Abstract
Background
The PPARG2 Pro12Ala (rs1801282) and IL6 -174G >C (rs1800795) have important function in body weight regulation and a potential role in obesity risk. We aimed to investigate the association between PPARG2 Pro12Ala and IL6 -174G >C variants and the genotypes interaction with body composition, metabolic markers, food consumption, and physical activity in severely obese patients.
Methods
150 severely obese patients (body mass index (BMI) ≥ 35 kg/m2) from Central Brazil were recruited. Body composition, metabolic parameters, physical activity, and dietary intake were measured. The genotype was determined by the qPCR TaqMan Assays System. Multiple linear regression and multiple logistic regression models were fitted adjusting for confounders.
Results
Ala carriers of the Pro12Ala polymorphism had higher adiposity measures (BMI: p=0.031, and fat mass: p=0.049) and systolic blood pressure (p=0.026) compared to Pro homozygotes. We found no important associations between the -174G >C polymorphism and obesity phenotypes. When genotypes were combined, individuals with genotypes ProAla + AlaAla and GC + CC presented higher BMI (p=0.029) and higher polyunsaturated fatty acids (PUFAs) consumption (p=0.045) compared to the ones with genotypes ProPro and GG, and individuals carriers of the PPARG2 Ala allele only (genotype ProAla + AlaAla and GG) had higher fat mass and systolic and diastolic blood pressure compared to the ones with genotypes ProPro and GG.
Conclusions
Severely obese individuals carrying the Ala allele of the PPARG2 Pro12Ala polymorphism had higher measures of adiposity and blood pressure, while no important associations were found for the IL6 -174G >C polymorphism.
1. Introduction
Studies about the genetic obesity susceptibility have investigated polymorphisms related to genes encoding factors involved in food and energy intake regulation, energy expenditure, and adipogenesis control [1]. In this context, the polymorphisms PPARG2 Pro12Ala (rs1801282) and IL6 -174G >C (rs1800795) have received attention for their possible influence in body weight regulation [1–3].
The PPARG2 has an important role in modulating the expression of genes involved in the regulation of adipose tissue differentiation and lipid metabolism [4, 5]. The mostly studied polymorphism of PPARG2, the Pro12Ala, may promote lower affinity of the PPAR-γ for the response element and about 50% lower transcriptional capacity [6]. The Ala allele has been associated with lower body mass index (BMI) [6, 7]; nevertheless, some meta-analyses have found contradictory results showing higher BMI in Ala carriers, especially in severely obese individuals [8–11]. Considering this potential association of the Ala allele and increased BMI, it is important to investigate whether this variant influences other parameters in severely obese individuals.
Regarding interleukin-6 (IL-6), one of the major proinflammatory cytokines, it is positively related to increased BMI [3, 12]. The association studies between obesity and the IL6 -174G >C polymorphism have shown higher BMI in the presence of C allele in cross-sectional and cohort studies [13–17], but it was not confirmed by meta-analysis [18, 19]. Thus, the role of Pro12Ala and -174G >C in modulating BMI is still to be confirmed and also the information regarding other body composition parameters such as fat mass, fat-free mass, percentage body fat, and lean mass.
Interaction of genes with environmental factors, such as diet and physical activity, may be involved in the discrepancies of associations between studies [1, 20, 21]. Gene-diet interaction studies with Pro12Ala and -174G >C polymorphisms have found that the energy content and composition of the diet may affect obesity phenotypes, showing the importance to assess the interactions among genotypes and potentially modifiable lifestyle factors [20, 22, 23]. Considering that the PPARG2 Pro12Ala and IL6 -174G >C polymorphisms have important functions in body weight regulation with a potential role in obesity risk and the alarming increase in severe obesity worldwide [24], they are promising single nucleotide polymorphisms (SNPs) for association studies of obesity phenotypes. Thus, we aimed to investigate the association between PPARG2 Pro12Ala and IL6 -174G >C variants and the genotypes interaction with body composition, metabolic markers, food consumption, and physical activity in severely obese patients.
2. Methods
2.1. Subjects
This study is an analysis of baseline data from participants of the clinical trial “Effect of Nutritional Intervention and Olive Oil in Severe Obesity: Randomized Controlled Trial” (DieTBra Trial) (registered at ClinicalTrials.gov: NCT02463435). A total of 150 severely obese patients (BMI ≥ 35 kg/m2) aged 18 to 65 years were recruited from primary care of the Brazilian Unified Health System at Goiânia, Goiás State, in Central Brazil. Patients were referred to the Nutrition in Severe Obesity Outpatient Clinic, and the study took place at the Clinical Research Unit of the Clinical Hospital/Federal University of Goiás. The study excluded individuals that had already underwent bariatric surgery, under actual nutritional treatment for weight loss or in the previous 2 years, using antiobesity or anti-inflammatory drugs, having HIV/AIDS, as well as heart/kidney/hepatic insufficiency, chronic obstructive pulmonary disease, cancer, and pregnancy. Patients were recruited from June to November 2015. All patients who agreed to participate in the study gave written consent. The study was approved by the Research Ethics Committee of Clinical Hospital of Federal University of Goiás (protocol number 747.792).
2.2. Anthropometric and Body Composition Measurements
Body weight and height were measured using standardized procedures [25]. BMI (kg/m2) was calculated dividing the body mass (kg) by the squared height (m2). Severe obesity was defined as BMI ≥ 35 kg/m2 [24].
Fat mass (kg), fat-free mass (kg), percentage body fat, and lean mass were measured using multifrequency bioelectrical impedance analysis (BIA). The measurement was performed with the InBody S10 device (Biospace Co., Ltd., Seoul, Korea) by using different frequencies (1, 5, 50, 250, 500, and 1000 kHz) at each segment (right arm, left arm, trunk, right leg, and left leg).
For BIA assessment, patients were instructed to fast for 12 h and avoid strenuous physical activity and alcohol, as well as food and drinks containing caffeine on the previous day [26]. BIA assessment was conducted according to the manufacturer's guidelines.
2.3. Dietary Intake
Food consumption was assessed using three 24 h records collected within seven days, being two face to face and one by phone. Data were assessed by trained registered dieticians. We used the multiple pass method (MPM) to collect the 24 h records [27], and the nutritional analysis was performed using Avanutri Online® (Avanutri Equipamentos de Avaliação Ltda, Rio de Janeiro, BR). Energy (kcal), proteins (%), carbohydrates (%), lipids (%), saturated fatty acids (SAFs) (%), monounsaturated fatty acids (MUFAs) (%), polyunsaturated fatty acids (PUFAs) (%), polyunsaturated : saturated fatty acids ratio (P : S ratio), cholesterol (g), and fiber (g) were obtained calculating the mean of the three 24 h records.
2.4. Physical Activity Assessment
Physical activity was assessed using a triaxial accelerometer ActiGraph wGT3X (ActiGraph, Pensacola, FL, USA) for movement registration. Patients were instructed to wear the accelerometer 24 h a day for six consecutive days over the nondominant wrist, even during shower and water activities, as the device was waterproof. The sampling frequency of the accelerometer was set at 30 Hz, and the data collection interval was set at one min. Accelerometers were set up and downloaded at ActiLife 6 software. Output data were processed using the R-package GGIR (http://cran.r-project.org). The outcome measures used in the present study were moderate-to-vigorous physical activity (MVPA) (>100 mg) defined as estimated time spent in ≥ 10 min per bout during a week and the sedentary time (<50 mg, without bouts) in min per day.
2.5. Blood Pressure and Comorbidities
Systolic and diastolic blood pressures were measured with the patient in the sitting position after resting for at least 5 min. Two measures were taken within the 3 min interval using the Omron HEM 742INT (Omron Healthcare Inc., Kyoto, Japan) automatic blood pressure monitor with an appropriately sized cuff, and the mean was calculated.
The presence of comorbidities was analyzed as a dichotomous variable (presence/absence). Subjects with systolic/diastolic blood pressure higher than 140/90 mmHg or under antihypertensive therapy were considered hypertensive [28]. Subjects with fasting glucose ≥ 126 mg/dL or under glucose-lowering therapy were considered diabetic [29]. Individuals with low-density lipoprotein (LDL) cholesterol ≥ 160 mg/dL and/or triglycerides ≥ 150 mg/dL or high-density lipoprotein (HDL) cholesterol < 40 mg/dL for men and < 50 mg/dL for women were classified as dyslipidaemic [30].
2.6. Laboratory Tests
Blood samples were collected for metabolic markers and genomic DNA extraction after 12 h overnight fasting. Serum glucose, total cholesterol, HDL cholesterol, LDL cholesterol, and triglycerides were measured by enzyme-colorimetric methods. Serum insulin was measured by chemiluminescence, and hemoglobin A1c (HbA1c) was measured by liquid chromatography. The homeostasis model assessment of insulin resistance (HOMA-IR) was calculated following the formula derived by Matthews et al. [31].
2.7. DNA Extraction and Genotyping
Genomic DNA was extracted from whole blood with the PureLink™ Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA). DNA concentration and purity were evaluated by spectrophotometric determination of the A260/280 ratio with NanoDrop® 2000c (Thermo Fisher Scientific, Waltham, MA, USA), and DNA quality was checked using agarose gel electrophoresis. Genotyping was performed using custom TaqMan SNP genotyping assays—ID c__1129864_10 for PPARG2 Pro12Ala (rs1801282) and ID c__1839697_20 for IL6 -174G >C (rs1800795) (Applied Biosystems, Foster City, CA, USA)—on a StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The standard real-time polymerase chain reaction (qPCR) was carried out using the TaqMan GTXpress™ Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) reagent kit in a 21 µL volume according to the manufacturer's instructions. Although DNA samples were extracted for all study participants, the qPCR amplification was only conducted for the PPARG2 Pro12Ala polymorphism on samples from 146 individuals and for the IL6 -174G >C polymorphism on samples from 148 individuals.
2.8. Statistical Analysis
The dataset was structured using EpiData 3.1, and double entry typing with validation was performed. The data were presented as mean ± SD for continuous variables and frequencies for categorical variables. The chi-squared test was used to analyze the agreement of genotype frequencies with the Hardy–Weinberg equilibrium expectation. Allele frequency was determined by manual counting. Normal distribution was tested for all measured variables using the Kolmogorov–Smirnov test and histograms; skewed variables were normalized by log transformation and then backtransformed for results presentation.
Individual genotype analysis and the combination of the two genotypes analysis (PPARG2 + IL6) were performed. For the combination of genotypes, patients were grouped as follows: no variants (genotypes ProPro and GG, n=78), IL6 only (genotypes ProPro and GC + CC, n=48), PPARG2 only (genotypes ProAla + AlaAla and GG, n=15), and both variants (genotypes ProAla + AlaAla and GC + CC, n=4). Comparisons were performed using Student's t-test or ANOVA and chi-squared test or Fisher's exact test. Due to the low frequency of the variant allele, we compared carriers versus noncarriers of the minor allele. We fitted multiple linear regression models to adjust the analysis for potential confounders (age, sex, BMI, sedentary time, and diabetes). For binary variables, odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated and multiple logistic regression models were fitted adjusting for the same confounders. Statistical analyses were performed in Stata 12.
3. Results
All the study participants (n=150) had DNA extracted; however, genetic samples were viable in 146 individuals for the PPARG2 Pro12Ala polymorphism and in 148 individuals for the IL6 -174G >C polymorphism. The genotype distribution for PPARG2 Pro12Ala was 86.9%, 12.4%, and 0.7% for ProPro, ProAla, and AlaAla, respectively. The minor allele frequency of the Ala allele was 0.065. For IL6 -174G >C, the frequencies of GG, GC, and CC genotypes were 65.3%, 31.3%, and 3.4%, respectively. The minor allele frequency of the C allele was 0.193. Observed genotype frequencies were in agreement with the Hardy–Weinberg equilibrium (p=0.689 for PPARG2 Pro12Ala and p=0.863 for IL6 -174G >C).
The characteristics of the study participants according to the PPARG2 Pro12Ala polymorphism are displayed in Table 1. Analysis of the Pro12Ala polymorphism showed higher BMI (p=0.031) and fat mass (p=0.049) for Ala carriers, even after adjustment for age, sex, sedentary time, and diabetes. Ala carriers presented significantly higher SBP and DBP, but after adjustments, only SBP (p=0.026) remained associated (Table 1).
Table 1.
Demographic and clinical characteristics of studied participants according to the PPARG2 Pro12Ala (rs1801282) polymorphism.
| Variables | Total | Pro12Ala polymorphism | p value∗ | Adjusted p value | |
|---|---|---|---|---|---|
| N=146 | ProPro (N=127) | Ala carriers (N=19) | |||
| Clinical, anthropometrical, and body composition variables | |||||
| Age (years) | 39.82 ± 8.70 | 40.00 ± 8.64 | 38.63 ± 2.12 | 0.524 | |
| Female/male, N (%) | 125 (85.6)/21 (14.4) | 108 (86.4)/19 (90.5) | 17 (13.6)/2 (9.5) | 1.000† | 0.427a |
| BMI (kg/m2) | 46.09 ± 6.42 | 45.62 ± 6.12 | 49.26 ± 1.75 | 0.020 | 0.026 b |
| Fat mass (kg)1 | 61.43 ± 13.02 | 60.52 ± 12.73 | 67.67 ± 13.67 | 0.029 | 0.046 b |
| Fat-free mass (kg)1 | 57.22 ± 8.86 | 57.00 ± 9.06 | 58.77 ± 7.43 | 0.431 | 0.227b |
| SBP (mmHg) | 128.66 ± 17.89 | 124.35 ± 13.82 | 133.92 ± 22.51 | 0.011 | 0.022 c |
| DBP (mmHg) | 85.92 ± 13.68 | 81.63 ± 9.65 | 87.60 ± 16.04 | 0.024 | 0.050c |
| Diabetes, N (%) | 40 (27.4) | 36 (28.4) | 4 (21.0) | 0.593‡ | 0.400d |
| Hypertension, N (%) | 96 (65.8) | 83 (65.4) | 13 (68.4) | 0.793† | 0.760c |
| Dyslipidemia, N (%) | 114 (78.1) | 102 (80.3) | 12 (63.2) | 0.099‡ | 0.122c |
| Log MVPA (min/week)2 | 44.54 ± 61.42 | 46.67 ± 63.73§ | 31.17 ± 43.27§ | 0.260 | 0.204e |
| Sedentary time (min/day)2 | 1175.25 ± 83.06 | 1174.10 ± 83.45 | 1182.44 ± 80.56 | 0.685 | 0.817e |
|
| |||||
| Biochemical parameters | |||||
| Fasting glucose (mg/dL) | 110.73 ± 45.38 | 112.12 ± 47.92 | 101.47 ± 20.15 | 0.342 | 0.350d |
| Fasting glucose range, N (%) | 0.596† | 0.441d | |||
| <100 mg/dL, N (%) | 85 (58.2) | 75 (88.2) | 10 (11.8) | ||
| ≥100 mg/dL, N (%) | 29 (19.9) | 52 (85.2) | 9 (14.8) | ||
| Fasting insulin (μU/mL) | 23.43 ± 14.88 | 23.04 ± 14.51 | 26.03 ± 17.34 | 0.415 | 0.413d |
| HOMA-IR | 6.44 ± 4.91 | 6.42 ± 4.86 | 6.56 ± 4.72 | 0.915 | 0.829d |
| GHb (%) | 6.29 ± 1.45 | 6.30 ± 1.47 | 6.23 ± 1.32 | 0.843 | 0.875d |
| Total cholesterol (mg/dL) | 188.68 ± 36.57 | 190.42 ± 36.86 | 177.05 ± 33.12 | 0.138 | 0.232c |
| HDL cholesterol (mg/dL) | 47.47 ± 11.11 | 47.69 ± 11.49 | 45.95 ± 8.20 | 0.525 | 0.501c |
| LDL cholesterol (mg/dL) | 119.20 ± 34.01 | 110.08 ± 34.68 | 103.42 ± 29.42 | 0.429 | 0.497c |
| Triglyceride (mg/dL) | 160.16 ± 78.60 | 163.42 ± 80.67 | 138.37 ± 60.34 | 0.196 | 0.331c |
|
| |||||
| Dietary intake | |||||
| Energy (kcal) | 1709.50 ± 704.50 | 1682.55 ± 695.05 | 1889.62 ± 759.68 | 0.233 | 0.139e |
| Proteins (%) | 17.42 ± 4.60 | 17.58 ± 4.61 | 16.34 ± 4.54 | 0.274 | 0.239e |
| Carbohydrates (%) | 51.64 ± 8.56 | 51.79 ± 8.72 | 50.64 ± 7.49 | 0.589 | 0.593e |
| Lipids (%) | 27.97 ± 6.59 | 27.74 ± 6.70 | 29.51 ± 5.70 | 0.278 | 0.245e |
| Saturated (%) | 8.38 ± 2.65 | 8.30 ± 2.70 | 8.98 ± 2.26 | 0.294 | 0.372e |
| Polyunsaturated (%) | 4.29 ± 1.66 | 4.25 ± 1.57 | 4.52 ± 2.24 | 0.515 | 0.452e |
| Monounsaturated (%) | 7.62 ± 2.60 | 7.54 ± 2.63 | 8.16 ±2.40 | 0.363 | 0.509e |
| P : S ratio | 0.56 ± 0.28 | 0.56 ± 0.27 | 0.55 ± 0.36 | 0.438 | 0.475e |
| Cholesterol (g) | 222.50 ± 120.74 | 217.38 ± 114.16 | 256.66 ± 157.45 | 0.187 | 0.206e |
| Fiber (g) | 15.28 ± 8.04 | 15.03 ± 8.21 | 16.52 ± 9.72 | 0.472 | 0.452e |
Data are presented as mean ± SD or N (%). BMI: body mass index; HOMA-IR: homeostatic model assessment for insulin resistance; GHb: glycated hemoglobin; HDL: high-density lipoprotein; LDL: low-density lipoprotein; SBP: systolic blood pressure; DBP: diastolic blood pressure; MVPA: moderate-to-vigorous physical activity; P : S ratio: polyunsaturated : saturated fatty acids ratio. ∗Student's t-test; †chi-squared test; ‡Fisher's exact test. 1N=141; 2N=138. aAdjusted for age, BMI, sedentary time, and diabetes; badjusted for age, gender, sedentary time, and diabetes; cadjusted for age, gender, sedentary time, BMI, and diabetes; dadjusted for age, gender, sedentary time, and BMI; eadjusted for age, gender, BMI, and diabetes; §values were presented backtransformed.
The characteristics of the study participants according to the IL6 -174G >C polymorphism are displayed in Table 2. Sex and MVPA were associated with the -174G >C polymorphism after adjustments (p=0.043 and p=0.024, respectively). Males had triple probability (OR: 3.60; 95% CI: 1.04–12.48) to be C carriers, and the C carriers spent lower amount of time in MVPA (Table 2).
Table 2.
Demographic and clinical characteristics of participants according to the IL6 -174G >C (rs1800795) polymorphism.
| Variables | Total | -174G >C polymorphism | p value∗ | Adjusted p value | |
|---|---|---|---|---|---|
| N=148 | GG (N=96) | C carriers (N=52) | |||
| Clinical, anthropometrical, and body composition variables | |||||
| Age (years) | 39.61 ± 8.62 | 39.54 ± 7.96 | 39.75 ± 9.81 | 0.889 | – |
| Female/male, N (%) | 126 (85.1)/22 (14.9) | 87 (69.0)/9 (40.9) | 39 (31.0)/13 (59.1) | 0.011 † | 0.022 a |
| BMI (kg/m2) | 45.97 ± 6.38 | 45.50 ± 6.28 | 46.84 ± 6.49 | 0.221 | 0.386b |
| Fat mass (kg)1 | 61.24 ± 12.93 | 60.02 ± 11.92 | 63.44 ± 14.45 | 0.130 | 0.219b |
| Fat-free mass (kg)1 | 57.32 ± 9.00 | 56.41 ± 8.03 | 58.95 ± 10.41 | 0.107 | 0.983b |
| SBP (mmHg) | 128.32 ± 17.92 | 125.52 ± 16.08 | 125.44 ± 14.46 | 0.975 | 0.198c |
| DBP (mmHg) | 85.72 ± 13.69 | 83.13 ± 11.28 | 81.00 ± 9.72 | 0.252 | 0.059c |
| Diabetes, N(%) | 41 (27.7) | 27 (28.1) | 14 (26.9) | 0.876† | 0.331d |
| Hypertension, N (%) | 96 (4.8) | 62 (64.6) | 34 (65.4) | 0.922† | 0.549c |
| Dyslipidemia, N (%) | 117 (79.0) | 75 (71.1) | 42 (80.8) | 0.706† | 0.319c |
| Log MVPA (min/week)2 | 44.52 ± 61.04 | 50.77 ± 63.42§ | 32.92 ± 55.11§ | 0.115 | 0.024 e |
| Sedentary time (min/day)2 | 1176.82 ± 83.26 | 1170.42 ± 84.64 | 1188.71 ± 80.14 | 0.216 | 0.230e |
|
| |||||
| Biochemical parameters | |||||
| Fasting glucose (mg/dL) | 110.14 ± 45.34 | 109.48 ± 42.48 | 111.35 ± 50.61 | 0.812 | 0.609d |
| Fasting glucose range, N (%) | 0.230 | 0.174d | |||
| <100 mg/dL | 87 (58.8) | 53 (60,9) | 34 (39.1) | ||
| ≥100 mg/dL | 61 (41.2) | 43 (70.5) | 18 (29.5) | ||
| Fasting insulin (μU/mL) | 23.38 ± 14.79 | 23.49 ± 15.64 | 23.17 ± 13.22 | 0.901 | 0.589d |
| HOMA-IR | 6.40 ± 4.90 | 6.45 ± 5.20 | 6.32 ± 4.33 | 0.887 | 0.464d |
| GHb (%) | 6.29 ± 1.44 | 6.26 ± 1.39 | 6.34 ± 1.54 | 0.755 | 0.792d |
| Total cholesterol (mg/dL) | 189.86 ± 37.98 | 188.82 ± 41.36 | 191.79 ± 31.06 | 0.652 | 0.966c |
| HDL cholesterol (mg/dL) | 47.57 ± 11.33 | 47.83 ± 11.65 | 47.08 ± 10.81 | 0.700 | 0.816c |
| LDL cholesterol (mg/dL) | 110.03 ± 35.40 | 109.57 ± 38.68 | 110.90 ± 28.50 | 0.830 | 0.943c |
| Triglyceride (mg/dL) | 161.46 ± 78.11 | 156.56 ± 73.11 | 170.50 ± 12.01 | 0.302 | 0.629c |
|
| |||||
| Dietary intake | |||||
| Energy (kcal) | 1699.44 ± 708.99 | 1652.17 ± 683.44 | 1786.71 ± 752.88 | 0.272 | 0.906e |
| Proteins (%) | 17.36 ± 4.59 | 16.97 ± 4.32 | 18.08 ± 5.00 | 0.163 | 0.177e |
| Carbohydrates (%) | 51.57 ± 8.71 | 52.23 ± 8.65 | 50.36 ± 8.78 | 0.216 | 0.186e |
| Lipids (%) | 27.91 ± 6.52 | 28.02 ± 6.20 | 27.71 ± 7.13 | 0.784 | 0.557e |
| Saturated (%) | 8.43 ± 2.71 | 8.51 ± 2.69 | 8.29 ± 2.77 | 0.630 | 0.459e |
| Polyunsaturated (%) | 4.27 ± 1.66 | 4.13 ± 1.65 | 4.53 ± 1.66 | 0.166 | 0.156e |
| Monounsaturated (%) | 7.61 ± 2.61 | 7.35 ± 2.54 | 8.12 ± 2.70 | 0.107 | 0.216e |
| P : S ratio | 0.56 ± 0.29 | 0.53 ± 0.30 | 0.59 ± 0.24 | 0.272 | 0.056e |
| Cholesterol (g) | 219.39 ± 120.29 | 208.14 ± 114.74 | 240.16 ± 128.48 | 0.122 | 0.434e |
| Fiber (g) | 15.17 ± 8.43 | 15.14 ± 7.72 | 15.22 ± 9.67 | 0.952 | 0.424e |
Data are presented as mean ± SD or N (%). BMI: body mass index; HOMA-IR: homeostatic model assessment for insulin resistance; HbA1c: hemoglobin A1c; HDL: high-density lipoprotein; LDL: low-density lipoprotein; SBP: systolic blood pressure; DBP: diastolic blood pressure; MVPA: moderate-to-vigorous physical activity; P : S ratio: polyunsaturated : saturated fatty acids ratio. ∗Student's t-test; †chi-squared test. 1N=140; 2N=143. aAdjusted for age, BMI, sedentary time, and diabetes; badjusted for age, gender, sedentary time, and diabetes; cadjusted for age, gender, sedentary time, BMI, and diabetes; dadjusted for age, gender, sedentary time, and BMI; eadjusted for age, gender, BMI, and diabetes; §values were presented backtransformed.
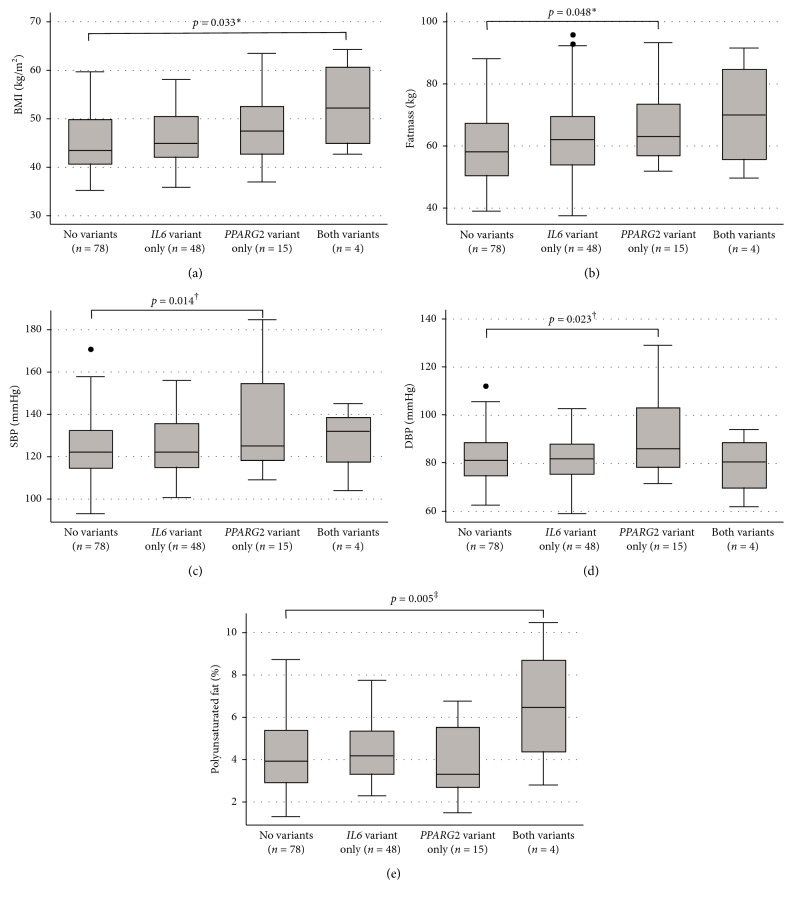
For combined genotypes, participants were grouped as follows: no variants (n=78), IL6 variant only (n=48), PPARG2 variant only (n=15), and both variants (n=4). The same variables of the individual genotype analysis were tested for the combined genotypes, but the results were presented in figures only for the variables with a significant statistical difference. Analysis of the combined effects of the two genotypes showed association with BMI, fat mass, SBP, DBP, and polyunsaturated fat consumption after adjustments. Individuals with both variants had higher BMI (p=0.023) compared to the ones with no variants. Fat mass, SBP, and DBP were higher for participants with the PPARG2 variant only compared to those with no variants (p=0.045, p=0.018, and p=0.030, respectively). Individuals with both variants presented higher consumption of PUFA compared to the ones with no variants (p=0.045) (Figure 1). The analysis of BMI in categories did not show association with the polymorphisms or the genotype combination (Table 3).
Figure 1.
Comparison of BMI (a), fat mass (b), SBP (c), DBP (d), and percentage of polyunsaturated fat consumption (e) between different genotype combinations in severely obese patients. BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure. Results are expressed as median (percentiles 25–75%). Dots represent outlier values. ∗Adjusted for age, gender, and sedentary time. †Adjusted for age, gender, sedentary time, BMI, and diabetes. ‡Adjusted for age, gender, BMI, and diabetes.
Table 3.
Association between BMI categories and the genotypes in severely obese patients.
| Genotypes | BMI (kg/m2) | p value∗ | Adjusted p valuea | |
|---|---|---|---|---|
| 35–49.9 | ≥50 | |||
| PPARG2 Pro12Ala (N=146) | 0.123 | 0.154 | ||
| ProPro (N=127) | 95 (74.8) | 32 (25.20) | ||
| Ala carriers (N=19) | 11 (57.9) | 8 (42.1) | ||
| IL6 -174G >C (N=148) | 0.907 | 0.851 | ||
| GG (N=96) | 71 (74.0) | 25 (26.0) | ||
| C carriers (N=52) | 38 (73.1) | 14 (26.9) | ||
| Genotype combination (N=145) | 0.361† | 0.257 | ||
| No variants (N=78) | 59 (75.6) | 19 (24.4) | ||
| PPARG2 variant only (N=48) | 36 (75.0) | 12 (25.0) | ||
| IL6 variant only (N=15) | 9 (60.0) | 6 (40.0) | ||
| Both variants (N=4) | 2 (50.0) | 2 (50.0) | ||
Data are presented as N (%). BMI: body mass index. ∗Chi-squared test; †Fisher's exact test; aadjusted for age, gender, sedentary time, and diabetes.
4. Discussion
The Ala allele of PPARG2 Pro12Ala (rs1801282) is potentially associated with higher BMI; however, few studies have investigated other key factors that may interact with the variant leading to severe obesity susceptibility such as metabolic, food consumption, and clinical parameters. To our knowledge, this is the first study to find association of the PPARG2 Pro12Ala (rs1801282) polymorphism with an obesity phenotype other than BMI and blood pressure in severely obese patients. We found higher measures of adiposity (BMI and fat mass) and higher SBP in Ala carriers of the Pro12Ala polymorphism compared to the ProPro individuals. These results may contribute to a better understanding of the pathophysiology of severe obesity and translate into more effective preventive and treatment measures to halt its increasing prevalence.
For the PPARG2 Pro12Ala polymorphism, we observed a genotype distribution (87% for ProPro and 13% for Ala carriers) similar to that in the few studies with Brazilians [32, 33]. Among studies with morbidly obese individuals (BMI ≥ 40 kg/m2), frequencies range from 72.5% to 86.0% for the ProPro genotype and from 14.0% to 27.5% for Ala carriers [34–38]. For the -174G >C polymorphism, frequencies vary widely. In the current study, we observed 64.9% of GG and 35.1% of GC + CC, while French morbidly obese individuals presented frequencies of 43.1% for GG and 56.9% for GC + CC [39].
Despite controversial results regarding the PPARG2 Pro12Ala polymorphism, meta-analyses have shown association of the Ala allele with higher BMI and fat mass, especially in severely obese individuals, corroborating our results [6–8, 10, 11, 40]. The controversies observed in other studies may be explained by a suggested interaction between dietary fat intake and the Pro12Ala polymorphism, showing that when the dietary P : S ratio is similar or lower than 0.66, Ala carriers present higher BMI than Pro homozygotes, while when the P : S ratio is higher than 0.66, the opposite occurs [41]. Our patients had the mean P : S ratio ≤ 0.66, possibly explaining the higher BMI among Ala carriers and demonstrating the effect of dietary fat intake on the phenotype determination.
PPAR-γ2 has a crucial role in modulating lipid metabolism and adipose tissue accumulation [4, 5]. The PPARG2 Pro12Ala polymorphism has been suggested to have a role in the variance of fat mass among analyses of candidate genes for adiposity changes [42]. Few studies have investigated the influence of the Pro12Ala polymorphism on body composition parameters other than body mass, BMI, and waist circumference. Association between the Ala allele and higher fat mass was found in Caucasians from the Québec Family Study [43] and in Italian women [44], corroborating the results of the current study. Thus, more detailed measurements of body composition are important to be addressed in studies of the Pro12Ala polymorphism.
We found an association between the Ala allele of the Pro12Ala polymorphism and higher blood pressure in severely obese individuals. Although the association between the Pro12Ala variants and blood pressure is controversial, a meta-analysis of eight studies with 3281 individuals (1865 cases and 1416 controls) suggested that the Ala allele might be protective for hypertension among East Asians, but not among Caucasians [45]. The mechanisms behind this association are unclear. Studies have suggested that the activation of the PPAR-γ2 inhibits processes related to inflammation and hypertension. The Ala allele has been associated with impaired function of the PPAR-γ2, leading to increased blood pressure [46–48].
We found higher probability of severely obese males to be C carriers of the IL6 -174G >C polymorphism after adjustments. Studies comparing elderly (octa/nonagenarians and centenarians) with younger individuals have found a tendency in the reduction of GG genotypes in males [49–52]. Regarding MVPA, we did not find any evidence that justifies this association. More investigation is needed to clarify the association of the higher frequency of C alleles in severely obese males and the -174G >C polymorphism and also the association of the C allele with lower time spent in MVPA.
When the Pro12Ala and the -174G >C genotypes were combined, association was found between the PPARG2 variant only and fat mass, SBP, and DBP compared to the ones with no variants, as demonstrated in the independent analysis of genotypes. We also observed higher BMI and higher PUFA consumption in the presence of both variants compared to the ones with no variants. Nevertheless, this association of the combined genotypes seems to be determined by the PPARG2 Pro12Ala polymorphism as the IL6 -174G >C polymorphism was not associated with adiposity indexes in the independent analysis of genotypes and also due to the low number of individuals with both variants.
Our study has limitations such as the small sample size, especially in the analysis of combined genotypes, and the impossibility to demonstrate causality due to the study design. However, we may address some strengths, such as the analysis of the association between SNPs and also the combination of genotypes, with a variety of parameters, such as anthropometrical, clinical, biochemical, physical activity, and dietary intake, in severely obese patients; the adjusted analysis for potential confounders, such as sociodemographic, anthropometrical, and physical activity measurements; and the high-quality methods used to assess physical activity (triaxial accelerometer) and body composition (BIA).
5. Conclusions
In summary, we found that severely obese individuals carrying the Ala allele of the PPARG2 Pro12Ala polymorphism have higher measures of adiposity (BMI and fat mass) and blood pressure, while no important associations were found for the IL6 -174G >C polymorphism. Further studies on gene-diet/gene-environment interactions are necessary to clarify the associations and underlying mechanisms between SNPs and severe obesity. This information may eventually be used to develop tailored interventions specific to the individual's genotype and enable more effective prevention and treatment measures to address this increasing public health problem.
Acknowledgments
We thank the patients who participated in the study. We also would like to thank the Clinical Research Unit of Clinical Hospital from Universidade Federal de Goiás and its technical staff for lending us the facilities to conduct this research and giving technical support. This study was partially funded by the Goiás State Research Support Foundation (FAPEG) (grant number 201310267000003).
Contributor Information
Ana Paula dos Santos Rodrigues, Email: anapsr@gmail.com.
Erika Aparecida Silveira, Email: erikasil@terra.com.br.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Razquin C., Marti A., Martinez J. A. Evidences on three relevant obesogenes: MC4R, FTO and PPARγ. Approaches for personalized nutrition. Molecular Nutrition and Food Research. 2011;55(1):136–149. doi: 10.1002/mnfr.201000445. [DOI] [PubMed] [Google Scholar]
- 2.Barbieri M., Rizzo M. R., Papa M., et al. Role of interaction between variants in the PPARG and interleukin-6 genes on obesity related metabolic risk factors. Experimental Gerontology. 2005;40(7):599–604. doi: 10.1016/j.exger.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Popko K., Gorska E., Demkow U. Influence of inteleukin-6 and G174C polymorphism in IL-6 gene on obesity and energy balance. European Journal of Medical Research. 2010;15(S2):123–127. doi: 10.1186/2047-783X-15-S2-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadian M., Suh J. M., Hah N., et al. PPARγ signaling and metabolism: the good, the bad and the future. Nature Medicine. 2013;19(5):557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feige J. N., Gelman L., Michalik L., Desvergne B., Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Progress in Lipid Research. 2006;45(2):120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Deeb S. S., Fajas L., Nemoto M., et al. A Pro12Ala substitution in PPAR gamma 2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature Genetics. 1998;20(3):284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 7.Stumvoll M., Häring H. The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism. Diabetes. 2002;51(8):2341–2347. doi: 10.2337/diabetes.51.8.2341. [DOI] [PubMed] [Google Scholar]
- 8.Masud S., Ye S. Effect of the peroxisome proliferator activated receptor-γ gene Pro12Ala variant on body mass index: a meta-analysis. Journal of Medical Genetics. 2003;40(10):773–780. doi: 10.1136/jmg.40.10.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galbete C., Toledo E., Martínez-González M. A., Martínez J. A., Guillén-Grima F., Marti A. Pro12Ala variant of the PPARG2 gene increases body mass index: an updated meta-analysis encompassing 49,092 subjects. Obesity. 2013;21(7):1486–1495. doi: 10.1002/oby.20150. [DOI] [PubMed] [Google Scholar]
- 10.Yao Y. S., Li J., Jin Y. L., Chen Y., He L. P. Association between PPAR-γ2 Pro12Ala polymorphism and obesity: a meta-analysis. Molecular Biology Reports. 2015;42(6):1029–1038. doi: 10.1007/s11033-014-3838-6. [DOI] [PubMed] [Google Scholar]
- 11.Mansoori A., Amini M., Kolahdooz F., Seyedrezazadeh E. Obesity and Pro12Ala polymorphism of peroxisome proliferator-activated receptor-gamma gene in healthy adults: a systematic review and meta-analysis. Annals of Nutrition and Metabolism. 2015;67(2):104–118. doi: 10.1159/000439285. [DOI] [PubMed] [Google Scholar]
- 12.Kubaszek A., Pihlajamäki J., Punnonen K., Karhapaa P., Vauhkonen I., Laakso M. The C-174G promoter polymorphism of the IL-6 gene affects energy expenditure and insulin sensitivity. Diabetes. 2003;52(2):558–561. doi: 10.2337/diabetes.52.2.558. [DOI] [PubMed] [Google Scholar]
- 13.Wernstedt I., Eriksson A. L., Berndtsson A., et al. A common polymorphism in the interleukin-6 gene promoter is associated with overweight. International Journal of Obesity. 2004;28(10):1272–1279. doi: 10.1038/sj.ijo.0802763. [DOI] [PubMed] [Google Scholar]
- 14.Berthier M., Paradis A., Tchernof A., et al. The interleukin 6 -174G/C Polymorphism is associated with indices of obesity in men. Journal of Human Genetics. 2003;48(1):14–19. doi: 10.1007/s100380300002. [DOI] [PubMed] [Google Scholar]
- 15.Strandberg L., Mellström D., Ljunggren O., et al. IL6 and IL1B polymorphisms are associated with fat mass in older men: the MrOS Study Sweden. Obesity. 2008;16(3):710–713. doi: 10.1038/oby.2007.95. [DOI] [PubMed] [Google Scholar]
- 16.Klipstein-Grobusch K., Möhlig M., Spranger J., et al. Interleukin-6 g. -174G > C promoter polymorphism is associated with obesity in the EPIC-potsdam study. Obesity. 2006;14(1):14–18. doi: 10.1038/oby.2006.3. [DOI] [PubMed] [Google Scholar]
- 17.Riikola A., Sipilä K., Kähönen M., et al. Interleukin-6 promoter polymorphism and cardiovascular risk factors: the Health 2000 Survey. Atherosclerosis. 2009;207(2):466–470. doi: 10.1016/j.atherosclerosis.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Qi L., Zhang C., van Dam R. M., Hu F. B. Interleukin-6 genetic variability and adiposity: associations in two prospective cohorts and systematic review in 26,944 individuals. Journal of Clinical Endocrinology and Metabolism. 2007;92(9):3618–3625. doi: 10.1210/jc.2007-0877. [DOI] [PubMed] [Google Scholar]
- 19.Huth C., Illig T., Herder C., et al. Joint analysis of individual participants’ data from 17 studies on the association of the IL6 variant -174G > C with circulating glucose levels, interleukin-6 levels, and body-mass index. Annals of Medicine. 2009;41(2):128–138. doi: 10.1080/07853890802337037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapice E., Vaccaro O. Interaction between Pro12Ala polymorphism of PPARγ2 and diet on adiposity phenotypes. Current Atherosclerosis Reports. 2014;16(12):1–11. doi: 10.1007/s11883-014-0462-9. [DOI] [PubMed] [Google Scholar]
- 21.Andreasen C. H., Andersen G. Gene-environment interactions and obesity—further aspects of genomewide association studies. Nutrition. 2009;25(10):998–1003. doi: 10.1016/j.nut.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Jourdan C., Kloiber S., Nieters A., et al. Gene-PUFA interactions and obesity risk. British Journal of Nutrition. 2011;106(8):1263–1272. doi: 10.1017/s0007114511001541. [DOI] [PubMed] [Google Scholar]
- 23.Anderson A. L., Harris T. B., Houston D. K., et al. Relationships of dietary patterns with body composition in older adults differ by gender and PPAR-γ Pro12Ala genotype. European Journal of Nutrition. 2010;49(7):385–394. doi: 10.1007/s00394-010-0096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Cesare M., Bentham J., Stevens G. A., et al. Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19.2 million participants. The Lancet. 2016;387(10026):1377–1396. doi: 10.1016/s0140-6736(16)30054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohman T. G., Roche A. F., Martorell R. Anthropometric Standardization Reference Manual. Champaing, IL, USA: Human Kinetics Books; 1988. [Google Scholar]
- 26.Kyle U. G., Bosaeus I., De Lorenzo A. D., et al. Bioelectrical impedance analysis—Part II: utilization in clinical practice. Clinical Nutrition. 2004;23(6):1430–1453. doi: 10.1016/j.clnu.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Moshfegh A. J., Rhodes D. G., Baer D. J., et al. The US department of agriculture automated multiple-pass method reduces bias in the collection of energy intakes. American Journal of Clinical Nutrition. 2008;88(2):324–332. doi: 10.1093/ajcn/88.2.324. [DOI] [PubMed] [Google Scholar]
- 28.James P. A., Oparil S., Carter B. L., et al. Evidence-based guideline for the management of high blood pressure in adults. Journal of the American Medical Association. 2013;1097(5):1–14. [Google Scholar]
- 29.American Diabetes Association. 2. Classification and diagnosis of diabetes. Diabetes Care. 2016;39(S1):S13–S22. doi: 10.2337/dc16-s005. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson T. A., Ito M. K., Maki K. C., et al. National lipid association recommendations for patient-centered management of dyslipidemia: Part 1—full report. Journal of Clinical Lipidology. 2015;9(2):129–169. doi: 10.1016/j.jacl.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Matthews D. R., Hosker J. P., Rudenski A. S., Naylor B. A., Treacher D. F., Turner R. C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/bf00280883. [DOI] [PubMed] [Google Scholar]
- 32.Caramori M. L., Canani L. H., Costa L. A., Gross J. L. The human peroxisome proliferator-activated receptor γ2 (PPARγ2) Pro12Ala polymorphism is associated with decreased risk of diabetic nephropathy in patients with type 2 diabetes. Diabetes. 2003;52(12):3010–3013. doi: 10.2337/diabetes.52.12.3010. [DOI] [PubMed] [Google Scholar]
- 33.Tavares V., Hirata R. D. C., Rodrigues A. C., et al. Association between Pro12Ala polymorphism of the PPAR-γ2 gene and insulin sensitivity in Brazilian patients with type-2 diabetes mellitus. Diabetes, Obesity and Metabolism. 2005;7(5):605–611. doi: 10.1111/j.1463-1326.2004.00453.x. [DOI] [PubMed] [Google Scholar]
- 34.Bracale R., Labruna G., Finelli C., et al. The absence of polymorphisms in ADRB3, UCP1, PPARγ, and ADIPOQ genes protects morbid obese patients toward insulin resistance. Journal of Endocrinology Investigation. 2012;35(1):2–4. doi: 10.1007/bf03345413. [DOI] [PubMed] [Google Scholar]
- 35.Kolehmainen M., Uusitupa M. I. J., Alhava E., Laakso M., Vidal H. Effect of the Pro12Ala polymorphism in the peroxisome proliferator-activated receptor (PPAR) gamma2 gene on the expression of PPARgamma target genes in adipose tissue of massively obese subjects. Journal of Clinical Endocrinology and Metabolism. 2003;88(4):1717–1722. doi: 10.1210/jc.2002-020603. [DOI] [PubMed] [Google Scholar]
- 36.Evans D., Mann W. A., de Heer J., et al. Variation in the gene for human peroxisome proliferator activated receptor gamma (PPARgamma) does not play a major role in the development of morbid obesity. International Journal of Obesity. 2000;24(5):647–651. doi: 10.1038/sj.ijo.0801214. [DOI] [PubMed] [Google Scholar]
- 37.Koumanis D. J., Christou N. V., Wang X. L., Gilfix B. M. Pilot study examining the frequency of several gene polymorphisms in a morbidly obese population. Obesity Surgery. 2002;12(6):759–764. doi: 10.1381/096089202320995529. [DOI] [PubMed] [Google Scholar]
- 38.Sesti G., Perego L., Cardellini M., et al. Impact of common polymorphisms in candidate genes for insulin resistance and obesity on weight loss of morbidly obese subjects after laparoscopic adjustable gastric banding and hypocaloric diet. Journal of Clinical Endocrinology and Metabolism. 2005;90(9):5064–5069. doi: 10.1210/jc.2005-0404. [DOI] [PubMed] [Google Scholar]
- 39.Poitou C., Lacorte J. M., Coupaye M., et al. Relationship between single nucleotide polymorphisms in leptin, IL6 and adiponectin genes and their circulating product in morbidly obese subjects before and after gastric banding surgery. Obesity Surgery. 2005;15(1):11–23. doi: 10.1381/0960892052993431. [DOI] [PubMed] [Google Scholar]
- 40.Galbete C., Toledo J., Martínez-González M. Á., Alfredo Martínez J., Guillén-Grima F., Marti A. Lifestyle factors modify obesity risk linked to PPARG2 and FTO variants in an elderly population: a cross-sectional analysis in the SUN Project. Genes and Nutrition. 2013;8(1):61–67. doi: 10.1007/s12263-012-0296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luan J., Browne P. O., Harding A., et al. Evidence for gene-nutrient interaction at the PPARγ locus. Diabetes. 2000;50(3):686–689. doi: 10.2337/diabetes.50.3.686. [DOI] [PubMed] [Google Scholar]
- 42.Bouchard L., Tremblay A., Bouchard C., Pérusse C. Contribution of several candidate gene polymorphisms in the determination of adiposity changes: results from the Québec Family Study. International Journal of Obesity. 2007;31(6):891–899. doi: 10.1038/sj.ijo.0803542. [DOI] [PubMed] [Google Scholar]
- 43.Robitaille J., Perusse L., Bouchard C., Vohl M. C. Genes, fat intake, and cardiovascular disease risk factors in the Quebec Family Study. Obesity. 2007;15(9):2336–2347. doi: 10.1038/oby.2007.277. [DOI] [PubMed] [Google Scholar]
- 44.Passaro A., Nora E. D., Marcello C., et al. PPARγ Pro12Ala and ACE ID polymorphisms are associated with BMI and fat distribution, but not metabolic syndrome. Cardiovascular Diabetology. 2011;10(1):p. 112. doi: 10.1186/1475-2840-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Liu C. Quantitative evaluation of common polymorphism (rs1801282) in the PPARγ2 gene and hypertension susceptibility. Gene. 2012;502(2):159–162. doi: 10.1016/j.gene.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 46.Sugawara A., Uruno A., Kudo M., Matsuda K., Yang C. W., Ito S. Effects of PPARγ on hypertension, atherosclerosis, and chronic kidney disease. Endocrinology Journal. 2010;57(10):847–852. doi: 10.1507/endocrj.k10e-281. [DOI] [PubMed] [Google Scholar]
- 47.Sugawara A., Takeuchi K., Uruno A., et al. Transcriptional suppression of type 1 angiotensin II receptor gene expression by peroxisome proliferator-activated receptor-gamma in vascular smooth muscle cells. Endocrinology. 2001;142(7):3125–3134. doi: 10.1210/endo.142.7.8272. [DOI] [PubMed] [Google Scholar]
- 48.Ji Y., Liu J., Wang Z., Liu N., Gou W. PPARgamma agonist, rosiglitazone, regulates angiotensin II-induced vascular inflammation through the TLR4-dependent signaling pathway. Laboratory Investigation. 2009;89(8):887–902. doi: 10.1038/labinvest.2009.45. [DOI] [PubMed] [Google Scholar]
- 49.Rea I. M., Ross O. A., Armstrong M., et al. Interleukin-6-gene C/G 174 polymorphism in nonagenarian and octogenarian subjects in the BELFAST study. Reciprocal effects on IL-6, soluble IL-6 receptor and for IL-10 in serum and monocyte supernatants. Mechanisms of Ageing and Development. 2003;124(4):555–561. doi: 10.1016/s0047-6374(03)00036-8. [DOI] [PubMed] [Google Scholar]
- 50.Ross O. A., Curran M. D., Meenagh A., et al. Study of age-association with cytokine gene polymorphisms in an aged Irish population. Mechanisms of Ageing and Development. 2003;124(2):199–206. doi: 10.1016/s0047-6374(02)00132-x. [DOI] [PubMed] [Google Scholar]
- 51.Bonafè M., Olivieri F., Cavallone L., et al. A gender-dependent genetic predisposition to produce high levels of IL-6 is detrimental for longevity. European Journal of Immunology. 2001;31(8):2357–2361. doi: 10.1002/1521-4141(200108)31:8<2357::aid-immu2357>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 52.Olivieri F., Bonafè M., Cavallone L., et al. The -174 C/G locus affects in vitro/in vivo IL-6 production during aging. Experimental Gerontology. 2002;37(2-3):309–314. doi: 10.1016/s0531-5565(01)00197-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.