Lymphoblastic lymphoma (LBL) is a rare subtype of non-Hodgkin lymphoma (NHL) seen primarily in children or young adults. The frequency of T-cell lymphoblastic lymphomas (T-LBL) are predominant and frequency of B-cell LBL (B-LBL) are only 10~20% of LBL. Therefore, B-LBL account for 1 ~11% of NHL.1–3
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is a cancer that originates from Pro-B to pre-B cells and proliferates in the bone marrow. The tumors originating from mature B cells proliferate at the extramedullary and are categorized as lymphoma. However, the origin of B-LBL is also the pro-B to pre-B cell stage. From a cell biology point of view, the differentiation stage of B-LBL and BCP-ALL are quite similar. The genetic and cell biological difference between B-LBL and BCP-ALL has not been fully elucidated. Therefore, the reason why B-LBL proliferate in the extramedullary and not in the bone marrow has not yet been determined.
BCP-ALL is one of the best-characterized neoplasms by cytogenetic and molecular genetic analysis. Molecular characterization of BCP-ALL results in important prognostic associations. However, due to the rarity of B-LBL, and limitation to the accessibility of tumor cells, cytogenetic and molecular genetic analysis are limited in B-LBL. The translocation t(1;19)(q23;p13) causing TCF3-PBX1 fusion gene is a well-known chromosome abnormality in childhood pre-B ALL, being present in 3–5% of all ALL. Historically, TCF3-PBX1 positive leukemia had been associated with a poor prognosis but has lost its prognostic significance in the context of modern ALL chemotherapy, especially that which contains high-dose methotrexate.4,5 Whilst little is known about the molecular cytogenetic signature of B-LBL, a few reports describe the hyperdiploidy as a universal abnormal karyotype in B-LBL.6,7 TCF3-PBX1 positive B-LBL has not been reported, and the clinical features and prognosis of LBL with this fusion is unclear.
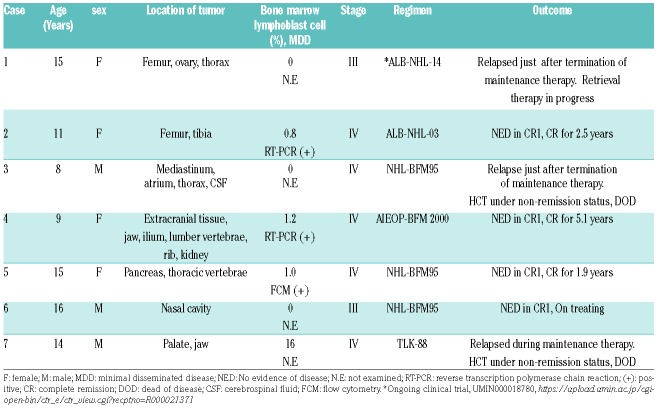
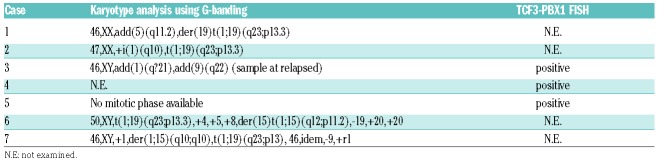
Although TCF3-PBX1 positive B-LBL is not unexpected to occur in B-LBL, the translocation appears to not have been previously described in literature and therefore is a novel finding worth reporting. Survey for TCF3-PBX1 positive B-LBL detected by the evidence of the t(1;19)(q23;p13.3) translocation, RT-PCR positivity, or presence of fusion mRNA detected by RNA sequencing was performed at fourteen local core hospitals with pediatric hematology-oncology patients in their care. Seven cases of TCF3-PBX1 positive B-LBL were identified in five institutes. Table 1 summarizes the clinical and radiologic features for 7 patients of B-LBL with TCF3-PBX1. The detailed clinical course, radiological findings and pathological findings of each patient are described in the Online Supplementary Material. The mean age at diagnosis was 12.5 years (range, 8-16 years). Three cases involved bone lesion. Although, microscopically, the number of blast cells in bone marrow was in the normal range in 6 cases, bone marrow minimal disseminated disease (MDD) was positive in 3 out of 3 evaluated cases. None of the patients had cytopenia or appearance of blast cells in the peripheral blood at the time of diagnosis. Laboratory findings showed only moderate to highly increased levels of lactate dehydrogenase (LDH) levels. Increased soluble IL2 receptor levels were mild to moderate. According to the St. Jude classification for NHL in children, 2 had stage III disease and 5 had stage IV. The karyotyping analysis was evaluable in 5 cases (Table 2). Although case 3 did not demonstrate t(1;19), case 1, 2, 6 and 7 were positive for t(1;19). There were no common additional abnormal cytogenetic changes in these 5 cases. TCF3-PBX1 fluorescence in situ hybridization (FISH) was performed in case 3, 4 and 5 and revealed the presence of the TCF3-PBX1 fusion gene. There were no specific common features regarding the cell surface marker expression between these cases (see Online Supplementary Material). All cases were treated with NHL-BFM95-based chemotherapeutic regimen, and 3 out of 7 cases relapsed during or immediately after completion of maintenance therapy. There were no common additional abnormal cytogenetic changes in these 3 relapsed cases. Two patients died due to tumor progression after receiving hematopoietic cell transplantation. It is interesting to compare the clinical similarities and differences between B-LBL with TCF3-PBX1 and preB-ALL with TCF3-PBX1. Compared with patients with other B-ALL, those with TCF3-PBX1 were relatively older (median 6.9 vs. 4.6 years).8 The age distribution of the children in our series suggests that this condition affects older children compared with TCF3-PBX1 positive PreB-ALL. It is also interesting to note that most of the children had bone or invasive bone involvement, and most had low level bone marrow disease detected at initial staging.
Table 1.
The cases and their clinical information.
Table 2.
Cytogenetic findings of the cases.
Recent advances in molecular analysis for leukemia gives us valuable information which is directly applicable to therapeutic intervention. However, due to the rarity of the disease, the molecular pathogenesis of B-LBL has not yet been elucidated. Cytogenetic analysis of 16 evaluable B-LBL cases revealed 6 normal karyotype, 5 hyperdiploidy and 5 other structural abnormalities such as t(12;17), t(4;11), t(9;12) or del 2p.6 Another cytogenetic analysis of 9 cases revealed 3 normal karyotype, 1 hyperdiploidy, and 4 other nonspecific structural abnormalities.7 Single-nucleotide polymorphism (SNP) microarray analysis revealed that 26% of cases carried CDKN2A/B deletions, 48% of cases were hyperdiploid, 13% of cases carried IKZF1 deletion, and 17% of cases carried PAX5 deletions, which was similar to the reported frequency in BCP-ALL. The gene expression profile analysis of T-ALL and T-LBL revealed that the T-ALL cluster was completely segregated with T-LBL, and immunohistochemical analysis based on gene expression profiles confirmed overexpression of MLL (KMT2A) in T-LBL tumor cells compared to T-ALL and CD47 in T-ALL tumors cells when compared to T-LBL.9 However, limited studies have been performed in B-LBL. Immunoglobulin light chain lambda (IGL) locus deletions consistent with normal light chain rearrangement were observed in 22% of B-LBL cases, compared with only 1% in BCP-ALL samples. None of the B-LBL cases showed abnormal, isolated VPREB1 deletion adjacent to IGL locus, which was identified in 25% of BCP-ALL.10 Other reports described that 71% of B-LBL was characterized by high-hyperdiploidy.11 However, comprehensive genomic or transcriptome analysis has not been performed for B-LBL. These observations suggest that hyperdiploidy is one of the major genomic signatures of B-LBL and BCP-ALL. Due to the similarity of cell biology phenotype and limited genomic analysis between B-LBL and BCP-ALL, it can easily be speculated that the fusion gene observed in BCP-ALL may also be observed in B-LBL. However, few reports have described the fusion gene in B-LBL. There are several case reports with KMT2A (MLL) rearranged B-LBL.12–17 BCR-ABL1 positive LBL has also been reported.17 TCF3-PBX1 positive B-LBL has not been reported. Until now, the reason why a limited case developed as an LBL and not as an ALL, whilst having the same genetic alteration, has not been determined. Genetic base comparison between LBL and ALL will give us further insight.
In patients with B-LBL, event fee survival (EFS) was almost 80~90%.1,18 In the case of T-LBL, 6q loss is significantly related to poor outcome.19 However, what affects the outcome in B-LBL is still unknown. In our study, 3 out of 7 patients relapsed after standard childhood LBL chemotherapy. Due to the limited number of cases, it is hard to predict a poor outcome of TCF3-PBX1 positive LBL; more cases therefore need to be studied. Historically, the prognosis of TCF3-PBX1 positive ALL was improved by intensification of chemotherapy, which is one of a number of options to improve survival of TCF3-PBX1 positive LBL until prognostic factors are revealed. To this end, routine cryptogenic analysis and comprehensive chimera message RNA analysis using biopsy specimen is required in LBL. Gene signature orientated therapy based on these results may contribute to improving the outcome of B-LBL, as has previously been successfully demonstrated in BCP-ALL.
Supplementary Material
Footnotes
Funding: this work is supported by operating cost of Department of Pediatrics and Developmental Biology, Tokyo Medical and Dental University.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Landmann E, Burkhardt B, Zimmermann M, et al. Results and conclusions of the European Intergroup EURO-LB02 trial in children and adolescents with lymphoblastic lymphoma. Haematologica. 2017; 102(12):2086–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkhardt B, Woessmann W, Zimmermann M, et al. Impact of cranial radiotherapy on central nervous system prophylaxis in children and adolescents with central nervous system-negative stage III or IV lymphoblastic lymphoma. J Clin Oncol. 2006;24(3):491–499. [DOI] [PubMed] [Google Scholar]
- 3.Horibe K, Saito AM, Takimoto T, et al. Incidence and survival rates of hematological malignancies in Japanese children and adolescents (2006-2010): based on registry data from the Japanese Society of Pediatric Hematology. Int J Hematol. 2013; 98(1):74–88. [DOI] [PubMed] [Google Scholar]
- 4.Pang L, Liang Y, Pan J, Wang JR, Chai YH, Zhao WL. Clinical features and prognostic significance of TCF3-PBX1 fusion gene in Chinese children with acute lymphoblastic leukemia by using a modified ALL-BFM-95 protocol. Pediatr Hematol Oncol. 2015; 32(3):173–181. [DOI] [PubMed] [Google Scholar]
- 5.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013; 381(9881):1943–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ducassou S, Ferlay C, Bergeron C, et al. Clinical presentation, evolution, and prognosis of precursor B-cell lymphoblastic lymphoma in trials LMT96, EORTC 58881, and EORTC 58951. Br J Haematol. 2011; 152(4):441–451. [DOI] [PubMed] [Google Scholar]
- 7.Maitra A, McKenna RW, Weinberg AG, Schneider NR, Kroft SH. Precursor B-cell lymphoblastic lymphoma. A study of nine cases lacking blood and bone marrow involvement and review of the liter ature. Am J Clin Pathol. 2001; 115(6):868–875. [DOI] [PubMed] [Google Scholar]
- 8.Yen HJ, Chen SH, Chang TY, et al. Pediatric acute lymphoblastic leukemia with t(1;19)/TCF3-PBX1 in Taiwan. Pediatr Blood Cancer. 2017; 64(10). [DOI] [PubMed] [Google Scholar]
- 9.Raetz EA, Perkins SL, Bhojwani D, et al. Gene expression profiling reveals intrinsic differences between T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Pediatr Blood Cancer. 2006; 47(2):130–140. [DOI] [PubMed] [Google Scholar]
- 10.Meyer JA, Zhou D, Mason CC, et al. Genomic characterization of pediatric B-lymphoblastic lymphoma and B-lymphoblastic leukemia using formalin-fixed tissues. Pediatr Blood Cancer. 2017; 64(7). [DOI] [PubMed] [Google Scholar]
- 11.Schraders M, van Reijmersdal SV, Kamping EJ, et al. High-resolution genomic profiling of pediatric lymphoblastic lymphomas reveals subtle differences with pediatric acute lymphoblastic leukemias in the B-lineage. Cancer Genet Cytogenet. 2009;191(1):27–33. [DOI] [PubMed] [Google Scholar]
- 12.Larish AM, Dolan M, Casey T, Perkins JL. Bilateral ovarian B-lineage lymphoblastic lymphoma with MLL gene rearrangement: a novel case in infancy. J Pediatr Hematol Oncol. 2015; 37(4):e215–217. [DOI] [PubMed] [Google Scholar]
- 13.Ahlmann M, Meyer C, Marschalek R, et al. Complex MLL rearrangement in non-infiltrated bone marrow in an infant with stage II precursor B-lymphoblastic lymphoma. Eur J Haematol. 2014; 93(4):349–353. [DOI] [PubMed] [Google Scholar]
- 14.Mater DV, Goodman BK, Wang E, Gaca AM, Wechsler DS. MLL duplication in a pediatric patient with B-cell lymphoblastic lymphoma. J Pediatr Hematol Oncol. 2012; 34(3):e120–123. [DOI] [PubMed] [Google Scholar]
- 15.Takachi T, Iwabuchi H, Imamura M, Imai C. Lymphoblastic lymphoma with mature b-cell immunophenotype and MLL-AF9 in a child. Pediatr Blood Cancer. 2011;57(7):1251–1252. [DOI] [PubMed] [Google Scholar]
- 16.Shafer D, Wu H, Al-Saleem T, et al. Cutaneous precursor B-cell lymphoblastic lymphoma in 2 adult patients: clinicopathologic and molecular cytogenetic studies with a review of the literature. Arch Dermatol. 2008; 144(9):1155–1162. [DOI] [PubMed] [Google Scholar]
- 17.Boddu P, Yin CC, Kanagal-Shamanna R, et al. An unsuspected finding of t(9;22): a rare case of Philadelphia chromosome-positive B-lymphoblastic lymphoma. Case Rep Hematol. 2017; 2017:2413587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunami S, Sekimizu M, Takimoto T, et al. Prognostic impact of intensified maintenance therapy on children with advanced lymphoblastic lymphoma: a report from the Japanese Pediatric Leukemia/Lymphoma Study Group ALB-NHL03 Study. Pediatr Blood Cancer. 2016;63(3):451–457. [DOI] [PubMed] [Google Scholar]
- 19.Bonn BR, Rohde M, Zimmermann M, et al. Incidence and prognostic relevance of genetic variations in T-cell lymphoblastic lymphoma in childhood and adolescence. Blood. 2013; 121(16):3153–3160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.