Abstract
Balancing self-renewal with differentiation is crucial for neural stem cells (NSC) functions to ensure tissue development and homeostasis. Over the last years, multiple studies have highlighted the coupling of either metabolic or epigenetic reprogramming to NSC fate decisions. Metabolites are essential as they provide the energy and building blocks for proper cell function. Moreover, metabolites can also function as substrates and/or cofactors for epigenetic modifiers. It is becoming more evident that metabolic alterations and epigenetics rewiring are highly intertwined; however, their relation regarding determining NSC fate is not well understood. In this review, we summarize the major metabolic pathways and epigenetic modifications that play a role in NSC. We then focus on the notion that nutrients availability can function as a switch to modify the epigenetic machinery and drive NSC sequential differentiation during embryonic neurogenesis.
Keywords: Neurogenesis, neural stem cells, cerebrospinal fluids, nutrients, lipid metabolism, glutamine, one carbon folate pathway, glycolysis, epigenetics
Introduction
The mammalian cortex is composed of an incredible diversity of neurons and glial cells which arise from the differentiation of neural stem cells (NSC) during embryonic stages and the first postnatal weeks. During this process, the transition from NSC to fully differentiated neurons and glia is called neurogenesis and gliogenesis, respectively. While most of the specialized stem cells are capable of producing several different lineages, NSC produce different cell types sequentially and their potentiality decreases with time in a phenomenon called sequential fate restriction.1,2 Indeed, NSC switch from proliferative symmetrical divisions to asymmetrical cell division to sequentially produce all cortical neurons which will populate the 6 different layers of the cortex. Finally, NSC switch to gliogenesis which persists after birth. The mechanisms regulating this temporal differentiation progression are not fully understood. In recent years, it has become clear that both extracellular factors as well as intracellular cues can control the proliferation/differentiation balance in NSC. Among the extracellular factors, nutrients modulate fundamental cellular processes including proliferation, secretion, and autophagy.3 In addition to their bioenergetics intracellular function, recent work showed that extracellular nutrients and intracellular metabolites can influence cell state by acting as signaling molecules affecting both signaling pathways and gene expression particularly through their effect on chromatin modifications. Indeed, most chromatin-modifying enzymes require substrates or cofactors that are intermediates of cellular metabolism.4 Thus, variation of these metabolic inputs will determine epigenome remodeling and transcription. This is of interest because epigenetics, which are defined as the heritable traits that involve chromatin changes rather than DNA sequence alterations, control gene expression which is at the heart of differentiation and development. In fact, long-term epigenetic silencing of key developmental genes that are associated with specific cell lineages is at the center of NSC fate restriction.5
Epigenetic landscape can be modified by DNA and histone modification enzymes following metabolic changes, and conversely, epigenetic mechanisms can regulate the cellular metabolome through modulating metabolic gene expression. Thus, understanding the interplay between metabolism and epigenetics has proven to be a difficult task. Extensive research during the past decades has focused on elucidating the roles of metabolic pathways in the control of stem cells fate decisions; however, the role they may play during neurogenesis has been largely understudied. In this review, we will summarize the recent research progress in the epigenetic and metabolic regulation of NSC cell fate and discuss how the understanding of the link between epigenetic and metabolism could identify new vintage points in the field of neurogenesis.
The Cerebrospinal Fluid and Neurogenesis
NSC are in permanent and direct contact with the cerebrospinal fluid (CSF) at the ventricular surface from the earliest stages of brain development. Indeed, the primitive cerebroventricular system emerges with the closure of the neural tube which entraps some of the amniotic fluid and serves as the initial CSF. The CSF is then actively regenerated throughout embryogenesis and adulthood from arterial blood by the choroid plexus tissues.6 The CSF has been shown to have age-dependent effect on NSC proliferation suggesting that its composition is relevant to normal corticogenesis.7 Due to the advancement in proteomics, the highly dynamic CSF proteome starts to be characterized and have revealed similarities between human and rodent proteomes at different time points during corticogenesis.8,9 While we are only beginning to uncover CSF composition, the range of factors present in the CSF known to be important for NSC already includes fibroblast growth factors (FGF), insulin-like growth factor (IGF), sonic hedgehog (Shh), and retinoic acid (RA). Interestingly, the presence of regulators of lipid metabolism, glucose, as well as folate and some of its derivatives has also been reported10; however, the metabolic profile of CSF during corticogenesis has yet to be established. Finally, whether variation of CSF composition can have great influence on NSC intracellular metabolites levels and how that will affect neurogenesis will require further investigation.
Metabolism and Epigenetic Modifications in NSC
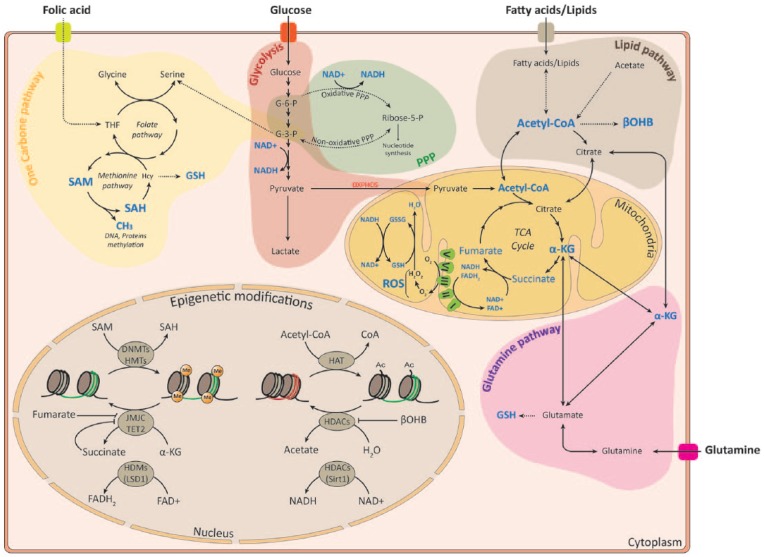
Early pioneering studies have linked metabolic gradients to developmental patterning indicating that metabolic differences are intrinsically and functionally linked to cell differentiation in different developmental contexts.11,12 Flexibility in energy metabolism supports stage-specific energetic demands. The most known metabolic rewiring is the glycolysis/oxidative phosphorylation (OXPHOS) switch where glycolysis maintains stemness through provision of energy and OXPHOS allows for more efficient energy production to match the needs of the differentiating progeny.13 For instance, in drosophila, this metabolic switch was shown to trigger neuroblasts terminal differentiation during metamorphosis.14 However, in mammals, while most studies have focused on the role of metabolites in adult NSC,15–17 much less is known regarding their function in embryonic NSC. Transcriptomic approaches have highlighted the temporal changes in both NSC metabolic gene expression and NSC epigenetic landscape during sequential generation of the different neuronal subtypes hinting toward their important role in corticogenesis.18,19 Given the fact that metabolism intermediates are often used as cofactors and substrates for epigenetic modifying enzymes (Figure 1), we focus on the link between the major metabolic pathways and NSC fate.
Figure 1.
Overview of metabolic pathways implicated in epigenetic modifications in NSC. Metabolites that are used as substrates and cofactors for reactions that coordinate epigenetic status are highlighted in blue color. Epigenetic modificatons are represented as methylation and acetylation marks on histones. α-KG indicates α-ketoglutarate; βOHB, β-hydroxybutyrate; Ac, acetyl; CoA, co-enzyme; DNMT, DNA methyltransferase; FAD, flavine adenine dinucleotide; FADH2, flavin adenine dinucleotide dihydride; G-6-P, glucose-6-phosphate; G-3-P, glyceraldehyde-3-phosphate; GSH, glutathione; GSSG, glutathione disulfide; HAT, histone acetyltransferases; Hcy, homocysteine; HDM, histone demethylases; HDAC, histone deacetylase; HMT, histone methyltransferase; JMJC, Jumonji domain demethylase; LSD1, lysine-specific demethylase 1; Me, methyl; NAD+, nicotinamide adenine dinucleotide; NADH, nicotinamide adenine dinucleotide hydride; PPP, pentose phosphate pathway; P, phosphate; ROS, reactive oxygen species; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; TCA, tricarboxylic acid; TET2, ten-eleven translocation 2; THF, tetrahydrofolate.
One-carbon pathway
One-carbon (1C) metabolism consists of a series of complex cyclical reactions in which a single carbon unit is transferred from donors to acceptors. It comprised 2 intertwined pathways: the folate and methionine pathways. The folate pathway is involved in DNA synthesis with the production of purine and pyrimidine via the metabolism of tetrahydrofolate (THF). However, the methionine pathway is involved in methylation reactions as it converts methionine to S-adenosyl methionine (SAM), which is a methyl donor.20 1C metabolism is implicated in NSC self-renewal and differentiation as its deficiency inhibited the proliferation of both embryonic NSC in vitro and adult hippocampal NSC.21,22 Furthermore, in vitro folate supplementation increases NSC proliferation and neuronal differentiation by enhancing DNA methyltransferases (DNMT).23,24 Finally, folate supplementation during pregnancy facilitates oligodendrocyte differentiation.25 Recent work from us has shown that inhibition of the folate pathway induced a depletion of embryonic NSC in vivo.26 NSC depletion was correlated to decreased methylation on lysine 4 of histone 3 (H3K4me3) at the promoter of key progenitor genes. Furthermore, we have identified a link between 1C metabolism and the cell-cell communication pathway Eph-Ephrins. This is of interest as it identifies metabolites of the folate pathway as signaling molecules connecting extracellular signals to cellular gene expression. Whether 1C metabolism–driven epigenetic alterations are involved throughout embryonic neurogenesis or are required at a specific stage to drive NSC specification warrants future investigation.
Lipids pathway
Lipids, which have been often considered as membrane components, can also function as signal transduction molecules.27 Lipids can act either by binding specific receptors and/or modulating the environment of receptors associated with lipid rafts; thus, lipid metabolism is important for the interaction between adult NSC and their niche.28 Lipid production is upregulated in adult NSC through the fatty acid synthase (FASN)-dependent de novo lipogenesis which is essential for NSC proliferation as well as neurogenesis.29 While lipid metabolism has been shown to regulate the relative proportions of asymmetric/symmetric divisions in adult NSC, not much is known on its role in NSC at the embryonic stages. Recent evidences have linked lipids breakdown through the fatty acid oxidation (FAO) to NSC activity during development. Indeed, FAO inhibition led to a reduced NSC pool due to their increased differentiation and reduced self-renewal.30 Lipid metabolism has also been associated with epigenetic reprogramming. Histone acetyltransferases (HAT) use lipid-derived acetyl-CoA as a major source for histone acetylation and have been shown to drive cellular growth by promoting histone acetylation and expression of growth-related genes.31 Furthermore, acetyl-CoA can be reduced to produce beta-hydroxybutyrate (βOHB) which is an endogenous inhibitor of histone deacetylases (HDAC).32 Finally, pharmacological inhibition of histone deacetylation promoted neuronal differentiation while decreasing astrocyte differentiation in vitro.33 While the tight connection between lipid metabolism and epigenetic regulations of gene expression has been consolidated in different stem cells as well as adult neurogenesis, whether it can exert similar functions in embryonic NSC remains to be elucidated.
Glutamine pathway
Glutamine (GLN) is the most abundant amino acid in plasma, up to 60% of the total free amino acid pool, and is the primary nutrient for maintaining and promoting cellular function.34 The function of GLN goes beyond that of a simple metabolic fuel or protein precursor as it is actively involved in several cell-specific processes including nucleotide synthesis, cell survival/proliferation, redox homeostasis, and fatty acid synthesis.35 In NSC, the GLN pathway is essential for their growth and long-term maintenance and is upregulated in astrocytic lineages.36,37 Moreover, removal of GLN from growth culture medium impairs the spontaneous differentiation of cortical neurons in vitro.38 Glutamate, which is produced by GLN hydrolysis, is a precursor in the synthesis of reduced glutathione and thus regulates NSC redox balance. This balance is important to control reactive oxygen species (ROS) levels which play a role in maintaining the proliferation of adult progenitor cells within this neurogenic niche.39
GLN undergoes deamination to produce α-ketoglutarate (α-KG) through 2 pathways, namely, Glutaminase I and II pathways, which is a tricarboxylic acid cycle (TCA)-Cycle intermediate and a substrate for demethylases that modify both proteins and DNA. Indeed, α-KG is used as a substrate by Jumonji-C domain containing histone demethylases (JMJC) and Tet-DNA demethylases. High α-KG levels promote naïve pluripotency by suppressing the accumulation of repressive histone modifications and DNA methylation in mouse embryonic stem cells.39 Even though many studies have highlighted the importance of GLN metabolism in stem cell maintenance and differentiation, its function in embryonic NSC needs to be further characterized.
Glycolysis and pentose phosphate pathway
Glucose is imported into cells via glucose transporters (GLUT) and is a pivotal source of fuel that is used in different metabolic pathways. Glucose is metabolized into pyruvate (glycolysis) which can either be fermented into lactate or shuttled into the mitochondria to be used in the tricarboxylic acid (TCA) cycle (OXPHOS).17 Glycolysis is critical for the function of embryonic NSC as neural tube abnormalities are detected in diabetic pregnancies.40 Neuronal differentiation from human embryonic stem cells–derived NSC is accompanied by metabolic reprogramming from aerobic glycolysis to neuronal OXPHOS.41 The TCA cycle provides several intermediates that are used in numerous other reactions. Among these intermediates, acetyl-CoA and α-KG are described above, as well as the reducing agents nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2). FAD, the oxidized form of FADH2, is a cofactor for the lysine-specific demethylase (LSD1) that regulates NSC proliferation in adult hippocampal dentate gyrus through modulation of histone methylation.42 Moreover, LSD1 repressed the expression of HEYL through demethylation of H3K4me3 to promote neuronal differentiation of human fetal NSC.43 NADPH is important to manage oxidative stress and ROS by maintaining reduced glutathione levels.44 Increase in NAD+, the oxidized form of NADH, to NADH ratio activates SIRT1, a NAD+-dependent deacetylase which has been shown to inhibit adult hippocampal NSC self-renewal and promote embryonic NSC neuronal differentiation.45,46 However, activation of SIRT1 by resveratrol to mimic early neural developmental stress triggered OCT6 deacetylation and increased neural tube defects in mouse embryos.47 Thus, tight regulation of NAD+/NADH ratio is essential for neurogenesis. Finally, fumarate and succinate which are TCA intermediates have been shown to inhibit α-KG-dependent histone and DNA demethylases.48 Several TCA intermediates are exported out of the mitochondria and contribute to epigenetic regulation of transcription (eg, α-KG, NADH, FADH2); however, the role of glycolysis as a whole in regulating embryonic gene expression is not well studied.
Glucose-6-phosphate (G-6-P), a glycolytic intermediate, is used either for glycolysis or the pentose phosphate pathway (PPP). PPP is divided into 2 branches, oxidative PPP and non-oxidative PPP. The oxidative PPP uses the G-6-P to produce the reducing agent NADH. In the non-oxidative PPP branch, ribose-5-phosphate and/or xylulose-5-phosphate are produced, which function as signaling molecules that regulate transcription.49 In vitro studies have shown the reliance of NSC on the PPP which is stimulated during neuronal differentiation.50 Furthermore, PPP dysfunction contributes to impaired adult hippocampal neurogenesis.51 Whether PPP is linked to embryonic neurogenesis and NSC epigenetic remodeling remains to be elucidated.
Concluding Remarks
While human brain maturation continues throughout life, the first 1000 days is a unique period where the foundation of optimum growth and neurodevelopment are established.52 Maternal malnutrition or placenta insufficiency leads to intrauterine growth restriction (IGUR) as the fetus fails to reach its genetic potential size. In IGUR, blood is redirected preferentially toward brain at the expense of other vital organs highlighting the importance of brain nutrition during the early developmental stages. While “brain-sparing” refers to the relative protection of the brain, it does not guarantee its normal development as recent studies have shown child behavioral problems and altered neurotransmitters’ profiles.53,54 Thus, understanding how nutrition and metabolites affect NSC proliferation and differentiation during embryonic neurogenesis has profound impacts not only in understanding basic processes of brain development but also in the field of neurological disorders.
Work in the past decade has uncovered the complexity of the connections between metabolism and chromatin dynamics and how that influences neurogenesis, mostly in the adult. Metabolites are now considered as signaling molecules that provide a link between the cellular environment and nuclear transcription. While systems biology approaches are helping to understand this complexity at an unprecedented pace, technological challenges are present that must be overcome to unravel the mystery at the next level. Most studies have focused on expression levels of each metabolic enzyme inside cells, but these may not completely reflect the actual level of enzymatic activity. To move forward, it is crucially important to understand how metabolic profiles shift within NSC during embryonic neurogenesis. We have highlighted in this review the recent discoveries as well as the gaps in our understanding of the process of metabolic reprogramming in embryonic neurogenesis. Future studies investigating the epigenetic landscape of NSC should include analyses of intracellular metabolites as a deeper understanding of this connection may help shed light on how NSC adapt to their environment to coordinate brain construction as well as the etiology of a variety of complex diseases.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: MF and AD conceived and designed the article. MF wrote the manuscript. MF and AD edited the manuscript.
References
- 1. Toma K, Hanashima C. Switching modes in corticogenesis: mechanisms of neuronal subtype transitions and integration in the cerebral cortex. Front Neurosci. 2015;9:274. doi: 10.3389/fnins.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci. 2013;14:823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Mol Cell. 2013;49:379–387. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Etchegaray JP, Mostoslavsky R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell. 2016;62:695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Podobinska M, Szablowska-Gadomska I, Augustyniak J, Sandvig I, Sandvig A, Buzanska L. Epigenetic modulation of stem cells in neurodevelopment: the role of methylation and acetylation. Front Cell Neurosci. 2017;11:23. doi: 10.3389/fncel.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bueno D, Garcia-Fernàndez J. Evolutionary development of embryonic cerebrospinal fluid composition and regulation: an open research field with implications for brain development and function. Fluids Barriers CNS. 2016;13:5. doi: 10.1186/s12987-016-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehtinen MK, Zappaterra MW, Chen X, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zappaterra MD, Lisgo SN, Lindsay S, Gygi SP, Walsh CA, Ballif BA. A comparative proteomic analysis of human and rat embryonic cerebrospinal fluid. J Proteome Res. 2007;6:3537–3548. doi: 10.1021/pr070247w. [DOI] [PubMed] [Google Scholar]
- 9. Feliciano DM, Zhang S, Nasrallah CM, Lisgo SN, Bordey A. Embryonic cerebrospinal fluid nanovesicles carry evolutionarily conserved molecules and promote neural stem cell amplification. PLoS ONE. 2014;9:e88810. doi: 10.1371/journal.pone.0088810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zappaterra MW, Lehtinen MK. The cerebrospinal fluid: regulator of neurogenesis, behavior, and beyond. Cell Mol Life Sci. 2012;69:2863–2878. doi: 10.1007/s00018-012-0957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyazawa H, Aulehla A. Revisiting the role of metabolism during development. Development. 2018;145:dev131110. doi: 10.1242/dev.131110. [DOI] [PubMed] [Google Scholar]
- 12. Bulusu V, Prior N, Snaebjornsson MT, et al. Spatiotemporal analysis of a glycolytic activity gradient linked to mouse embryo mesoderm development. Dev Cell. 2017;40:331.e4–341.e4. doi: 10.1016/j.devcel.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Homem CCF, Steinmann V, Burkard TR, Jais A, Esterbauer H, Knoblich JA. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014;158:874–888. doi: 10.1016/j.cell.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 15. Rafalski VA, Brunet A. Energy metabolism in adult neural stem cell fate. Prog Neurobiol. 2011;93:182–203. doi: 10.1016/j.pneurobio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 16. Knobloch M, Jessberger S. Metabolism and neurogenesis. Curr Opin Neurobiol. 2017;42:45–52. doi: 10.1016/j.conb.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 17. Kim DY, Rhee I, Paik J. Metabolic circuits in neural stem cells. Cell Mol Life Sci. 2014;71:4221–4241. doi: 10.1007/s00018-014-1686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okamoto M, Miyata T, Konno D, et al. Cell-cycle-independent transitions in temporal identity of mammalian neural progenitor cells. Nat Commun. 2016;7:11349. doi: 10.1038/ncomms11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Albert M, Kalebic N, Florio M, et al. Epigenome profiling and editing of neocortical progenitor cells during development. EMBO J. 2017;36:2642–2658. doi: 10.15252/embj.201796764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X, Liu H, Cong G, et al. Effects of folate on notch signaling and cell proliferation in neural stem cells of neonatal rats in vitro. J Nutr Sci Vitaminol (Tokyo). 2008;54:353–356. [DOI] [PubMed] [Google Scholar]
- 22. Kruman II, Mouton PR, Emokpae R, Cutler RG, Mattson MP. Folate deficiency inhibits proliferation of adult hippocampal progenitors. Neuroreport. 2005;16:1055–1059. [DOI] [PubMed] [Google Scholar]
- 23. Luo S, Zhang X, Yu M, et al. Folic acid acts through DNA methyltransferases to induce the differentiation of neural stem cells into neurons. Cell Biochem Biophys. 2013;66:559–566. doi: 10.1007/s12013-012-9503-6. [DOI] [PubMed] [Google Scholar]
- 24. Liu H, Cao J, Zhang H, et al. Folic acid stimulates proliferation of transplanted neural stem cells after focal cerebral ischemia in rats. J Nutr Biochem. 2013;24:1817–1822. doi: 10.1016/j.jnutbio.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 25. Weng Q, Wang J, Tan B, et al. Folate metabolism regulates oligodendrocyte survival and differentiation by modulating AMPKα activity. Sci Rep. 2017;7:1705. doi: 10.1038/s41598-017-01732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fawal MA, Jungas T, Kischel A, Audouard C, Iacovoni JS, Davy A. Cross talk between one-carbon metabolism, Eph signaling, and histone methylation promotes neural stem cell differentiation. Cell Rep. 2018;23:2864.e7–2873.e7. doi: 10.1016/j.celrep.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 27. Tracey TJ, Steyn FJ, Wolvetang EJ, Ngo ST. Neuronal lipid metabolism: multiple pathways driving functional outcomes in health and disease. Front Mol Neurosci. 2018;11:10. doi: 10.3389/fnmol.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamilton LK, Dufresne M, Joppé SE, et al. Aberrant lipid metabolism in the forebrain niche suppresses adult neural stem cell proliferation in an animal model of Alzheimer’s disease. Cell Stem Cell. 2015;17:397–411. doi: 10.1016/j.stem.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 29. Knobloch M, Braun SM, Zurkirchen L, et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2013;493:226–230. doi: 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xie Z, Jones A, Deeney JT, Hur SK, Bankaitis VA. Inborn errors of long-chain fatty acid β-oxidation link neural stem cell self-renewal to autism. Cell Rep. 2016;14:991–999. doi: 10.1016/j.celrep.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 32. Shimazu T, Hirschey MD, Newman J, et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Balasubramaniyan V, Boddeke E, Bakels R, et al. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–951. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 34. Smith RJ, Wilmore DW. Glutamine nutrition and requirements. JPEN. 1990;14:94S-99S. doi: 10.1177/014860719001400412. [DOI] [PubMed] [Google Scholar]
- 35. Curi R, Newsholme P, Procopio J, Lagranha C, Gorjão R, Pithon-Curi TC. Glutamine, gene expression, and cell function. Front Biosci. 2007;12:344–357. [DOI] [PubMed] [Google Scholar]
- 36. Yeo H, Lyssiotis CA, Zhang Y, et al. FoxO3 coordinates metabolic pathways to maintain redox balance in neural stem cells. EMBO J. 2013;32:2589–2602. doi: 10.1038/emboj.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sá JV, Kleiderman S, Brito C, et al. Quantification of metabolic rearrangements during neural stem cells differentiation into astrocytes by metabolic flux analysis. Neurochem Res. 2017;42:244–253. doi: 10.1007/s11064-016-1907-z. [DOI] [PubMed] [Google Scholar]
- 38. Velletri T, Romeo F, Tucci P, et al. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle. 2013;12:3564–3573. doi: 10.4161/cc.26771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Renault VM, Rafalski VA, Morgan AA, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paddock M, Akram R, Jarvis DA, et al. The assessment of fetal brain growth in diabetic pregnancy using in utero magnetic resonance imaging. Clin Radiol. 2017;72:427.e1–427.e8. doi: 10.1016/j.crad.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 41. Zheng X, Boyer L, Jin M, et al. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun G, Alzayady K, Stewart R, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hirano K, Namihira M. LSD1 mediates neuronal differentiation of human fetal neural stem cells by controlling the expression of a novel target gene, HEYL. Stem Cells. 2016;34:1872–1882. doi: 10.1002/stem.2362. [DOI] [PubMed] [Google Scholar]
- 44. Dringen R. Metabolism and functions of glutathione in brain. Prog Neurobiol. 2000;62:649–671. [DOI] [PubMed] [Google Scholar]
- 45. Hisahara S, Chiba S, Matsumoto H, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci U S A. 2008;105:15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma CY, Yao MJ, Zhai QW, Jiao JW, Yuan XB, Poo MM. SIRT1 suppresses self-renewal of adult hippocampal neural stem cells. Development. 2014;141:4697–4709. doi: 10.1242/dev.117937. [DOI] [PubMed] [Google Scholar]
- 47. Li G, Jiapaer Z, Weng R, et al. Dysregulation of the SIRT1/OCT6 axis contributes to environmental stress-induced neural induction defects. Stem Cell Reports. 2017;8:1270–1286. doi: 10.1016/j.stemcr.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiao M, Yang H, Xu W, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krüger A, Grüning NM, Wamelink MM, et al. The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid Redox Signal. 2011;15:311–324. doi: 10.1089/ars.2010.3797. [DOI] [PubMed] [Google Scholar]
- 50. Candelario KM, Shuttleworth CW, Cunningham LA. Neural stem/progenitor cells display a low requirement for oxidative metabolism independent of hypoxia inducible factor-1alpha expression. J Neurochem. 2013;125:420–429. doi: 10.1111/jnc.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao Y, Pan X, Zhao J, Wang Y, Peng Y, Zhong C. Decreased transketolase activity contributes to impaired hippocampal neurogenesis induced by thiamine deficiency. J Neurochem. 2009;111:537–546. doi: 10.1111/j.1471-4159.2009.06341.x. [DOI] [PubMed] [Google Scholar]
- 52. Cusick SE, Georgieff MK. The role of nutrition in brain development: the golden opportunity of the “first 1000 days.” J Pediatr. 2016;175:16–21. doi: 10.1016/j.jpeds.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. García-Contreras C, Valent D, Vázquez-Gómez M, et al. Fetal growth-retardation and brain-sparing by malnutrition are associated to changes in neurotransmitters profile. Int J Dev Neurosci. 2017;57:72–76. doi: 10.1016/j.ijdevneu.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 54. Roza SJ, Steegers EA, Verburg BO, et al. What is spared by fetal brain-sparing? Fetal circulatory redistribution and behavioral problems in the general population. Am J Epidemiol. 2008;168:1145–1152. doi: 10.1093/aje/kwn233. [DOI] [PubMed] [Google Scholar]