked M1-linked ubiquitination of cFLIP compared with WT cFLIP (Fig. 6A). Because we proposed that M1-linked ubiquitination of cFLIP could compete Lys-48–linked ubiquitination, we detected M1-ubiquitination and Lys-48–linked ubiquitination of WT and mutated cFLIP in reconstituted Jurkat cells. After TNFα/CHX treatment, the M1-linked ubiquitination of cFLIP was significantly inhibited in cFLIP-K351R/K353R cells, but Lys-48–linked ubiquitination was significantly increased (Fig. 6B). Together, these results indicate that M1-linked ubiquitination of cFLIP on Lys-351 and Lys-353 competes with Lys-48-linked ubiquitination, which contributes to protect cells from TNFα-induced apoptosis (Fig. 7).
Figure 6.
Lys-351 and Lys-353 in cFLIP are the two main M1-linked ubiquitination sites. A, Western blot analysis of immunoprecipitates (IP) from 293T cells transiently transfected with the indicated constructs using anti-FLAG beads. B, Western blot analysis of immunoprecipitates from lysates of cFLIP-WT and cFLIP-K351/353R mutant Jurkat cells after pretreatment with MG132 (10 μm) for 1.5 h followed by treatment with TNFα and CHX (20 ng/ml and 10 μg/ml, respectively) for the indicated times using anti-FLAG beads. Lys-48–linked and linear ubiquitin chains were distinguished by antibodies specifically against Lys-48–linked or linear ubiquitin chains.
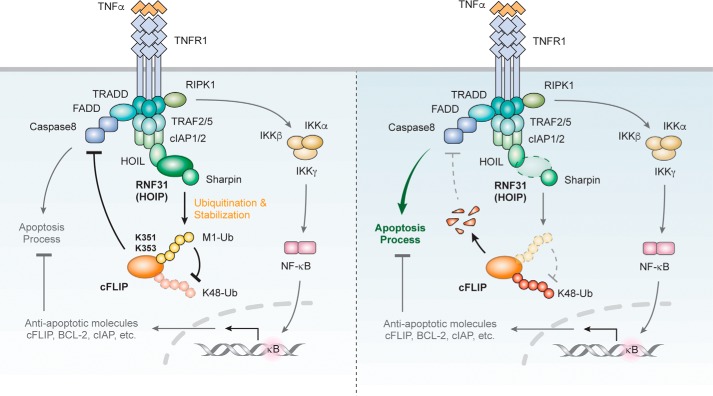
Figure 7.
Proposed model of LUBAC-mediated ubiquitination on cFLIP in the TNFα receptor signaling pathway. In normal conditions, linear ubiquitination of cFLIP induced by LUBAC inhibits Lys-48–linked ubiquitination of cFLIP, thereby suppressing caspase 8 activity and apoptosis process. When LUBAC is deficient, cFLIP cannot be ubiquitinated by LUBAC. Instead, cFLIP degrades through Lys-48–linked ubiquitination in a dependent manner, thereby promoting apoptosis process.
Discussion
In summary, we found that cFLIP was a new substrate of M1-linked ubiquitination and identified the target sites of this modification, revealing the mechanism of how LUBAC and M1-linked ubiquitination directly regulate apoptosis. Our findings also demonstrated that M1-linked ubiquitination stabilizes cFLIP by competing with the Lys-48 ubiquitination chains, suggesting dynamic regulation of posttranslational modifications by M1-linked ubiquitination. The present study provides insight into the role of M1-linked ubiquitination in the apoptosis pathway.
Previous studies suggested that LUBAC regulates the NF-κB signaling pathway by conjugating M1-linked ubiquitination chains to NEMO and RIPK1. But NF-κB activation was only slightly reduced in LUBAC-deficient mice or cells (18, 19, 28), indicating that LUBAC may directly regulate cell death upstream of the NF-κB signaling through affecting formation of the TNFR1 complex II. Consistently, deficiency of RNF31 led to more complex II formation (28). Besides, loss of complex II components in mice, including FADD, caspase 8, and cFLIP, could result in embryonic lethality around E10.5 (35–37), which were similar to RNF31 or HOIL-1 KO mice. In contrast, deficiency of NF-κB activation related molecules results in a much later lethality than RNF31 or HOIL-1 KO mice. For example, IKKβ- and IKKγ-deficient mice die around E14.5 (38), whereas IKKα-deficient mice die perinatally (39, 40). In this study, we reveal a new mechanism by which LUBAC directly regulates apoptosis through inducing M1-linked ubiquitination of cFLIP. Our study further supported that LUBAC can be an upstream regulator of cell death. Moreover, another study reported that M1-linked ubiquitination of NEMO and FADD are decreased in apoptosis process (41), and this may explain the limited function of the cFLIP-K351R/K353R mutant to promote TNFα-induced apoptosis in our study (Fig. 5, B and D). Therefore, LUBAC may directly regulate apoptosis by targeting multiple proteins, and our study identifies cFLIP as one of LUBAC targets that contributing to apoptosis process.
cFLIP is an anti-apoptotic molecule that forms a complex with caspase 8 and suppresses its protease activity (9). cFLIP can be cleaved by activated caspase 8 during apoptosis (9), and it is also actively modified by posttranslational modifications, such as ubiquitination, which leads to proteasome-dependent degradation. Proteasome inhibitors could protect cells from death receptor-mediated apoptosis via stabilization of cFLIP (42). Thus far, researchers have identified two different ubiquitination sites, Lys-167 and Lys-192/Lys-195 (33, 34), which are targets of Lys-48–linked ubiquitination. Our present results indicated that Lys-351 and Lys-353 in cFLIP are the targeted residues for M1-linked ubiquitination. M1-linked ubiquitination of these residues regulates cFLIP stability by competing with Lys-48-linked ubiquitination. However, we do not know whether both Lys-48- and M1-linked ubiquitination have identical cFLIP sites or which type of modification is more dominant. Furthermore, phosphorylation of cFLIP is a pre-requisite for Lys-48-linked ubiquitination; phosphorylation of threonine 166 is a pre-requisite for the ubiquitination of Lys-167 (34), and phosphorylation of serine 193 is a pre-requisite for ubiquitination of Lys-192/ Lys-195 (33). Therefore, phosphorylation of cFLIP may also regulate its M1-linked ubiquitination and stability. Further studies are required to address these questions.
cFLIP is highly expressed in the basal layer of the epidermis, and plays a pivotal role in the epidermis (43). Mice with conditional deletion of cFLIP in the epidermis display severe skin inflammation originating from TNFα-mediated keratinocyte apoptosis (44, 45). Treatment with the proteasome inhibitor bortezomib actually alleviated dermatitis in Sharpin-deficient mice (32). These studies suggested that degradation of cFLIP is the key process, which could explain the cause of the skin phenotype in Sharpin-deficient mice. Interestingly, our recent findings found that RNF31 KO primary keratinocytes had a lower basal expression level of cFLIP, and cFLIP also degraded faster compared with WT keratinocytes after TNFα stimulation (46). Another study showed that epidermal loss of cFLIP in keratinocytes may be an important prerequisite for the increased caspase 8-mediated epidermal cell death seen in patients with TEN/SJS (44). Besides, deficiency of LUBAC components results in severe human autoinflammatory diseases (47–49). So, our new findings may be helpful to develop the novel therapeutic approach for related diseases.
Experimental procedures
Cell cultures and transfection
HEK293T and HeLa (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS), 1% penicillin, and streptomycin. Jurkat cells (American Type Culture Collection) were maintained in RPMI 1640 medium supplemented with 10% FBS and 1% antibiotics. For the induction of apoptosis, cells were starved for 16 h with Dulbecco's modified Eagle's medium and 0.5% FBS before treatment with the indicated inducers. The calcium phosphate transfection method was used for 293T cell transfection in transfection experiments. In these experiments, 293T cells (7.5 × 105) were plated onto a 6-well plate, and CaCl2/Hanks' balanced salt solution/DNA precipitates were added to wells after 16 h (1–4 μg of DNA/well). Cells were lysed for further experiments 24 h after transfection.
Reagents and plasmids
The human TNFα recombinant protein RTNFAI was purchased from Thermo Scientific. The de novo protein synthesis inhibitor CHX (ALX-380-269) were purchased from Enzo Life Sciences. Proteasome inhibitor MG132 (S2619) was purchased from Selleck. Pan-caspase inhibitor Z-VAD-FMK (S1748) was purchased from Beyotime Biotechnology.
Antibodies against RNF31 (ab85294) and Sharpin (ab125188) were purchased from Abcam. Antibodies specific for PARP (9542), caspase 3 (9665), caspase 8 (9746), caspase 9 (9508), BclxL (2764), Mcl-1 (5453), Bcl2 (2870), and cFLIP (56343) were obtained from Cell Signaling Technology. Antibodies against cFLIPS/L (SC-5276), actin (sc-8432), β-tubulin (sc-5274), Myc (sc-7899), ubiquitin (SC9133), Noxa (sc-30209), and cIAP2 (sc-7944) were obtained from Santa Cruz Biotechnology. FLAG antibody (M20008) and HA antibody (M20003) were purchased from Abmart. An antibody specific for RIPK1 (610459) was obtained from BD Biosciences. A Lys-48 linkage-specific polyubiquitin antibody (05-1307) was purchased from Millipore. An anti-linear polyubiquitin antibody was provided by Genentech.
WT RNF31, HOIL-1, and Sharpin were cloned from Jurkat cDNA. All mutants of RNF31 (including WT, C885S, NT, CT, ΔRL, RL, and RBR) were generated using PCR with WT cDNA and verified via sequencing. Phosphorylated EF4-FLAG-cFLIP was provided by Dr. Jianke Zhang (Thomas Jefferson University). Mutant constructs were generated using PCR and cloned into pcDNA3.1 plasmid. shRNA clones for human RNF31 and Sharpin were obtained from Open Biosystems through MD Anderson's shRNA and ORFeome Core. The RNF31 clone number was V2LHS_284762 (GACAATAACGTCATGTTTA).
KO of cFLIP and RNF31 using the CRISPR-Cas9 system
Single guide RNAs were designed using CRISPR Design and inserted into a LentiCRISPR vector expressing hSpCas9 and guide RNA. The single guide RNA sequence for human RNF31 was TTGACACCACGCCAGTACCG, and for human cFLIP was ATGAAGGATTACATGGGCCG.
Viral production and infection
A lentiviral supernatant was collected 48 h after co-transfection of pGIPZ (for shRNA) or LentiCRISPR (for the CRISPR system) with packaging plasmids (pCMV-VSV-G and pCMV-dR8.2 dvpr) into HEK293T cells. Plasmids for rescue experiments were transfected into Phoenix cells with pCMV-VSV-G for retroviral production. Viral supernatants were collected after 48 h, and target cells were incubated with the supernatant in the presence of Polybrene for 8–12 h. After infection with virus, viral supernatants were replaced with fresh medium. After 24 h, infected cells were selected using puromycin (2 ng/ml) or sorted using flow cytometry. The efficiency of the infection was determined using Western blot analysis.
Ubiquitination assay
Twenty-four hours after transfection of FLAG-cFLIP, FLAG-RIPK1, and myc-LUBAC, 293T cells were lysed with 2% SDS lysis buffer (2% SDS, 150 mm NaCl, 10 mm Tris-HCl, pH 8.0) containing 2 mm sodium orthovanadate, 5 mm sodium fluoride, and 1 mm N-ethylmaleimide under denaturing conditions. After 10 min of boiling and sonication, samples were diluted with a dilution buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton) up to 10 times and incubated on ice for 30 min. Lysates were then immunoprecipitated with prewashed anti-FLAG M2 affinity gel (A2220; Sigma). Immunoprecipitates were analyzed using a Western blot assay.
For an in vitro ubiquitination assay, cFLIP-FLAG and LUBAC-FLAG were expressed in 293T cells, and FLAG antibody-conjugated beads were used to pulldown the recombinant cFLIP and LUBAC proteins. The recombinant cFLIP protein (1 μg) was incubated with or without lysine KO ubiquitin (500 ng), E1 (200 ng, Boston Biochem), E2 (500 ng, Boston Biochem), or LUBAC proteins (1 μg) at 37 °C for 2 h in in vitro ubiquitination buffer (20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP). The reaction was stopped by adding 2× loading buffer to the incubation followed by boiling, and the level of ubiquitination was monitored using a Western blot assay.
Western blot analysis
Cells were lysed in lysis buffer (50 mm Hepes, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1 mm sodium fluoride, 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture) (Roche Applied Science). For immunoprecipitation experiments, cell lysates were incubated with anti-FLAG M2 Affinity Gel (A2220; Sigma) overnight at 4 °C. The immunoprecipitates were washed with lysis buffer four times and eluted with 2× SDS loading buffer. Cell lysates and immunoprecipitates were then subjected to SDS-PAGE and transferred onto a nitrocellulose membrane (Bio-Rad). The membrane was sequentially probed with primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Enhanced chemiluminescence substrates (Pierce) were used to visualize the specific bands on the membrane. Signals were quantitated using ImageJ (NIH) and plotted relative to control.
Annexin V-staining apoptosis assay
HeLa cells (3 × 105) were seeded on six-well plates and incubated in starvation medium (0.5% FBS) for 16 h or without starvation. After treatment of the cells with the indicated inducers of apoptosis (TNF-α alone or with CHX), suspended cells in the supernatant were collected, and adherent cells on the plates were combined after detaching them using 0.05% trypsin (Sigma). Also, 1 × 106 Jurkat cells were stimulated with apoptosis inducers. After washing with PBS, the collected cells were stained with APC Annexin V (550474; BD Biosciences) and analyzed using flow cytometry.
Liquid chromatography-tandem MS
cFLIP immunocomplexes were immunoprecipitated using FLAG antibody beads, and then the immunocomplexes were separated by SDS-PAGE gel. After Coomassie Blue staining, the target gel band was excised and then analyzed by LC-electrospray ionization tandem MS. For LC-MS/MS analysis, peptides were separated by a 120-min gradient elution at a flow rate 0.300 μl/min with a Thermo-Dionex Ultimate 3000 HPLC system, which was directly interfaced with a Thermo Orbitrap Fusion Lumos mass spectrometer. The analytical column was a homemade fused silica capillary column (75 μm inner diameter, 150 mm length; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 A, 5 μm; Varian, Lexington, MA). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid. An LTQ-Orbitrap mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 4.0.27.10 software and is a single full-scan mass spectrum in the Orbitrap (300–1500 m/z, 120,000 resolution) followed by 3-s data-dependent MS/MS scans in an Ion Routing Multipole at 30% normalized collision energy (high-energy collision–induced dissociation). MS/MS spectra from each LC-MS/MS run were searched against the cFLIP sequence using the Proteome Discoverer (Version 1.4) searching algorithm. The search criteria were as follows: full tryptic specificity was required; two missed cleavages were allowed; carbamidomethylation was set as fixed modification; oxidation (M) and GlyGly (K) were set as variable modifications; precursor ion mass tolerance was 10 ppm for all MS acquired in the Orbitrap mass analyzer; and fragment ion mass tolerance was 0.02 Da for all MS2 spectra acquired in the Orbitrap. High confidence score filter (FDR < 1%) was used to select the “hit” peptides and their corresponding MS/MS spectra were manually inspected.
Statistics
All values in this article were given as mean ± S.E., unless stated otherwise. All experiments were reproduced at least three independent times, and results shown are representative. Statistical significance was calculated by two-tailed unpaired t test using GraphPad Prism software. Statistical significance was set based on p values; NS, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Author contributions
Y. T., D. J., J. J., M.-C. H., and X. L. conceptualization; Y. T. and D. J. data curation; Y. T., D. J., G. L., H. T., J. Y., X. Z., and X. L. formal analysis; Y. T., D. J., G. L., H. T., J. Y., and X. L. investigation; Y. T., D. J., G. L., H. T., and J. Y. methodology; Y. T. and D. J. writing-original draft; Y. T. and X. Z. project administration; J. J. resources; X. Z., M.-C. H., and X. L. supervision; X. Z., M.-C. H., and X. L. writing-review and editing; M.-C. H. and X. L. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Vishva Dixit (Genentech) for providing anti-linear ubiquitin antibodies. We thank the Protein Chemistry Facility at the Center for Biomedical Analysis of Tsinghua University for sample analysis.
This work was supported by Grants 81570211 and 81630058 from National Natural Science Foundation of China (to X. L.) and Grant R01AI11672211 from the National Institutes of Health (to M.-C. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
- TNFα
- tumor necrosis factor α
- LUBAC
- linear ubiquitin chain assembly complex
- cFLIP
- cellular FLICE-like inhibitory protein
- RIPK1/3
- receptor-interacting serine/threonine-protein kinase 1/3
- TRAFs
- TNF receptor-associated factors
- IKK
- IκB kinase
- FADD
- FAS-associated death domain protein
- BCL2
- B-cell lymphoma 2
- cIAP
- inhibitor of apoptosis protein
- CHX
- cycloheximide
- RBR
- RING-IBR-RING domain
- DED1
- death effector domain 1
- shRNA
- short hairpin RNA
- PARP
- poly(ADP-ribose) polymerase
- FMK
- fluoromethyl ketone
- Z
- benzyloxycarbonyl
- FBS
- fetal bovine serum.
References
- 1. Baud V., and Karin M. (2001) Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11, 372–377 10.1016/S0962-8924(01)02064-5 [DOI] [PubMed] [Google Scholar]
- 2. Ofengeim D., and Yuan J. (2013) Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat. Rev. Mol. Cell Biol. 14, 727–736 10.1038/nrm3683 [DOI] [PubMed] [Google Scholar]
- 3. Brenner D., Blaser H., and Mak T. W. (2015) Regulation of tumour necrosis factor signalling: live or let die. Nat. Rev. Immunol. 15, 362–374 10.1038/nri3834 [DOI] [PubMed] [Google Scholar]
- 4. Wajant H., Pfizenmaier K., and Scheurich P. (2003) Tumor necrosis factor signaling. Cell Death Differ. 10, 45–65 10.1038/sj.cdd.4401189 [DOI] [PubMed] [Google Scholar]
- 5. Micheau O., and Tschopp J. (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 10.1016/S0092-8674(03)00521-X [DOI] [PubMed] [Google Scholar]
- 6. Cho Y. S., Challa S., Moquin D., Genga R., Ray T. D., Guildford M., and Chan F. K. (2009) Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 10.1016/j.cell.2009.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He S., Wang L., Miao L., Wang T., Du F., Zhao L., and Wang X. (2009) Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137, 1100–1111 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 8. Zhang D. W., Shao J., Lin J., Zhang N., Lu B. J., Lin S. C., Dong M. Q., and Han J. (2009) RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 10.1126/science.1172308 [DOI] [PubMed] [Google Scholar]
- 9. Budd R. C., Yeh W. C., and Tschopp J. (2006) cFLIP regulation of lymphocyte activation and development. Nat. Rev. Immunol. 6, 196–204 10.1038/nri1787 [DOI] [PubMed] [Google Scholar]
- 10. Karin M., and Lin A. (2002) NF-κB at the crossroads of life and death. Nat. Immunol. 3, 221–227 10.1038/ni0302-221 [DOI] [PubMed] [Google Scholar]
- 11. Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., et al. (2007) IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell 131, 682–693 10.1016/j.cell.2007.10.037 [DOI] [PubMed] [Google Scholar]
- 12. Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., and Barker P. A. (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 10.1016/j.molcel.2008.05.014 [DOI] [PubMed] [Google Scholar]
- 13. Ghosh S., May M. J., and Kopp E. B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16, 225–260 10.1146/annurev.immunol.16.1.225 [DOI] [PubMed] [Google Scholar]
- 14. Lee E. W., Kim J. H., Ahn Y. H., Seo J., Ko A., Jeong M., Kim S. J., Ro J. Y., Park K. M., Lee H. W., Park E. J., Chun K. H., and Song J. (2012) Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat. Commun. 3, 978 10.1038/ncomms1981 [DOI] [PubMed] [Google Scholar]
- 15. Skaug B., Jiang X., and Chen Z. J. (2009) The role of ubiquitin in NF-κB regulatory pathways. Annu. Rev. Biochem. 78, 769–796 10.1146/annurev.biochem.78.070907.102750 [DOI] [PubMed] [Google Scholar]
- 16. Iwai K., and Tokunaga F. (2009) Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep. 10, 706–713 10.1038/embor.2009.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwai K., Fujita H., and Sasaki Y. (2014) Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508 10.1038/nrm3836 [DOI] [PubMed] [Google Scholar]
- 18. Gerlach B., Cordier S. M., Schmukle A. C., Emmerich C. H., Rieser E., Haas T. L., Webb A. I., Rickard J. A., Anderton H., Wong W. W., Nachbur U., Gangoda L., Warnken U., Purcell A. W., Silke J., and Walczak H. (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 10.1038/nature09816 [DOI] [PubMed] [Google Scholar]
- 19. Ikeda F., Deribe Y. L., Skånland S. S., Stieglitz B., Grabbe C., Franz-Wachtel M., van Wijk S. J., Goswami P., Nagy V., Terzic J., Tokunaga F., Androulidaki A., Nakagawa T., Pasparakis M., Iwai K., et al. (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 10.1038/nature09814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tokunaga F., Nakagawa T., Nakahara M., Saeki Y., Taniguchi M., Sakata S., Tanaka K., Nakano H., and Iwai K. (2011) SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 10.1038/nature09815 [DOI] [PubMed] [Google Scholar]
- 21. Yang Y. K., Yang C., Chan W., Wang Z., Deibel K. E., and Pomerantz J. L. (2016) Molecular determinants of scaffold-induced linear ubiquitinylation of B cell lymphoma/leukemia 10 (Bcl10) during T cell receptor and oncogenic caspase recruitment domain-containing protein 11 (CARD11) signaling. J. Biol. Chem. 291, 25921–25936 10.1074/jbc.M116.754028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teh C. E., Lalaoui N., Jain R., Policheni A. N., Heinlein M., Alvarez-Diaz S., Sheridan J. M., Rieser E., Deuser S., Darding M., Koay H. F., Hu Y., Kupresanin F., O'Reilly L. A., Godfrey D. I., et al. (2016) Linear ubiquitin chain assembly complex coordinates late thymic T-cell differentiation and regulatory T-cell homeostasis. Nat. Commun. 7, 13353 10.1038/ncomms13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rieser E., Cordier S. M., and Walczak H. (2013) Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem. Sci. 38, 94–102 10.1016/j.tibs.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 24. Park Y., Jin H. S., Lopez J., Lee J., Liao L., Elly C., and Liu Y. C. (2016) SHARPIN controls regulatory T cells by negatively modulating the T cell antigen receptor complex. Nat. Immunol. 17, 286–296 10.1038/ni.3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rodgers M. A., Bowman J. W., Fujita H., Orazio N., Shi M., Liang Q., Amatya R., Kelly T. J., Iwai K., Ting J., and Jung J. U. (2014) The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J. Exp. Med. 211, 1333–1347 10.1084/jem.20132486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumari S., Redouane Y., Lopez-Mosqueda J., Shiraishi R., Romanowska M., Lutzmayer S., Kuiper J., Martinez C., Dikic I., Pasparakis M., and Ikeda F. (2014) Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. Elife 3, e03422 10.7554/eLife.03422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rickard J. A., Anderton H., Etemadi N., Nachbur U., Darding M., Peltzer N., Lalaoui N., Lawlor K. E., Vanyai H., Hall C., Bankovacki A., Gangoda L., Wong W. W., Corbin J., Huang C., et al. (2014) TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. Elife 3, e03464 10.7554/eLife.03464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peltzer N., Rieser E., Taraborrelli L., Draber P., Darding M., Pernaute B., Shimizu Y., Sarr A., Draberova H., Montinaro A., Martinez-Barbera J. P., Silke J., Rodriguez T. A., and Walczak H. (2014) HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9, 153–165 10.1016/j.celrep.2014.08.066 [DOI] [PubMed] [Google Scholar]
- 29. Peltzer N., Darding M., Montinaro A., Draber P., Draberova H., Kupka S., Rieser E., Fisher A., Hutchinson C., Taraborrelli L., Hartwig T., Lafont E., Haas T. L., Shimizu Y., Böiers C., et al. (2018) LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature 557, 112–117 10.1038/s41586-018-0064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujita H., Tokunaga A., Shimizu S., Whiting A. L., Aguilar-Alonso F., Takagi K., Walinda E., Sasaki Y., Shimokawa T., Mizushima T., Ohki I., Ariyoshi M., Tochio H., Bernal F., Shirakawa M., and Iwai K. (2018) Cooperative domain formation by homologous motifs in HOIL-1L and SHARPIN plays a crucial role in LUBAC stabilization. Cell Rep. 23, 1192–1204 10.1016/j.celrep.2018.03.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Du C., Fang M., Li Y., Li L., and Wang X. (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42 10.1016/S0092-8674(00)00008-8 [DOI] [PubMed] [Google Scholar]
- 32. Liang Y., Seymour R. E., and Sundberg J. P. (2011) Inhibition of NF-κB signaling retards eosinophilic dermatitis in SHARPIN-deficient mice. J. Invest. Dermatol 131, 141–149 10.1038/jid.2010.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poukkula M., Kaunisto A., Hietakangas V., Denessiouk K., Katajamäki T., Johnson M. S., Sistonen L., and Eriksson J. E. (2005) Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J. Biol. Chem. 280, 27345–27355 10.1074/jbc.M504019200 [DOI] [PubMed] [Google Scholar]
- 34. Wilkie-Grantham R. P., Matsuzawa S., and Reed J. C. (2013) Novel phosphorylation and ubiquitination sites regulate reactive oxygen species-dependent degradation of anti-apoptotic c-FLIP protein. J. Biol. Chem. 288, 12777–12790 10.1074/jbc.M112.431320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Varfolomeev E. E., Schuchmann M., Luria V., Chiannilkulchai N., Beckmann J. S., Mett I. L., Rebrikov D., Brodianski V. M., Kemper O. C., Kollet O., Lapidot T., Soffer D., Sobe T., Avraham K. B., Goncharov T., Holtmann H., Lonai P., and Wallach D. (1998) Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9, 267–276 10.1016/S1074-7613(00)80609-3 [DOI] [PubMed] [Google Scholar]
- 36. Zhang J., Cado D., Chen A., Kabra N. H., and Winoto A. (1998) Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature 392, 296–300 10.1038/32681 [DOI] [PubMed] [Google Scholar]
- 37. Yeh W. C., Itie A., Elia A. J., Ng M., Shu H. B., Wakeham A., Mirtsos C., Suzuki N., Bonnard M., Goeddel D. V., and Mak T. W. (2000) Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity 12, 633–642 10.1016/S1074-7613(00)80214-9 [DOI] [PubMed] [Google Scholar]
- 38. Tanaka M., Fuentes M. E., Yamaguchi K., Durnin M. H., Dalrymple S. A., Hardy K. L., and Goeddel D. V. (1999) Embryonic lethality, liver degeneration, and impaired NF-κB activation in IKK-β-deficient mice. Immunity 10, 421–429 10.1016/S1074-7613(00)80042-4 [DOI] [PubMed] [Google Scholar]
- 39. Takeda K., Takeuchi O., Tsujimura T., Itami S., Adachi O., Kawai T., Sanjo H., Yoshikawa K., Terada N., and Akira S. (1999) Limb and skin abnormalities in mice lacking IKKα. Science 284, 313–316 10.1126/science.284.5412.313 [DOI] [PubMed] [Google Scholar]
- 40. Schmidt-Supprian M., Bloch W., Courtois G., Addicks K., Israël A., Rajewsky K., and Pasparakis M. (2000) NEMO/IKK γ-deficient mice model incontinentia pigmenti. Mol. Cell 5, 981–992 10.1016/S1097-2765(00)80263-4 [DOI] [PubMed] [Google Scholar]
- 41. Goto E., and Tokunaga F. (2017) Decreased linear ubiquitination of NEMO and FADD on apoptosis with caspase-mediated cleavage of HOIP. Biochem. Biophys. Res. Commun. 485, 152–159 10.1016/j.bbrc.2017.02.040 [DOI] [PubMed] [Google Scholar]
- 42. Sohn D., Totzke G., Essmann F., Schulze-Osthoff K., Levkau B., and Jänicke R. U. (2006) The proteasome is required for rapid initiation of death receptor-induced apoptosis. Mol. Cell. Biol. 26, 1967–1978 10.1128/MCB.26.5.1967-1978.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bachmann F., Buechner S. A., Wernli M., Strebel S., and Erb P. (2001) Ultraviolet light downregulates CD95 ligand and TRAIL receptor expression facilitating actinic keratosis and squamous cell carcinoma formation. J. Invest. Dermatol 117, 59–66 10.1046/j.0022-202x.2001.01380.x [DOI] [PubMed] [Google Scholar]
- 44. Panayotova-Dimitrova D., Feoktistova M., Ploesser M., Kellert B., Hupe M., Horn S., Makarov R., Jensen F., Porubsky S., Schmieder A., Zenclussen A. C., Marx A., Kerstan A., Geserick P., He Y. W., and Leverkus M. (2013) cFLIP regulates skin homeostasis and protects against TNF-induced keratinocyte apoptosis. Cell Rep. 5, 397–408 10.1016/j.celrep.2013.09.035 [DOI] [PubMed] [Google Scholar]
- 45. Weinlich R., Oberst A., Dillon C. P., Janke L. J., Milasta S., Lukens J. R., Rodriguez D. A., Gurung P., Savage C., Kanneganti T. D., and Green D. R. (2013) Protective roles for caspase-8 and cFLIP in adult homeostasis. Cell Rep. 5, 340–348 10.1016/j.celrep.2013.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang Y., Tu H., Liu G., Zheng G., Wang M., Li L., Zhao X., and Lin X. (2018) RNF31 regulates skin homeostasis by protecting epidermal keratinocytes from cell death. J. Immunol. 200, 4117–4124 10.4049/jimmunol.1800172 [DOI] [PubMed] [Google Scholar]
- 47. Aksentijevich I., and Zhou Q. (2017) NF-κB pathway in autoinflammatory diseases: dysregulation of protein modifications by ubiquitin defines a new category of autoinflammatory diseases. Front. Immunol. 8, 399 10.3389/fimmu.2017.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boisson B., Laplantine E., Dobbs K., Cobat A., Tarantino N., Hazen M., Lidov H. G., Hopkins G., Du L., Belkadi A., Chrabieh M., Itan Y., Picard C., Fournet J. C., Eibel H., Tsitsikov E., et al. (2015) Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J. Exp. Med. 212, 939–951 10.1084/jem.20141130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boisson B., Laplantine E., Prando C., Giliani S., Israelsson E., Xu Z., Abhyankar A., Israël L., Trevejo-Nunez G., Bogunovic D., Cepika A. M., MacDuff D., Chrabieh M., Hubeau M., Bajolle F., et al. (2012) Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13, 1178–1186 10.1038/ni.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.