Abstract
Quantification of regional cerebral blood flow (CBF) using [15O]H2O positron emission tomography (PET) requires the use of an arterial input function. Arterial sampling, however, is not always possible, for example in ill-conditioned or paediatric patients. Therefore, it is of interest to explore the use of non-invasive methods for the quantification of CBF. For validation of non-invasive methods, test–retest normal and hypercapnia data from 15 healthy volunteers were used. For each subject, the data consisted of up to five dynamic [15O]H2O brain PET studies of 10 min and including arterial sampling. A measure of CBF was estimated using several non-invasive methods earlier reported in literature. In addition, various parameters were derived from the time-activity curve (TAC). Performance of these methods was assessed by comparison with full kinetic analysis using correlation and agreement analysis. The analysis was repeated with normalization to the whole brain grey matter value, providing relative CBF distributions. A reliable, absolute quantitative estimate of CBF could not be obtained with the reported non-invasive methods. Relative (normalized) CBF was best estimated using the double integration method.
Keywords: Cerebral blood flow, method comparison, non-invasive, positron emission tomography
Introduction
Regional cerebral blood flow (CBF) represents the amount of blood that perfuses a volume of tissue, i.e. mL blood per mL of tissue per min. To date, the accepted unit for CBF is mL·cm−3·min−1, where mL·cm−3 is used to indicate the transfer from blood to tissue.1 Several modalities can be used to measure perfusion,2 but positron emission tomography (PET) with oxygen-15 labelled water is considered to be the reference standard method.
Over the years, various methods have been developed for deriving CBF from a dynamic [15O]H2O PET scan.3–14 Ultimately, all these methods are based on Kety's compartment model for (inert) H2O.15 Solving the differential equation leads to a convolution of the tissue response with the arterial input function (AIF). Quantitative studies therefore require the measurement of the AIF, which is obtained most reliably through continuous arterial sampling.16 However, this is a somewhat invasive procedure, which is less suitable for routine clinical studies. In some cases, arterial sampling is clinically not feasible, for example in ill-conditioned patients or in children with Moyamoya disease. In case arterial sampling is not possible, it may be of interest to apply non-invasive methods that can estimate CBF or relative CBF distributions across the brain to identify regions with reduced perfusion. However, before using non-invasive methods, it is important to investigate their accuracy and precision against full-quantitative kinetic approaches.
One non-invasive approach is the use of an image-derived input function (IDIF).17 The main challenge for this approach is the limited spatial resolution inherent to PET. This affects the quality of the measured input function due to partial volume effects. This particularly affects CBF studies because, in contrast to myocardial blood flow studies, there are no large vascular structures within the field of view.17 The IDIF approach seems very promising, but requires complex and accurate methodology for partial volume correction and delineation of the arteries. As a consequence, use of IDIF for CBF measurements is not widely used and had limited success so far.
Instead, this study focuses on validation of simplified methods independent of a measured AIF. As far as we know, the methods described below are all that have been reported for [15O]H2O PET.10,11,13 We also included the integral count approach from early brain activation studies.18 In MR perfusion research, often additional parameters describing the time intensity curve are reported, such as the time-to-peak (TTP), wash-in slope and the peak height. Their equivalents for PET have not been evaluated, because typically in dynamic PET studies, frame times are not shorter than 5 s and data are noisy making it difficult to estimate these parameters reliably. These parameters were included to confirm to this hypothesis.
The aim of this study was to select the best method based on their performance to estimate (relative) CBF, initiated by our interest in studying CBF changes in children suffering from Moyamoya disease for whom arterial sampling is clinically not feasible. In a head-to-head comparison with the reference kinetic method and including a report on the test–retest variability, this paper should provide clarity on the best non-invasive method for obtaining (relative) CBF.
Material and methods
Subjects and study protocol
PET scans were acquired on a Gemini TF-64 PET/CT system (Philips Healthcare, Cleveland, TN, USA). The research participants were 16 healthy volunteers. All participants gave written informed consent for this study prior to inclusion. The study has been approved by the medical ethical review committees of both participating centers; the Amsterdam Medical Center and the VU University Medical Center. The study was conducted in accordance with the Declaration of Helsinki. Characteristics of the participants and scanning protocol have been described previously.19 In brief, each person underwent five [15O]H2O scans in two separate scanning sessions. During the first scan session, people were scanned twice under baseline cerebrovascular conditions and once during hypercapnia. During the second session, planned 28 days later, a single baseline scan was followed by a hypercapnia scan. Hypercapnia was induced using 5% CO2 enriched air.
Each person received an arterial line for blood sampling and a venous line in the opposite arm for administration of [15O]H2O. Scanning sessions started with a 1 min low-dose CT scan, which served for attenuation and scatter correction of the subsequent PET acquisitions. Emission scans were acquired in list mode for 10 min. A bolus of 800 MBq [15O]H2O was administered at the start of each scan. Arterial blood was measured continuously using an automatic blood sampler.16 The resulting AIF was calibrated using three manual arterial samples collected at 5.5, 8 and 10 min post injection.
The scans were acquired in list mode and reconstructed into 26 frames of 1 × 10 s, 8 × 5 s, 4 × 10 s, 2 × 15 s, 3 × 20 s, 2 × 30 s, 6 × 60 s. The row action maximum likelihood algorithm (RAMLA) as provided by the scanner manufacturer was used for reconstruction of the scans with an isotropic voxel size of 2 mm. Thereafter, the dynamic images were smoothed using an isotropic 5 mm FWHM Gaussian kernel.
Brain region time-activity curves
T1-weighted MR images were acquired on a Philips 3 T Intera System (Philips Healthcare, Best, the Netherlands) at the Amsterdam Medical Center. This anatomical reference scan of each subject was co-registered to the emission scans using the SPM12 software package (Functional Imaging Laboratory, 2014, London, UK). For this purpose, emission scans were summed over all time frames. After co-registration, the anatomical scans were segmented using PVELab (Neurobiology Research Unit, 2010, Copenhagen, Denmark) into grey matter (GM), white matter (WM) and cerebrospinal fluid (CSF) and divided into 67 brain regions using the Hammers brain atlas.20,21 Segmentations were then applied to the dynamic [15O]H2O images to generate regional GM time-activity curves (TACs). Furthermore, the union of GM and WM was used as whole brain mask for some methods; this will further be referred to as whole brain.
CBF reference methods
CBF was calculated using two reference methods: (a) full kinetic analysis of brain region TACs using non-linear regression (NLR) and (b) the basis function method (BFM)14 for voxel wise (=parametric) calculations. Thus, NLR gives the CBF per region, and BFM gives a parametric map of CBF. Both methods used the single tissue compartment model with additional arterial blood volume parameter:
| (1) |
Here is the tissue concentration of the tracer over time, the arterial blood volume fraction, the AIF, f the cerebral blood flow (f = CBF) and the volume of distribution. For NLR, the AIF is estimated by the measured arterial tracer concentration corrected for delay. Dispersion correction was omitted in favour of fitting the arterial blood volume parameter. Blood flow estimated with this parameter is near equivalent to blood flow estimated with dispersion correction, as noted by Bol et al.22 For BFM, the measured arterial tracer concentration is corrected for both delay and dispersion as described previously.14
Simplified methods
Implementation of the methods described in this paper used the following calculations as published in the original papers. All methods are based on Kety's differential equation for the one-tissue reversible compartment model, see equation (2), where is the tissue concentration of the tracer over time, is the arterial tracer concentration over time, f is the CBF and the volume of distribution.
| (2) |
Mejia et al.10 introduced the double integration method (DIM),10 which eliminates the need for the AIF by using the whole brain as a reference and assuming the global CBF to be the normal average value. The method is based on the double integration of equation (2), leading to equation (3), where end time T is 3 min (in accordance with the original method).
| (3) |
The double integration of the arterial tracer concentration is substituted by A, which is estimated using the TAC of the whole brain and an assumed global CBF , as described in equation (4).
| (4) |
A is calculated with mL·cm−3·min−1 and . With A substituted in equation (3), the flow is calculated for every voxel, fixing at 0.86. Note that in the original publication, and were fixed at unity; however, a later (1992) recommended value is used here.23
In 1996, the DIM was extended by Watabe et al.11 using a two-step calculation strategy to estimate the global CBF and volume of distribution instead of fixing them to the normal average value. In Watabe's method, the same substitution is performed, and the TAC of a second reference region is introduced, yielding equation (5). This second region was defined as the 10% of voxels with the highest number of total counts.
| (5) |
Fixing at 0.86, NLR is performed using a trust-region reflective algorithm to fit the values for , and . A is then calculated using the fitted values for and and substituted in equation (3) to calculate f for every voxel, again fixing at 0.86.
Another approach was published by Treyer et al.. It is based on Alpert's weighted integration method24 to estimate both CBF and the washout parameter k2, but uses a standard AIF.13 This standard input function is corrected for delay and dispersion (Meyer's method8). Because CBF depends on the amplitude of the AIF, and k2 does not, estimated CBF values were then scaled using the estimated k2 by making their averages in GM equal. However, as Treyer at al. note in their discussion, this causes a bias because the washout parameter is defined as . Therefore, in this study, the estimated CBF values were scaled by making the estimate GM value equal to the average k2 times , fixing at 0.86. Unlike in the original study, we did not use a separate set of measured input functions to create a standard input function. Instead, the measured input functions of the subjects were used. However, to ensure that the used input function was independent of the subject, a ‘standard’ input function was produced per subject by averaging the input functions of all other subjects. Before averaging, the functions were normalized by their integral and the time of the peaks was aligned, as was done in the original study.13
For the above methods, the first 3 min of scan data were used; the following TAC-derived parameters used all or part of the 10-min scans. The definitions of the TAC-derived parameters are illustrated in the supplementary material. These CBF-related parameters were derived from the TAC: the area under the time activity curve (AUC); the AUC for a 60-s interval (AUC60) after the beginning of the wash-in (t0); peak height (peak); time between the beginning of the wash-in and the peak (TTP); the maximum wash-in slope (slope); the wash-out curve fitted with an exponential function (washoutEXP) and a power function (washoutpowerlaw). The parameters were calculated using Matlab 7.10 (R2010a) (The MathWorks, Inc., Natick, MA, USA).
Evaluation of simplified methods
The various parameters associated with CBF were compared with the reference method using both correlation and agreement analysis. Firstly, using the simplified CBF estimates per brain region and NLR as reference, Pearson correlation coefficients were calculated. The five highest correlating simplified parameters were included for further investigation.
Secondly, Bland–Altman analysis25 was used to investigate agreement. The results are reported as the mean difference (an estimate of the bias) and 1.96 times the standard deviation of the differences (an estimate of the precision). The results do not focus on individual regions, but rather report the average agreement for brain regions. To include the parameters with different units, the CBF estimates of each method were converted to a percentage of the sample average—the average over all subjects—and the standard deviation of the differences were reported in percentage points. Note that the mean difference is now zero and hence not reported. In addition, brain region values of all methods were normalized to their whole brain value providing relative CBF, and the agreement of the parameters relative to whole brain was also investigated. Using BFM as the reference, the agreement analyses were repeated using the parametric maps.
Repeatability of all methods was investigated using the brain region values of the repeated baseline measurements. The repeatability performance is reported by the repeatability index (RI), the repeatability coefficient25 as percentage of the sample average,19 again allowing to compare the repeatability of metrics with different units. The repeatability was re-calculated after normalizing each scan to the whole brain value. Intra-session test–retest performance was investigated using the two consecutive baseline scans of the first scanning session. For inter-session repeatability, the baseline scan of the second session was used in combination with the first baseline scan of the first session. The 95% confidence interval of the repeatability indices is also reported.
Finally, agreement analysis was performed on hypercapnia-induced differences estimated by each method. The relative changes between the baseline and hypercapnia scans, see equation (6), were calculated with each method for all brain regions. For this, differences between scan 3 and scan 1 and between scan 5 and scan 4 were used. These differences were compared to the differences found by the reference method (NLR) and the average difference between them is reported in percentage points.
| (6) |
Simulations
Simulations were performed in order to better understand the behaviour of the CBF methods. TACs were generated without noise to investigate bias as function of simulated CBF. The TACs were generated with the single tissue compartment model, as described in equation (1), and a representative input function. The representative input function was constructed from all measured input functions after normalization by their integral and aligning the peaks temporally. The volume of distribution VT and the arterial blood volume fraction Va were kept constant and simulated CBF parameter f ranged from 0.2 to 1.0 in 128 steps. In addition, noise was added to the TACs to investigate the noise characteristics of the methods. The noise level ranged from 0% to 16% coefficient of variation (COV) and was increased in 128 steps. Details of the noise simulation have been described earlier.26 For every combination of noise and CBF, 500 noisy TACs were generated.
The simulated data were then analysed with the BFM and simplified methods to estimate CBF. The whole brain reference TAC for the DIM was a noise free TAC generated with mL·cm−3·min−1. Watabe's extension of the DIM was not investigated. The scale factor for Treyer's method was calculated with the average k2 and K1 of all generated TACs.
The mean observed CBF errors and their standard deviations are reported in error maps. The errors as percentage of simulated CBF are reported as relative error maps. Error plots are shown for the noise free TACs and the TACs with 8% noise level.
Results
Average time between sessions was 34 days (25–45 days). From the total of 80 scans, 70 scans were successfully evaluated. Acquisition failed for 10 scans of 5 volunteers: tracer production failure twice, inadequate arterial blood sampling twice, and acute nausea once. Details were reported elsewhere.19
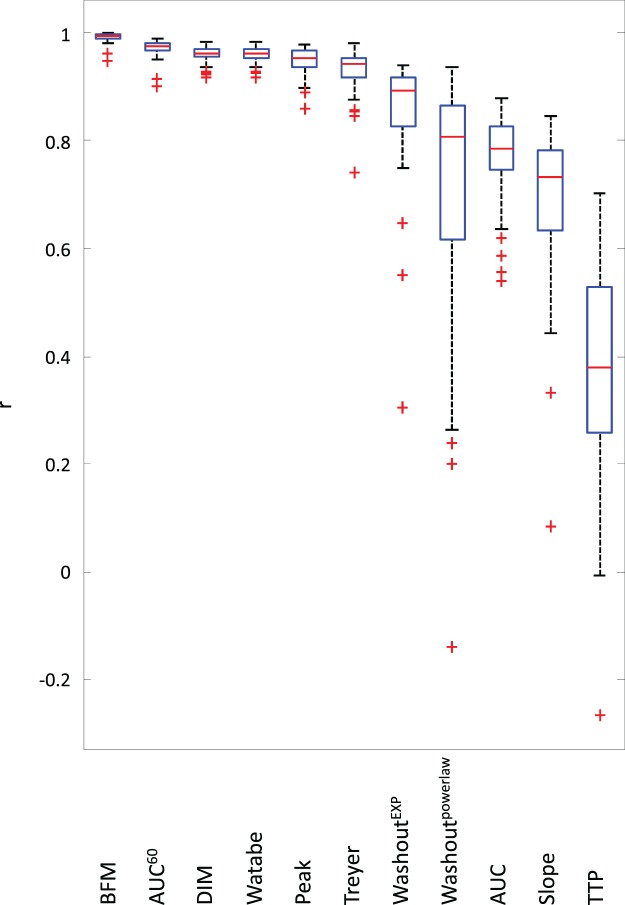
The Pearson correlation coefficients per scan between regional CBF values of simplified metrics and the reference method (NLR) are presented in Figure 1. Results obtained with BFM were added for comparison. Highest correlations were found for DIM, Watabe, Treyer, AUC60, and the peak height. The other measures (washoutEXP, washoutpowerlaw, AUC, slope and TTP) showed lower correlation and were excluded from further analysis.
Figure 1.
Correlation per scan of the investigated methods with full kinetic analysis using NLR. NLR: non-linear regression.
The results of the agreement analysis using brain regions are summarized in Table 1. The mean difference was zero for the DIM and Watabe's method. Treyer's method showed a mean difference that was significantly different from zero (α<.001), indicating bias. The standard deviations of the differences are an estimation of precision and show that for the absolute estimation of CBF, the DIM and Treyer's method are most precise, followed by the AUC60 and peak height. For estimation of relative CBF, the DIM and the AUC60 are most precise, followed by the Treyer's method and peak height. Watabe's method was most imprecise for estimation of both absolute and relative CBF. Table 2 shows the same results, but for parametric comparison. The standard deviations of the differences are larger than for regional comparison. The DIM seems least affected by this.
Table 1.
Regional agreement with NLR.
| Relative CBF |
||||||||
|---|---|---|---|---|---|---|---|---|
| Method | Mean | (95% CI) | 1.96 SD | 1.96 SD (% of sample average) | 1.96 SD (% of global CBF) | |||
| BFM | 0.00 | (−0.01 – 0.01) | 0.08 | (0.06–0.10) | 16.6 | (13.1–20.1) | 5.3 | (5.1–5.4) |
| DIM | 0.01 | (−0.01 – 0.03) | 0.18 | (0.14–0.21) | 36.6 | (28.9–44.4) | 10.4 | (10.2–10.7) |
| Watabe | 0.00 | (−0.03 – 0.03) | 0.27 | (0.22–0.33) | 56.7 | (44.7–68.6) | 15.4 | (15.0–15.8) |
| Treyer | 0.12 | (0.09–0.14) | 0.21 | (0.17–0.25) | 37.5 | (29.6–45.4) | 13.0 | (12.7–13.3) |
| AUC60 | N/A | N/A | 44.0 | (34.7–53.3) | 10.2 | (10.0–10.5) | ||
| peak | N/A | N/A | 45.2 | (35.7–54.7) | 13.2 | (12.8–13.5) | ||
Note: Average over all brain regions and 95% confidence interval between brackets (95% CI). The third column is converted to percentages by dividing over the average of all subjects, relative CBF is a percentage of the global CBF per scan.
BFM: basis function method; CBF: cerebral blood flow; DIM: double integration method; SD: standard deviation.
Table 2.
Parametric agreement with BFM. Average over all brain regions and 95% confidence interval between brackets (95% CI).
| Relative CBF |
||||||||
|---|---|---|---|---|---|---|---|---|
| Method | Mean | 1.96 SD | 1.96 SD (% of sample average) | 1.96 SD (% of global CBF) | ||||
| DIM | 0.01 | (−0.02–0.04) | 0.23 | (0.19–0.27) | 45.4 | (37.7–53.1) | 27.0 | (22.4–31.6) |
| Watabe | 0.03 | (−0.03–0.10) | 0.52 | (0.43–0.61) | 96.7 | (80.3–113.1) | 29.0 | (24.1–33.9) |
| Treyer | 0.10 | (0.06–0.13) | 0.29 | (0.24–0.34) | 50.7 | (42.1–59.3) | 34.7 | (28.8–40.6) |
| AUC60 | N/A | N/A | 52.0 | (43.2–60.8) | 28.1 | (23.3–32.9) | ||
| Peak | N/A | N/A | 55.0 | (45.6–64.3) | 35.4 | (29.4–41.4) | ||
BFM: basis function method; CBF: cerebral blood flow; DIM: double integration method; SD: standard deviation; N/A: not applicable.
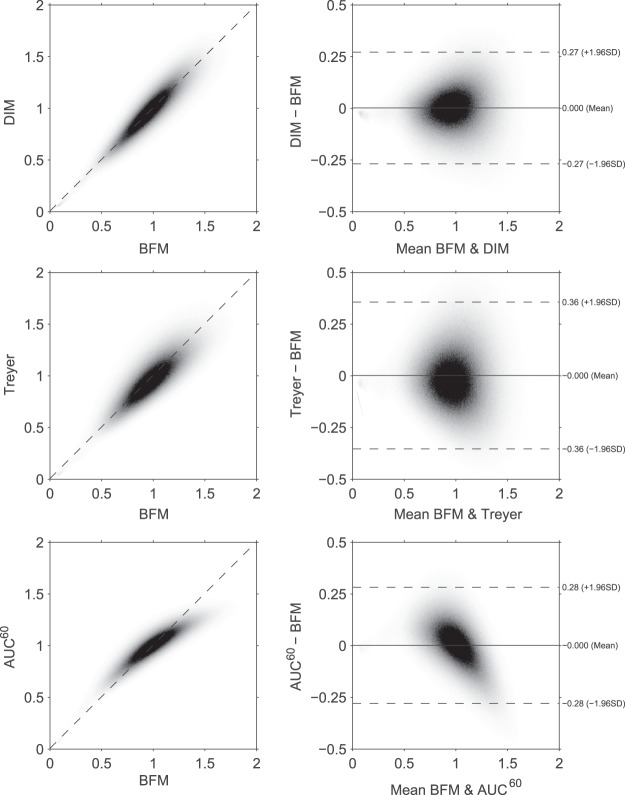
Figure 2 shows voxel-wise scatter and Bland–Altman plots of relative CBF calculated with the DIM, Treyer's and AUC60 methods using BFM as reference. Mean differences were zero due to normalization to the whole brain average. An example of parametric maps calculated with the various methods can be found in the supplementary material.
Figure 2.
Voxel wise scatter and Bland–Altman plots of the methods vs. BFM after normalization to whole brain (relative CBF). BFM: basis function method; CBF: cerebral blood flow.
Repeatability performance is shown in Table 3 for intrasession (n = 14 subjects) and Table 4 for intersession (n = 14 subjects). The DIM shows the same results, both with and without normalisation to the whole brain. Results of Watabe's method show the largest RI for intrasession repeatability. The TAC-derived parameters (AUC60 and peak height) show the best reproducibility indices for relative CBF estimation. The intersession RIs are approximately twice as large as the intrasessions RIs for Treyer's method, AUC60 and peak height.
Table 3.
Intrasession test–retest repeatability from data of 14 subjects.
| Relative CBF |
||||
|---|---|---|---|---|
| Method | RI (%) | RI (%) | ||
| NLR | 27.3 | (25.7–28.9) | 14.4 | (13.6–15.3) |
| BFM | 26.1 | (24.6–27.6) | 15.2 | (14.3–16.0) |
| DIM | 12.8 | (12.1–13.5) | 12.8 | (12.1–13.5) |
| Watabe | 37.2 | (35.1–39.3) | 14.4 | (13.6–15.2) |
| Treyer | 21.5 | (20.2–22.7) | 15.6 | (14.7–16.5) |
| AUC60 | 23.5 | (22.2–24.8) | 10.3 | (9.7–10.8) |
| Peak | 23.2 | (21.9–24.5) | 12.3 | (11.6–13.0) |
Note: Average over all brain regions and 95% confidence interval between brackets (95% CI).
BFM: basis function method; CBF: cerebral blood flow; DIM: double integration method; SD: standard deviation; RI: repeatability index.
Table 4.
Intersession test–retest repeatability from data of 14 subjects.
| Relative CBF |
||||
|---|---|---|---|---|
| Method | RI (%) | RI (%) | ||
| NLR | 29.5 | (27.8–31.3) | 14.4 | (13.5–15.2) |
| BFM | 31.8 | (30.0–33.6) | 15.0 | (14.1–15.8) |
| DIM | 14.3 | (13.5–15.2) | 14.3 | (13.5–15.2) |
| Watabe | 36.9 | (34.8–39.0) | 17.2 | (16.2–18.2) |
| Treyer | 43.9 | (41.4–46.4) | 16.6 | (15.7–17.5) |
| AUC60 | 49.8 | (46.9–52.6) | 11.7 | (11.0–12.3) |
| peak | 47.5 | (44.8–50.1) | 12.9 | (12.2–13.7) |
Average over all brain regions and 95% confidence interval between brackets (95% CI).
BFM: basis function method; CBF: cerebral blood flow; DIM: double integration method; SD: standard deviation; RI: repeatability index.
Agreement on hypercapnia induced differences is presented in Table 5. Scatter and Bland–Altman plots of these differences are shown in the supplementary material. The DIM and Watabe's method show a clear disagreement with the reference method for estimating differences between the hypercapnia scans and the baseline scans. Treyer's method shows best performance among the simplified methods.
Table 5.
Regional agreement with NLR of hypercapnia induced differences.
| Method | Mean | 1.96 SD | ||
|---|---|---|---|---|
| BFM | 2.4 | (−1.6 – 6.3) | 32.7 | (25.8–39.6) |
| DIM | −24.5 | (−29.5 – −19.5) | 41.1 | (32.5–49.8) |
| Watabe | −31.9 | (−37.7 – −26.2) | 47.2 | (37.3–57.2) |
| Treyer | 1.6 | (−3.0 – 6.3) | 38.0 | (30.0–46.1) |
| AUC60 | −3.7 | (−11.0 – 3.6) | 59.9 | (47.2–72.5) |
| Peak | −4.1 | (−11.5 – 3.3) | 60.9 | (48.0–73.7) |
Average over all brain regions and 95% confidence interval between brackets (95% CI).
BFM: basis function method; CBF: cerebral blood flow; DIM: double integration method; SD: standard deviation; NLR: non-linear regression.
The error maps and plots showing the simulation results are available in the supplementary material. BFM shows the least bias, only slightly overestimating CBF for simulated CBFs >0.8 mL·cm−3·min−1. The sawtooth pattern visible on the BFM error graph for the simulation without noise is caused by the use of the limited number of basis functions. The DIM shows slight overestimation at low CBF and slight underestimation at high CBF. The AUC60 and Peak methods largely underestimate and overestimate CBF at low and high simulated CBF values. The bias of the peak method also shows dependence on noise, which is visible in the error maps, whereas the bias of all other methods is independent on noise. Treyer's method shows overestimation of CBF over the entire simulated CBF range, but increases with simulated higher CBF values
The error precision maps show that precision of the AUC60 method is constant at different CBF and all other methods show declining precision with increasing CBF. All methods show worse precision with increasing noise levels. For BFM and the DIM, the precision is proportional to CBF, which can be seen in the relative precision maps.
Discussion
This study compared a wide range of simplified methods for estimating (absolute and relative) cerebral perfusion, independent of measurement of the AIF, in healthy volunteers. Their performance was investigated against reference kinetic methods, which use an AIF. Moreover, our study included assessment of repeatability performance of all metrics and methods tested; both intra- and inter-session.
Most TAC-derived parameters (washoutEXP, washoutpowerlaw, AUC, slope and TTP) showed poor correlation with NLR-derived CBF. As expected, these parameters are thus of little value for estimating CBF. Two TAC-derived parameters, the peak height and the AUC60, do show high correlation (see Figure 1). The AUC60 showed better results than the peak height: higher correlation, lower RIs and smaller standard deviation of the differences. Peak height does show a linear relationship, yet its performance is worse.
Relative CBF distributions can be estimated with reasonable precision using the DIM and AUC60 methods. However, it is known that the relationship between integral counts and CBF is nonlinear. This can also be seen in Figure 2 and this causes the lower contrast which can be seen in the parametric maps (supplementary material). This is probably also why the AUC60 has a lower RI for both inter- and intra-session repeatability. Because the DIM does not show worse agreement performance, it should be the method of choice for estimating relative CBF. However, because the global CBF is always assumed to be the normal average, this method cannot estimate absolute CBF and should only be considered when studying relative CBF changes between subjects or longitudinally. This is exemplified by the disagreement of this method for absolute longitudinal changes, as presented in Table 5.
In our study, we observed that none of the non-invasive methods are able to estimate absolute CBF reliably. Watabe's method estimates global CBF using NLR. However, as they clearly explain in the original paper, there exists a very shallow error surface around the optimal solution. Hence, the method is very sensitive to get trapped in local minima, and Treyer et al. indeed report this as well. Furthermore, Watabe et al.11 mention in their discussion that “From the simulation it follows that a 2-min administration period performs better than a 1-min period.” Perhaps this explains the disparity in the results. In this study (and in the study of Treyer et al. too), a bolus injection was used, which had an even shorter administration period of 15 s (20 s in the study of Treyer et al.). Clearly, the method performs worse on bolus injection data and cannot be recommended for estimation of CBF for our imaging procedure.
In comparison with Watabe's method, Treyer's method shows better precision for estimating absolute CBF. However, Treyer's method showed overestimations of CBF in this dataset. The reason for this is unclear, but could have to do with the presumed volume of distribution. If we look at the precision in percentage points, it is clear that the precision of Treyer's method is similar to the DIM's precision, whereas the TAC-derived parameters have a worse precision. For the assessment of CBF and relative CBF changes, most simplified methods show similar RIs the reference methods (NLR or BFM with arterial sampling) for intra-session CBF and relative CBF data and somewhat worse RIs for inter-session CBF values. When short-term longitudinal changes in CBF need to be assessed, Treyer's method may be considered.
The simulations largely confirmed the observations seen in the clinical data. Generally, the BFM provided most accurate and robust CBF estimates, while several simplified non-invasive methods suffer from substantial bias and poor precision. In line with the clinical studies, the DIM seems to be able to estimate CBF accurately and with high precision over a large range of simulated CBF values and noise levels and comparable to those seen with BFM. It should be noted that we did not simulate deviations in volume of distribution or input functions and some observations for the simplified methods may therefore be more optimistic than seen in the clinical data. Yet, in general, the simulations show the same trends as seen in the clinical studies.
Conclusion
In this study, we evaluated the performance of a wide range of non-invasive methods for quantifying CBF and/or relative CBF which can be applied in studies where the collection of an AIF is clinically not feasible (e.g. in children with Moyamoya disease). Performance of these methods was compared with quantitative CBF derived using a kinetic model including an AIF. The double integration method showed the best performance for measuring relative cerebral perfusion (and its change) without arterial sampling. The main disadvantage of this method is the inability to estimate global CBF. Therefore, it is concluded that among the non-invasive methods tested, the double integration method seems to be most optimal method for measuring relative CBF. None of the non-invasive methods were able to measure absolute CBF accurately, but Treyer's method may be considered when studying changes in CBF within the same subject in a longitudinal setting.
Supplementary Material
Acknowledgements
The authors would like to thank the volunteers.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was financially supported by the Netherlands Organisation for Health Research and Development, grant 10-10400-98-14002.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' contributions
TK drafted the manuscript. TK and MY contributed to analysis of the data. TK, MY, AAL and RB contributed to the interpretation of the data and design of the study. RB conceived the concept of the study. DFRH contributed to acquisition of the data. All authors TK, MY, DFRH, AAL, AJN, BNMB, and RB approved the final manuscript.
Supplementary material
Supplementary material for this paper can be found at the journal website: http://journals.sagepub.com/home/jcb
References
- 1.Innis RB, Cunningham VJ, Delforge J, et al. Consensus Nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 2007; 27: 1533–1539. [DOI] [PubMed] [Google Scholar]
- 2.Wintermark M, Sesay M, Barbier E, et al. Comparative overview of brain perfusion imaging techniques. Stroke 2005; 36: e83–e99. [DOI] [PubMed] [Google Scholar]
- 3.Frackowiak RS, Lenzi GL, Jones T, et al. Quantitative measurement of regional cerebral blood flow and oxygen metabolism in man using 15O and positron emission tomography: theory, procedure, and normal values. J Comput Assist Tomogr 1980; 4: 727–736. [DOI] [PubMed] [Google Scholar]
- 4.Herscovitch P, Markham J and Raichle ME. BrainBloodFlow Measuredwith IntravenousH2'50. I. TheoryandErrorAnalysis, www.researchgate.net/profile/Peter_Herscovitch/publication/16592454_Brain_blood_flow_measured_with_intravenous_H2(15)O._I._Theory_and_error_analysis/links/0c9605292697be3570000000.pdf (1983, accessed 25 May 2016).
- 5.Raichle ME, Martin WR, Herscovitch P, et al. Brain blood flow measured with intravenous H2(15)O. II. Implementation and validation. J Nucl Med Off Publ Soc Nucl Med 1983; 24: 790–798. [PubMed] [Google Scholar]
- 6.Huang S-C, Carson RE, Hoffman EJ, et al. Quantitative measurement of local cerebral blood flow in humans by positron computed tomography and 15O-water. J Cereb Blood Flow Metab 1983; 3: 141–153. [DOI] [PubMed] [Google Scholar]
- 7.Kanno I, Lammertsma AA, Heather JD, et al. Measurement of cerebral blood flow using bolus inhalation of C15O2 and positron emission tomography: description of the method and its comparison with the C15O2 continuous inhalation method. J Cereb Blood Flow Metab 1984; 4: 224–234. [DOI] [PubMed] [Google Scholar]
- 8.Meyer E. Simultaneous correction for tracer arrival delay and dispersion in CBF measurements by the H215O autoradiographic method and dynamic PET. J Nucl Med 1989; 30: 1069–1078. [PubMed] [Google Scholar]
- 9.Lammertsma AA, Cunningham VJ, Deiber MP, et al. Combination of dynamic and integral methods for generating reproducible functional CBF images. J Cereb Blood Flow Metab 1990; 10: 675–686. [DOI] [PubMed] [Google Scholar]
- 10.Mejia MA, Itoh M, Watabe H, et al. Simplified nonlinearity correction of oxygen-15-water regional cerebral blood flow images without blood sampling. J Nucl Med 1994; 35: 1870–1877. [PubMed] [Google Scholar]
- 11.Watabe H, Itoh M, Cunningham V, et al. Noninvasive quantification of rCBF using positron emission tomography. J Cereb Blood Flow Metab 1996; 16: 311–319. [DOI] [PubMed] [Google Scholar]
- 12.Iida H, Law I, Pakkenberg B, et al. Quantitation of regional cerebral blood flow corrected for partial volume effect using O-15 water and PET: I. theory, error analysis, and stereologic comparison. J Cereb Blood Flow Metab 2000; 20: 1237–1251. [DOI] [PubMed] [Google Scholar]
- 13.Treyer V, Jobin M, Burger C, et al. Quantitative cerebral H2 15O perfusion PET without arterial blood sampling, a method based on washout rate. Eur J Nucl Med Mol Imaging 2003; 30: 572–580. [DOI] [PubMed] [Google Scholar]
- 14.Boellaard R, Knaapen P, Rijbroek A, et al. Evaluation of basis function and linear least squares methods for generating parametric blood flow images using 15O-water and positron emission tomography. Mol Imaging Biol 2005; 7: 273–285. [DOI] [PubMed] [Google Scholar]
- 15.Kety SS, Schmidt CF. The nitrous oxide method for the quantitative determination of cerebral blood flow in man: theory, procedure and normal values. J Clin Invest 1948; 27: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boellaard R, van Lingen A, van Balen SCM, et al. Characteristics of a new fully programmable blood sampling device for monitoring blood radioactivity during PET. Eur J Nucl Med 2001; 28: 81–89. [DOI] [PubMed] [Google Scholar]
- 17.Zanotti-Fregonara P, Chen K, Liow J-S, et al. Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab 2011; 31: 1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lammertsma AA. Noninvasive estimation of cerebral blood flow. J Nucl Med 1994; 35: 1878–1879. [PubMed] [Google Scholar]
- 19.Heijtel DFR, Mutsaerts HJMM, Bakker E, et al. Accuracy and precision of pseudo-continuous arterial spin labeling perfusion during baseline and hypercapnia: a head-to-head comparison with 15O H2O positron emission tomography. Neuroimage 2014; 92: 182–192. [DOI] [PubMed] [Google Scholar]
- 20.Svarer C, Madsen K, Hasselbalch SG, et al. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage 2005; 24: 969–979. [DOI] [PubMed] [Google Scholar]
- 21.Hammers A, Allom R, Koepp MJ, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp 2003; 19: 224–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bol A, Vanmelckenbeke P, Michel C, et al. Measurement of cerebral blood flow with a bolus of oxygen-15-labelled water: comparison of dynamic and integral methods. Eur J Nucl Med Mol Imaging 1990; 17: 234–241. [DOI] [PubMed] [Google Scholar]
- 23.Lammertsma AA, Martin AJ, Friston KJ, et al. In vivo measurement of the volume of distribution of water in cerebral grey matter: effects on the calculation of regional cerebral blood flow. J Cereb Blood Flow Metab 1992; 12: 291–295. [DOI] [PubMed] [Google Scholar]
- 24.Alpert NM, Eriksson L, Chang JY, et al. Strategy for the measurement of regional cerebral blood flow using short-lived tracers and emission tomography. J Cereb Blood Flow Metab 1984; 4: 28–34. [DOI] [PubMed] [Google Scholar]
- 25.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet Lond Engl 1986; 1: 307–310. [PubMed] [Google Scholar]
- 26.Yaqub M, Boellaard R, Kropholler MA, et al. Optimization algorithms and weighting factors for analysis of dynamic PET studies. Phys Med Biol 2006; 51: 4217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.