Abstract
As fast terminators of G-protein coupled receptor (GPCR) signaling, regulators of G-protein signaling (RGS) serve critical roles in fine-tuning second messenger levels and, consequently, cellular responses to external stimuli. Here, we report the creation of an optogenetic RGS2 (opto-RGS2) that suppresses agonist-evoked calcium oscillations by the inactivation of Gαq protein. In this system, cryptochrome-mediated hetero-dimerization of the catalytic RGS2-box with its N-terminal amphipathic helix reconstitutes a functional membrane-localized complex that can dynamically suppress store-operated release of calcium. Engineered opto-RGS2 cell lines were used to establish the role of RGS2 as a key inhibitory feedback regulator of the stochasticity of the Gαq-mediated calcium spike timing. RGS2 reduced the stochasticity of carbachol-stimulated calcium oscillations, and the feedback inhibition was coupled to the global calcium elevation by calmodulin/RGS2 interactions. The identification of a critical negative feedback circuit exemplifies the utility of optogenetic approaches for interrogating RGS/GPCR biology and calcium encoding principles through temporally precise molecular gain-of-function.
Keywords: Regulators of G-protein signaling (RGS), Optogenetics, Calcium signaling, Stochasticity
INTRODUCTION:
Regulators of G-protein signaling (RGS) are the primary fast terminators of G-protein coupled receptor (GPCR) signaling. They exert their inhibitory function through GAP (GTPase accelerating protein) activity on Gα protein to promote reformation of the heterotrimeric G-protein complex, thereby terminating signal transduction1–4. By fine-tuning the timing and intensity of GPCR signaling, RGS critically shape dynamic cellular responses to external stimuli. However, temporally precise technologies for probing the regulatory roles of specific RGS are lacking; subtype-selective pharmacological agents are challenging to create, and traditional genetic manipulation techniques are prone to compensatory confounds from RGS of non-interest, and from signaling changes that maintain basal second messenger load5–9. With the ability to dynamically recapitulate protein function in response to light, optogenetic approaches are ideal for fulfilling such technological needs.
Here, we engineer an optogenetic RGS2 (opto-RGS2) for temporally precise termination of Gαq-signaling and downstream release of calcium from the endoplasmic reticulum (ER)10–15. RGS2 serves key dynamic roles in regulating the frequency of resting and agonist-induced intracellular calcium oscillations9, 16, 17. Opto-RGS2 suppressed intracellular calcium oscillations in a rapidly light-inducible manner, and was used to establish a critical calcium encoding role for RGS2 in negative feedback control over the stochasticity of ligand-induced oscillations18, 19. By emulating brief pharmacological stimulation of RGS2 in the absence of available selective agonists, this work expands the optogenetic toolbox for GPCR signaling and reveals how RGS impact cellular dynamics through inhibitory regulation of oscillatory calcium signals.
RESULTS AND DISCUSSION:
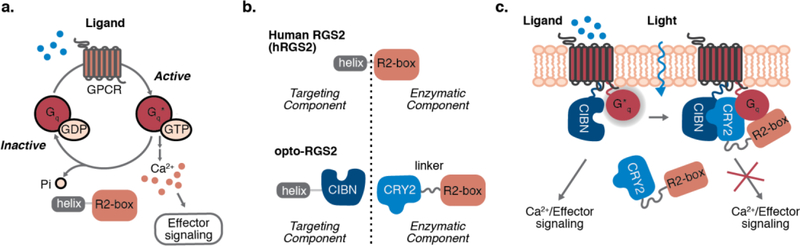
RGS signaling specificity (Figure 1a) predominately relies on terminal conserved domains to guide sub-cellular localization and target receptor co-localization of the catalytic “RGS box” domain. In the case of human RGS2 (hRGS2), its N-terminal amphipathic helix is critical for efficient inhibition of Gαq signaling12–15, 20, 21. As both components are involved in signaling function, opto-RGS2 was designed such that the split components co-localize upon light-induced hetero-dimerization, where the R2-box is fused to Arabidopsis cryptochrome 2 (CRY2-PHR)22 and the R2-helix is fused to the CRY2 interaction partner, CIBN (Figure 1b). CRY2-R2box is physically sequestered in the cytoplasm and unable to bind Gαq in the dark, and upon illumination, hetero-dimerizes with R2helix-CIBN to reconstitute an active protein that terminates the induction of calcium release (Figure 1c). A 15-residue glycine/serine-rich (GS) linker was placed between the RGS2-box and CRY2 to promote proper folding and improve expression (Supplementary Figure 1).
Figure 1. Design of optogenetic RGS2 (opto-RGS2).
(a) RGS2 terminates activated Gαq signaling as a GAP (GTPase accelerating protein). (b) Opto-RGS2 splits the catalytic box and N-terminal amphipathic helix of human RGS2, and fuses them respectively to cryptochrome (CRY2-PHR) and its interaction partner, CIBN. (c) CRY2-R2box is sequestered in the cytosol in the dark. Optogenetic hetero-dimerization reconstitutes a functional membrane-localized complex.
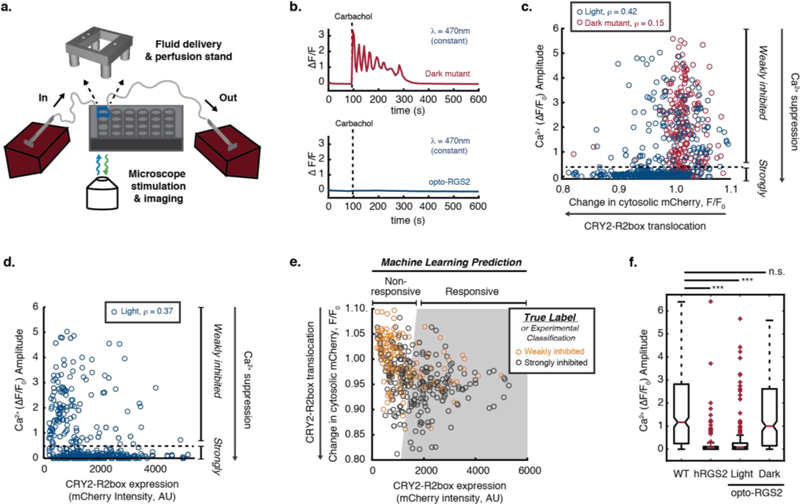
To assess the efficiency of opto-RGS2 in suppressing calcium signaling and identify its determinant factors, intracellular calcium elevations were induced through carbachol-stimulation (30 uM CCh) of endogenous M3 muscarinic acetylcholine receptors (M3R) in HEK cells and monitored by single-cell GCaMP6f imaging (Figure 2a–b). A CRY2 “dark mutant” or “blind mutant”, which harbors a D387A mutation to prevent uptake of the optically active flavin cofactor necessary for photo-induced signaling23, was used as a control to account for the effects of metabolic load and illumination. The degree of calcium amplitude suppression positively correlated with translocation efficiency (inverse of change in cytosolic mCherry (F/F0)) and with the expression level of CRY2-R2box (mCherry tag, F0), with Spearman’s rank correlation coefficients, ρ = 0.42 and 0.37, respectively (Figure 2c–d). The CRY2-R2box translocation of the “dark mutant” (red) is minimal and, in contrast, shows poor correlation with calcium suppression (ρ = 0.15) (Figure 2c).
Figure 2. Opto-RGS2 functional determinants in transfected HEK cells.
(a) Experimental setup for single-cell functional assays of optogenetic suppression of carbachol-induced oscillations. (b) GCamp6f imaging traces of exemplar 30 uM carbachol (CCh)-induced calcium oscillations and optogenetic suppression. (Top) Red = Illuminated opto-RGS2 dark mutant or optically insensitive apoprotein control. (Bottom) Blue = Illuminated opto-RGS2 or flavin holoprotein. (c) Correlation analysis between oscillation suppression and CRY2-R2box translocation (inverse of change in cytosolic mCherry intensity (F/F0)). ρ = Spearman’s rank correlation coefficient. Cells with strong calcium suppression (calcium amplitude lower than 0.5, beneath the dashed line) are considered “strongly inhibited”. (d) Correlation analysis between oscillation suppression and CRY2-R2box expression. (e) Machine learning-predicted decision boundary for discriminating cells likely to suppress calcium levels robustly (predicted responsive, gray-shaded area) based on translocation efficiency and CRY2-R2box expression level. True Label = Classification from experimental data shown in panels c-d as training set. The boundary was optimized to maximize the true positive prediction with 80% precision, where 80% of “predicted responsive” cells are “strongly inhibited” experimentally. (f) Box-and whisker plot of calcium oscillation peak amplitude in wild-type HEK cells, constitutively active hRGS2 cells, opto-RGS2 dark mutant, and opto-RGS2 cells selected by the machine learning criteria from panel e (dataset distinct from the training set). N = 174–367.
Given the cell-to-cell variability in transiently transfected cells, clonal cell lines are required to ensure that opto-RGS2 function is reliable. Thus, we sought quantitative criteria that would enable systematic selection of high-performing clones, while maintaining tractable cell line selection throughput. Machine learning (logistic regression) was performed to identify such a parameter space that predicts reliable calcium suppression based on CRY2-R2box translocation and expression level (Figure 2e), which are more facile screening parameters to obtain than calcium dynamics. The model was able to predict the magnitude of optogenetic response with a true positive rate as high as 80%, meaning that cells within the gray-shaded “predicted responsive” region are 80% likely to be “strongly inhibited” (defined in Figures 2c–d) experimentally. Note that as the model was built to maximize the true positive prediction for efficient cell line generation, the minimization of false negative predictions was not of interest.
To test the prediction criteria, calcium suppression assays were performed using transfected opto-RGS2 cells that were distinct from the machine learning training set. As expected in “predicted responsive” cells, opto-RGS2 activation efficiently suppressed calcium elevations and to a similarly near-complete extent as over-expressed hRGS2, whereas the dark mutant showed no inducible suppression, identical to the wild-type HEK cells (Figure 2f). The validated criteria were then deemed suitable for clonal selection.
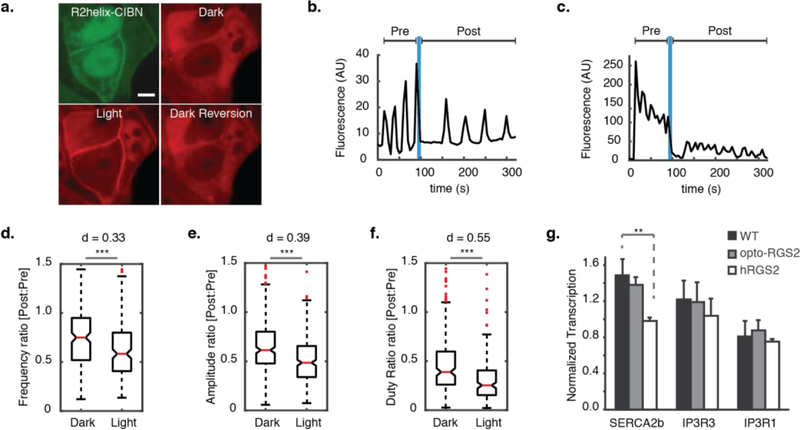
Opto-RGS2 cell lines (Figure 3a) were then created by lentivirus-mediated co-transduction of the hetero-dimerizing components and clonal selection based on the machine learning-identified parameters. Experiments were performed where opto-RGS2 cells were illuminated after the oscillation was established (Figure 3b–c, not simultaneously as in Figure 1). This single-cell paradigm enabled the light-induced dampening of the oscillation to be normalized to its own initial dark response to account for variability. A single illumination epoch was sufficient for opto-RGS2 activation because the CRY2 off-kinetics was long relative to the imaging duration (τoff = 396s ± 45 s, τon = 19.7s ± 2.0 s, Supplementary Figure 2). Oscillations (100 uM CCh-evoked) were dampened by a single illumination epoch, with significant reductions in frequency, amplitude, and duty ratio, the latter as a general suppression measure that encompasses the other two parameters (p < 0.001 for all; Figure 3d–f). The Cohen’s d effect size (d) was calculated to contextualize the significance, and the duty ratio suppression was of medium effect size (dduty = 0.55). As seen in the example traces (Figure 3b–c), individual cells varied in their respective responses, where the suppression often manifested itself as a change primarily in amplitude (damp = 0.33) or in frequency (dfreq = 0.39), and, thus, duty ratio effects were more pronounced across the population as a parameter.
Figure 3. Inhibitory regulation of calcium oscillations in opto-RGS2 cell lines.
(a) Fluorescence micrographs of light-induced translocation of mCherry-tagged CRY2-R2box (red) in opto-RGS2 cell lines. (top left) GFP-tagged R2helix-CIBN is membrane- and nuclear-localized, consistent with known R2helix distribution patterns. (top right) CRY2-R2box in the cytoplasm of the same cell, (bottom left) membrane localization 30 seconds after brief (1 s) blue light stimulation, and (bottom right) thermal reversion back to the cytoplasm in the dark (900 s post-illumination). Scale = 5 um. (b) Example X-rhodamine calcium imaging traces of dynamic suppression of 100 uM CCh-induced oscillations in response to a single epoch of blue light. Optogenetic dampening primarily decreased the frequency. (c) Same as panel b, except where optogenetic dampening primarily decreased the amplitude. (d) Single epoch of blue light dynamically reduced oscillation frequency. Post-illumination parameter values were normalized to pre-illumination values of the same oscillation in the same cell, as in panel a. (*** p < 0.001, d = Cohen’s effect size) (e) Same as panel d, except for amplitude. (f) Same as panel d, except for duty ratio. (g) Quantitative PCR measurements of SERCA2b and IP3R transcripts. SERCA2b level was altered in hRGS2 cells to compensate for long-term reduction in calcium load (p < 0.01), but not in opto-RGS2 cells.
A consequence of reversibility is that opto-RGS2 expression should not induce compensatory physiological changes to maintain calcium load (vs. wild-type) that would otherwise confound calcium encoding studies. Such changes are known to occur with traditional genetic manipulation, where knockout of hRGS2 alters IP3R (inositol triphosphate receptor) and SERCA2b (sarco/endoplasmic reticulum calcium ATPase) levels to maintain the intracellular calcium load9. In contrast, mRNA levels of these proteins measured here by qPCR were not altered by opto-RGS2, thus leaving basal transcriptional profiles undistorted unlike the over-expression of constitutively active hRGS2, which altered SERCA2b levels in cell lines (Figure 3g). Given that opto-RGS2 permits light vs. dark comparisons with less genetic variability than comparisons made between multiple populations of traditional genetic mutants vs. wild-type cells, it is valuable for defining cell circuits and their dynamic features.
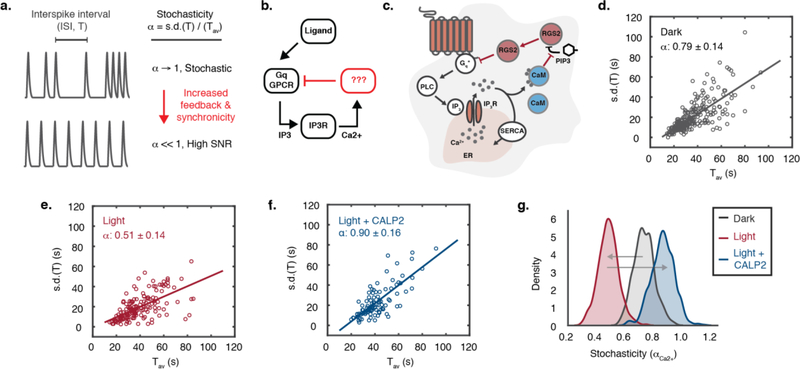
One such undefined circuit in calcium encoding is the feedback circuit that regulates the synchronicity of oscillations, which is critical to coordinating multiple calcium-dependent processes evoked by ligand-stimulation of Gαq-coupled GPCRs18, 19, 24, 25 (Figure 4a–b). It is known that the timing of an intracellular calcium oscillation spike does not depend on the previous one, and is thus stochastic, where stochasticity (αCa2+) is defined as the ratio of the standard deviation of the inter-spike interval (ISI, T) over the average period (Tav)18, 19. The oscillation signal-to-noise ratio (SNR, 1/αCa2+) can be enhanced by a negative feedback loop26. It has been suggested that the oscillation feedback occurs at the GPCR-source level because the stochasticity is unchanged by manipulation of IP3R, phospholipase C, and SERCA, which respectively operate at the ER-level or before it18.
Figure 4. Opto-RGS2 inhibitory feedback reduces stochasticity of calcium oscillation spike timing.
(a) Interspike intervals of ligand-induced oscillations become less stochastic with feedback. (b) The negative feedback regulator and its coupling mechanism to global elevations in calcium are still unknown as reflected in the lump circuit diagram. (c) Proposed RGS2-mediated negative feedback loop, where inhibition of Gαq-signaling is coupled to global calcium elevations through relief of PIP3 inhibition of RGS2 by calcium-activated calmodulin (CaM). (d) Stochasticity (α) of 100 uM CCh-induced calcium oscillations in non-illuminated opto-RGS2 cell lines. (e) Opto-RGS2 activation decreased the stochasticity in response to a single blue light epoch (as in Figure 3). (f) The Inhibitory feedback is uncoupled from the global calcium elevation by CaM-antagonism by CALP2 peptide. (g) Statistical bootstrapping or resampling analysis of variance (N = 1000 trials) for panels d-f.
We hypothesized that RGS2 may be this regulator (Figure 4c), given that it (i) provides GPCR-source level inhibition, (ii) is coupled to global calcium elevations via Ca2+-bound calmodulin-mediated (Ca2+/CaM) relief of RGS2 inhibition by phosphatidylinositol-(3,4,5)-triphosphate (PIP3)9, 27, and (iii) is important for generating oscillations9, 16. Analysis of the oscillation suppression data in Figure 4 showed that the spike timing was highly stochastic in the dark, where αdark = 0.79 ± 0.14 (α = 1 is fully stochastic) (Figure 4d), and became more synchronized with light-activation of opto-RGS2, αlight = 0.51 ± 0.14 (Figure 4e). This finding showed that increasing RGS2-mediated inhibitory drive enhanced the regularity and SNR of the oscillation. RGS2 likely limits the degree of negative regulation in the proposed circuit diagram, because if the activated Ca2+/CaM were not in excess, the increased inhibitory drive from opto-RGS2 activation would be poorly feedback coupled to the global calcium elevation and would not decrease αCa2+.
Accordingly, to confirm that RGS2 is feedback coupled via CaM, the assays were performed in the presence of the CaM peptide antagonist, CALP2, which we hypothesized would uncouple the inhibition from the calcium elevation. Pharmacological interference of the putative coupling mechanism indeed increased the stochasticity in illuminated cells, αlight/CALP2 = 0.90 ± 0.16 (Figure 4f), and thus, RGS2 inhibitory feedback is calmodulin-dependent. Variance of αCa2+ was estimated by statistical resampling or bootstrapping, and the results were significant at the 95% confidence level (Figure 4g). Thus, this optogenetic analysis defines a negative feedback circuit for modulating the stochasticity of calcium oscillations, and consequently, the reliability of the signals they encode.
The ability to dynamically recapitulate RGS2 function is important because the creation of selective agonists is hindered by the conservation of the catalytic RGS-box across subtypes5–8. The R2-helix was chosen here to direct membrane localization based on its reported roles in RGS2 signaling12–15, 20, 21, but it should be noted that an isoprenylated peptide-CIBN fusion has also successfully been used to create optogenetic variants of another RGS subtype28. While protein-level design could greatly influence signaling performance, as evident by the improved expression of the CRY2-R2box chimera by introducing a long linker between the conserved domains, not all performance-related issues were a function of protein-level design alone. The performance of hetero-dimerization systems exhibits great cell-to-cell variability, but proper selection of a homogenous population with respect to screening parameters of expression and translocation efficiency provided the requisite reliability for calcium encoding studies. Thus, this work emphasizes the cellular engineering techniques required to successfully engineering a molecular tool, beyond the transgene sequence itself.
Opto-RGS2 is a key addition to the optogenetic toolbox for studying GPCRs and second messenger signaling. The ability to potently suppress calcium is absent from the toolbox, and thus, the functionality complements existing capabilities in GPCR-level elevation of calcium and elevation/suppression of cyclic AMP28–32. The functionality is powerful given the ubiquitous importance of GPCRs and calcium signaling in eukaryotes, and it led to the elucidation of a feedback circuit that regulates the stochasticity of agonist-evoked oscillations. This increase in oscillation synchronicity by up-regulated RGS2 suggests that RGS2-feedback has coordinative roles in calcium encoding, important for coincidence detection or multiplexing of the downstream calcium effectors33, 34. In summary, opto-RGS2 may advance the understanding of calcium encoding principles and RGS/GPCR signaling through temporally precise gain-of-function.
MATERIALS AND METHODS:
Cryptochrome and RGS-related plasmids (denoted as P0-P9) used during this study are summarized in Supplementary Table 1. Plasmids are available at Addgene (#112257 and #112258) (http://www.addgene.org/Brian_Chow/).
Genetic constructs
The plasmids were constructed using Gibson assembly and standard restriction enzyme cloning methods. Refer to Supplementary Information for detailed procedures.
Cell culture
Human embryonic kidney 293 (HEK293t) cells (from the Lazzara Lab, UPenn) were cultured in Dulbecco’s modified eagle medium (DMEM) with Glutamax (ThermoFisher, 10566016), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Sigma, F2442) and 1% penicillin-streptomycin (ThermoFisher, 15140122). Cells were maintained in a sterile water-jacketed mammalian cell culture incubator (Thermo/Forma, 3110), maintained at 5% CO2 and 37 °C. HEK cell authentication by STR and mycoplasma testing by Myco-Alert were performed by Biosynthesis, Inc.
Transient transfection
DNA plasmid (24-well plate, 250 ng/well; 6-well plate, 1.25 ug/well) was mixed with TransIT-293 transfection reagent (24-well plate, 0.75 uL/well; 6-well plate, 3.75 uL/well) (Mirus Bio, MIR2700) in Opti-MEM I Reduced-Serum Medium (24-well plate, 100 uL/well; 6-well plate, 500 uL/well) (ThermoFisher, 31985070). The mix was incubated at room temperature for 15 minutes and was subsequently added to seeded cells. Gene expression was assessed 18–24 hours post-transfection.
Linker analysis
The respective expressions of pCRY2-mCherry-linker-RGS2box constructs and pNtermRGS2-GFP-CIBN [P5] were assessed by flow cytometry. Cells were seeded onto a poly-D-lysine (PDL)-treated 6-well plate 24 hours prior to co-transfection. Cells were trypsinized 24 hours post-transfection and washed three times with phosphate-buffered saline (PBS) (ThermoFisher, 14190250). Cells were re-suspended in flow buffer containing 1X Ca2+/Mg2+-free PBS, 5 mM EDTA, 25 mM HEPES pH 7.0, and 1% FBS. Cells were subsequently strained with a 70 μm cell strainer (Sigma, Z742103) and collected in a 5 mL polystyrene round-bottom FACS tube (Corning, 352054). The flow analyses were performed on 4-laser BD LSRII Cell Analyzer (mCherry detection: green laser with 610/20 emission filter; GFP detection: blue laser with 515/20 emission filter), at a rate of approximately 4000 events/s, with 100,000 events collected for each condition. Three gates were applied to select for live single cells with expression of both cryptochrome components.
Fluorescence microscopy and hardware
Cells were imaged with a Metamorph-automated Leica DMI6000B fluorescence microscope, equipped with a sCMOS camera (PCO.edge) and LED illuminator (Lumencor Spectra-X). The excitation illumination was filtered at the light source-level, and the microscope filter cubes (Chroma) used were: EGFP, λdichroic < 495 nm and λem = 525/50 nm; mCherry, λdichroic < 585 nm and λem = 630/75 nm). Pharmacological agents were delivered using a custom automated perfusion system consisting of a 3D-printed well-aligning scaffold and programmable syringe pumps (Brain Scientific, BS-8000), triggered by MetaMorph.
Fluorescence micrographs of light-induced opto-RGS2 translocation shown in Figure 3a were acquired using a Leica DMI8 inverted microscope (courtesy of the Hammer Lab, UPenn) equipped with confocal spinning disk module from Spectral Applied Research and Hamamatsu ImagEM X2 EM-CCD camera.
Membrane localization kinetics
Cells were seeded onto a poly-D-lysine coated 24-well glass bottom plate (Cellvis, P24–1.5H-N) and were co-transfected with CRY2-R2box and R2helix-CIBN [P3 + P5]. After 24 hours, the culture media was replaced with phenol-free CO2-independent imaging media (phenol-free HBSS supplemented with 1% L-glutamine, 1% Penn/Strep, 2% essential amino acids, 1% non-essential amino acids, 2.5% HEPES pH 7.0, and 10% serum).
Cells were imaged with a 40X objective (0.5 s exposure time, 675 s total, using the mCherry filter cube with yellow excitation: λ = 580 nm, 24.20 mW/mm2). CRY2-R2box imaging was first performed without blue illumination for 15 s to establish baseline before blue light induction (0.5 s every 5 s for 60 s, λ = 475 nm, 7.37 mW/mm2). Blue light stimulation was then removed to determine dark reversion kinetics. The localization of R2helix-CIBN was acquired during the blue illumination period. Line sections of consistent length were manually drawn across selected segments of cell boundaries. The degree of CRY2-R2box membrane localization was calculated as the mCherry intensity at the pixels representing the cell membrane, normalized by the total intensity of the perimembrane region. Rate constants were determined by an exponential fit.
Calcium suppression assays in transfected cells
Calcium signals in cells expressing opto-RGS2 [P3 + P6], opto-RGS2 dark mutant [P4 + P6], and hRGS2 [P1] were evaluated against the negative control cell17 [P2 + P6]. Intracellular calcium signals were monitored using GCaMP6f in glass-bottom 24-well plates. (Addgene, Plasmid No. 40755). Seeded cells were co-transfected with GCaMP6f plasmid and one of the plasmid combinations above. After 24 hours post-transfection, the culture media was replaced with the phenol-free CO2 independent media. The cells were perfused with 30 uM carbachol (CCh) (Sigma, C4382) to stimulate the M3 muscarinic acetylcholine receptor (M3R). GCaMP6f fluorescence signals were monitored every 5 s for 900 s. mCherry fluorescence was imaged before GCaMP6f imaging, and again once every 60 s.
Image processing, manual cell segmentation and intensity quantifications were performed using ImageJ. The intensity data were imported to MATLAB for signal processing, peak identification, and peak prominence measurements of amplitude. Signal prominence was defined as the extent to which the peak stands out due to its intrinsic height, using the MATLAB “findpeaks()” function. The signal plotted is the relative change in fluorescence signal (ΔF/F0). Samples were considered outliers if they were greater than q3 + 1.5(q3 - q1) or less than q1 – 1.5(q3 - q1) where q1 and q3 represent the 25th and 75th percentiles of the sample data, respectively.
Correlation analysis and Machine learning prediction
Spearman’s rank correlation analysis was performed using the Python library pandas (pandas.Dataframe.corr()) to identify predictive parameters affecting the strength of the calcium suppression (defined as inverse of calcium prominence). Correlation between calcium suppression and the following parameters were analyzed: translocation efficiency, expression of CRY2-R2box, and expression of R2helix-CIBN. No correlation was observed for R2helix-CIBN level.
The parameter space at which cells completely inhibited the calcium signals were identified using a machine learning method logistic regression (scikit-learn library, L2 regularization, C = 1.0, “balanced” class weight); the model parameters were optimized using GridSearchCV function to maximize prediction precision (true-positive rate) (precision = 80%). The calcium signals considered strongly inhibited had normalized prominence values of ΔF/F0 < 0.5.
Lentivirus production and cell line generation
Opto-RGS2 lentivirus was generated by transfecting HEK293t cells with either pLJM1-hRGS2-mCherry-(GGGGS)3-hRGS2box [P8] or pLJM1-NtermRGS2-GFP-CIBN [P9] with 3rd generation helper plasmids. The media was replaced one day after transfection, and virus-containing supernatant was collected on the second and third days. After centrifugation (1000 rpm, 5 minutes), the supernatant containing the lentivirus was filtered (0.4 um PES) and used immediately.
To generate an opto-RGS2 stable cell line, HEK293t cells were co-infected with the generated lentiviruses carrying CRY2-R2box and R2helix-CIBN. The infected cells were grown and selected in culture media containing 0.125 ug/mL puromycin (Clontech, 631305). Individual clones expressing both components of opto-RGS2 were picked under a sterile fluorescence microscope (courtesy of the Cremins Lab, Penn) and grown in 24-well plates. Light-induced translocation of the CRY2-R2box was evaluated for each clone to select suitably responsive ones (i.e. based on correlation analysis-defined threshold). The hRGS2 stable cell line was generated similarly using P7 plasmid.
Calcium dynamics of stable cell lines
Opto-RGS2 stable cells seeded on a PDL-coated glass-bottom 24-well plate were loaded with 2 uM X-Rhod-1 calcium indicator dye (ThermoFisher, X14210). Cells were incubated with dye at 37 °C, and washed after 30 minutes, after which the media was replaced with CO2-independent media. Cells were incubated at 37 °C for another 30 minutes for de-esterificaition. To ensure oscillatory activity, the media was replaced with 5.2 mM calcium-containing CO2-independent media. Cells were perfused with 100 uM carbachol, and time-lapse images of XRhod1 signals were acquired. After 90 s, cells were exposed to brief blue light pulses (10 s total, 0.5 s ON, 0.5 s OFF).
Image processing, manual cell segmentation, and intensity quantification were performed in ImageJ. The intensity measurements were passed into automated custom MATLAB scripts for signal processing, peak identification (20% above baseline threshold), and to compute reported oscillation parameters. The frequency ratio was calculated by dividing post-illumination (t = 100 – 250s) oscillation frequency by pre-illumination (t = 0 – 100s) frequency. Student t-test, and Cohen’s d test were performed to evaluate the significance and the effect size of the light-induced change. Identical analyses were performed with amplitude and duty ratio.
Quantitative PCR (qPCR)
RNA was extracted using standard TRIzol RNA extraction procedures. The extracted RNA was transcribed into complementary DNA (cDNA) using SuperScript III reverse transcriptase (200 units/u, ThermoFisher, 18080093). The cDNA was subsequently used as a template for the SYBR-based qPCR. Refer to Supplementary Information for the detailed procedures and the list of qPCR primers.
Stochasticity analysis
Assay and data processing were performed as described under “Calcium dynamics of stable cell lines” section. 50 uM of CALP2 (Tocris, 2319) was added to the media 1 hour prior to calcium imaging to inhibit calmodulin activity. Stochasticity was assessed based on the variability of the interspike intervals (ISI, or T) within a signal relative to its mean (Tav). Linear regression analysis was performed using the MATLAB function LinearModel.fit to derive the estimated stochasticity. 450–1000 cells were analyzed for each experimental condition. The 95% bootstrap confidence interval was calculated using bias corrected and accelerated percentile methods, with 1000 bootstrap samples.
Supplementary Material
ACKNOWLEDGMENTS:
The authors thank Spencer Glantz for constructing the fluidic delivery system, Michael Magaraci for flow cytometry analysis, Ben Schuster for confocal analysis, Orion Weiner for helpful discussion on cell line development, Skip Brass for helpful discussion on RGS assays, Arjun Raj and Danielle Bassett for helpful discussion in statistical analysis methods, and Jennifer Phillips-Cremins and Dan Hammer for generously sharing equipment. B.Y.C. acknowledges the support of NSF Biophotonics (CBET 126497), NSF CAREER (MCB 652003), W.W. Smith Charitable Trust for the Heart, Penn Medicine Neuroscience Center, NIH/NIDA (1R21 DA040434–01), and NIH/NINDS (R01NS101106). P.H. was on partial fellowship support from the Thai Ministry of Science and Technology Scholarship.
ABBREVIATIONS:
- CaM
Calmodulin
- CCh
Carbachol
- CIBN
Cryptochrome 2 interaction partner
- CRY2 or CRY2-PHR
Cryptochrome 2
- ER
Endoplasmic reticulum
- GAP
GTPase accelerating protein
- GPCR
G-protein coupled receptor
- HEK
Human embryonic kidney
- hRGS
Human regulator of G-protein signaling
- IP3R
Inositol triphosphate receptor
- ISI
Interspike intervals
- Opto-RGS2
Optogenetic regulator of G-protein signaling 2
- PIP3
Phosphatidylinositol-(3,4,5)-triphosphate
- R2-helix
N-terminal amphipathic helix of human RGS2
- R2-box
C-terminal catalytic “RGS box” domain of human RGS2
- RGS
Regulator of G-protein signaling
- SERCA2b
Sarco/endoplasmic reticulum calcium ATPase
- SNR
Signal-to-noise ratio
- SOR
Store-operated release
Footnotes
SUPPLEMENTARY INFORMATION: Detailed experimental procedures on genetic constructs and qPCR and supplemental data, including: Supplementary Figure 1: Linker analysis; Supplementary Figure 2: Membrane translocation and dissociation kinetics; Supplementary Table 1: Plasmids constructed for this study; Supplementary Table 2: Primers used for qPCR experiments.
REFERENCES:
- [1].Berman DM, and Gilman AG (1998) Mammalian RGS Proteins: Barbarians at the Gate, Journal of Biological Chemistry 273, 1269–1272. [DOI] [PubMed] [Google Scholar]
- [2].Neubig RR, and Siderovski DP (2002) Regulators of G-protein signalling as new central nervous system drug targets, Nat Rev Drug Discov 1, 187–197. [DOI] [PubMed] [Google Scholar]
- [3].Hollinger S, and Hepler JR (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling, Pharmacological reviews 54, 527–559. [DOI] [PubMed] [Google Scholar]
- [4].Riddle EL, Schwartzman R. a., Bond M, and Insel P. a. (2005) Multi-tasking RGS proteins in the heart: the next therapeutic target?, Circulation research 96, 401–411. [DOI] [PubMed] [Google Scholar]
- [5].Sjogren B, and Neubig RR (2010) Thinking outside of the “RGS box”: new approaches to therapeutic targeting of regulators of G protein signaling, Mol Pharmacol 78, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, and Neubig RR (2007) Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay, Mol Pharmacol 71, 169–175. [DOI] [PubMed] [Google Scholar]
- [7].Kaur K, Kehrl JM, Charbeneau RA, and Neubig RR (2011) RGS-insensitive Galpha subunits: probes of Galpha subtype-selective signaling and physiological functions of RGS proteins, Methods in molecular biology (Clifton, N.J.) 756, 75–98. [DOI] [PubMed] [Google Scholar]
- [8].Kimple AJ, Bosch DE, Giguère PM, and Siderovski DP (2011) Regulators of G-Protein Signaling and Their Gα Substrates: Promises and Challenges in Their Use as Drug Discovery Targets, Pharmacological Reviews 63, 728–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang X, Huang G, Luo X, Penninger JM, and Muallem S (2004) Role of Regulator of G Protein Signaling 2 (RGS2) in Ca2+ Oscillations and Adaptation of Ca2+ Signaling to Reduce Excitability of RGS2-/- Cells, Journal of Biological Chemistry 279, 41642–41649. [DOI] [PubMed] [Google Scholar]
- [10].Kehrl JH, and Sinnarajah S (2002) RGS2: a multifunctional regulator of G-protein signaling, The international journal of biochemistry & cell biology 34, 432–438. [DOI] [PubMed] [Google Scholar]
- [11].Ingi T, Krumins AM, Chidiac P, Brothers GM, Chung S, Snow BE, Barnes CA, Lanahan AA, Siderovski DP, Ross EM, Gilman AG, and Worley PF (1998) Dynamic Regulation of RGS2 Suggests a Novel Mechanism in G-Protein Signaling and Neuronal Plasticity, The Journal of Neuroscience 18, 7178–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Heximer SP, Srinivasa SP, Bernstein LS, Bernard JL, Linder ME, Hepler JR, and Blumer KJ (1999) G protein selectivity is a determinant of RGS2 function, The Journal of biological chemistry 274, 34253–34259. [DOI] [PubMed] [Google Scholar]
- [13].Heximer SP, Watson N, Linder ME, Blumer KJ, and Hepler JR (1997) RGS2/G0S8 is a selective inhibitor of Gqalpha function, Proceedings of the National Academy of Sciences of the United States of America 94, 14389–14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Heximer SP, Lim H, Bernard JL, and Blumer KJ (2001) Mechanisms governing subcellular localization and function of human RGS2, The Journal of biological chemistry 276, 14195–14203. [DOI] [PubMed] [Google Scholar]
- [15].Bernstein LS, Ramineni S, Hague C, Cladman W, Chidiac P, Levey AI, and Hepler JR (2004) RGS2 Binds Directly and Selectively to the M1 Muscarinic Acetylcholine Receptor Third Intracellular Loop to Modulate Gq/11α Signaling, Journal of Biological Chemistry 279, 21248–21256. [DOI] [PubMed] [Google Scholar]
- [16].Luo X, Popov S, Bera a. K., Wilkie TM, and Muallem S (2001) RGS proteins provide biochemical control of agonist-evoked [Ca2+]i oscillations, Molecular cell 7, 651–660. [DOI] [PubMed] [Google Scholar]
- [17].Bernhardt ML, Lowther KM, Padilla-Banks E, McDonough CE, Lee KN, Evsikov AV, Uliasz TF, Chidiac P, Williams CJ, and Mehlmann LM (2015) Regulator of G-protein signaling 2 (RGS2) suppresses premature calcium release in mouse eggs, Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Thurley K, Tovey SC, Moenke G, Prince VL, Meena A, Thomas AP, Skupin A, Taylor CW, and Falcke M (2014) Reliable Encoding of Stimulus Intensities Within Random Sequences of Intracellular Ca2+ Spikes, Science Signaling 7, ra59–ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Thurley K, Skupin A, Thul R, and Falcke M (2012) Fundamental properties of Ca2+ signals, Biochim Biophys Acta 1820, 1185–1194. [DOI] [PubMed] [Google Scholar]
- [20].Nance MR, Kreutz B, Tesmer VM, Sterne-Marr R, Kozasa T, and Tesmer JJ (2013) Structural and functional analysis of the regulator of G protein signaling 2-galphaq complex, Structure 21, 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kimple AJ, Soundararajan M, Hutsell SQ, Roos AK, Urban DJ, Setola V, Temple BR, Roth BL, Knapp S, Willard FS, and Siderovski DP (2009) Structural determinants of G-protein alpha subunit selectivity by regulator of G-protein signaling 2 (RGS2), The Journal of biological chemistry 284, 19402–19411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, and Tucker CL (2010) Rapid blue-light-mediated induction of protein interactions in living cells, Nat Meth 7, 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, and Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis, Science 322, 1535–1539. [DOI] [PubMed] [Google Scholar]
- [24].Perc M, Green AK, Dixon CJ, and Marhl M (2008) Establishing the stochastic nature of intracellular calcium oscillations from experimental data, Biophysical Chemistry 132, 33–38. [DOI] [PubMed] [Google Scholar]
- [25].Rüdiger S (2014) Stochastic models of intracellular calcium signals, Physics Reports 534, 39–87. [Google Scholar]
- [26].Brandman O, and Meyer T (2008) Feedback Loops Shape Cellular Signals in Space and Time, Science 322, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Popov SG, Krishna UM, Falck JR, and Wilkie TM (2000) Ca2+/Calmodulin Reverses Phosphatidylinositol 3,4,5-Trisphosphate-dependent Inhibition of Regulators of G Protein-signaling GTPase-activating Protein Activity, Journal of Biological Chemistry 275, 18962–18968. [DOI] [PubMed] [Google Scholar]
- [28].O’Neill PR, and Gautam N (2014) Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration, Molecular Biology of the Cell 25, 2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Airan RD, Thompson KR, Fenno LE, Bernstein H, and Deisseroth K (2009) Temporally precise in vivo control of intracellular signalling, Nature 458, (7241)-1029. [DOI] [PubMed] [Google Scholar]
- [30].Hannanta-Anan P, and Chow BY (2016) Optogenetic Control of Calcium Oscillation Waveform Defines NFAT as an Integrator of Calcium Load, Cell systems 2, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kyung T, Lee S, Kim JE, Cho T, Park H, Jeong YM, Kim D, Shin A, Kim S, Baek J, Kim J, Kim NY, Woo D, Chae S, Kim CH, Shin HS, Han YM, Kim D, and Heo WD (2015) Optogenetic control of endogenous Ca(2+) channels in vivo, Nat Biotechnol 33, 1092–1096. [DOI] [PubMed] [Google Scholar]
- [32].Bailes HJ, Zhuang L-Y, and Lucas RJ (2012) Reproducible and Sustained Regulation of Gαs Signalling Using a Metazoan Opsin as an Optogenetic Tool, PLOS ONE 7, e30774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lin Y, Sohn CH, Dalal CK, Cai L, and Elowitz MB (2015) Combinatorial gene regulation by modulation of relative pulse timing, Nature 527, 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wollman R, and Meyer T (2012) Coordinated oscillations in cortical actin and Ca2+ correlate with cycles of vesicle secretion, Nat Cell Biol 14, 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.