Abstract
Inherited bone marrow failure syndromes are experiments of nature characterized by impaired hematopoiesis with cancer and leukemia predisposition. The mutations associated with inherited bone marrow failure syndromes affect fundamental cellular pathways, such as DNA repair, telomere maintenance, or proteostasis. How these disturbed pathways fail to produce sufficient blood cells and lead to leukemogenesis are not understood. The rarity of inherited cytopenias, the paucity of affected primary human hematopoietic cells, and the sometime inadequacy of murine or induced pluripotential stem cell models mean it is difficult to acquire a greater understanding of them. Zebrafish offer a model organism to study gene functions. As vertebrates, zebrafish share with humans many orthologous genes involved in blood disorders. As a model organism, zebrafish provide advantages that include rapid development of transparent embryos, high fecundity (providing large numbers of mutant and normal siblings), and a large collection of mutant and transgenic lines useful for investigating the blood system and other tissues during development. Importantly, recent advances in genomic editing in zebrafish can speedily validate the new genes or novel variants discovered in clinical investigation as causes for marrow failure. Here we review zebrafish as a model organism that phenocopies Fanconi anemia, Diamond-Blackfan anemia, dyskeratosis congenita, Shwachman-Diamond syndrome, congenital amegakaryocytic thrombocytopenia, and severe congenital neutropenia. Two important insights, provided by modeling inherited cytopenias in zebrafish, widen understanding of ribosome biogenesis and TP53 in mediating marrow failure and non-hematologic defects. They suggest that TP53-independent pathways contribute to marrow failure. In addition, zebrafish provide an attractive model organism for drug development.
Introduction
The inherited bone marrow failure syndromes (IBMFs) comprise a diverse group of rare monogenic disorders that are phenotypically heterogeneous. They may involve a single or multiple lineage(s). The classic disorders are: Fanconi anemia (FA), Diamond-Blackfan anemia (DBA), Shwachman-Diamond syndrome (SDS), dyskeratosis congenita (DC), severe congenital neutropenia (SCN), and congenital amegakaryocytic thrombocytopenia (CAMT). Besides their phenotypic characterizations, these syndromes correlate strongly with mutations involving a specific pathway. FA results from mutations in genes encoding components of the DNA damage response,1 DC in telomere maintenance,2 and DBA in ribosome function.3 SDS is emerging as a disorder in proteostasis and ribosome maturation (Table 1).4 The molecular basis for how these phenotypically and genotypically heterogeneous conditions result in single or multiple cytopenias remains poorly understood. No common pathway has yet been established, but zebrafish studies have suggested TP53 responses. Activation of the TP53 pathway in mediating marrow failure has been reported for DC,5 FA,6 and a novel bone marrow failure syndrome.7 The TP53 pathway has been suggested to mediate marrow failure for other inherited neutropenias such as SCN and SDS.8 Environmental exposures can accelerate marrow failure, for example, aldehydes producing DNA crosslinks in FA.9 How epigenetics and genetic co-modifiers contribute to these diseases is even less understood. Investigating the molecular basis of the IBMFS will lead to a greater understanding of hematopoiesis, and development and maintenance of non-hematologic tissues. Since the IBMFS constitute leukemia or cancer predisposition syndromes, insights into their pathophysiology will also benefit our understanding, prevention, and perhaps treatment of cancer and age-related genetic changes.
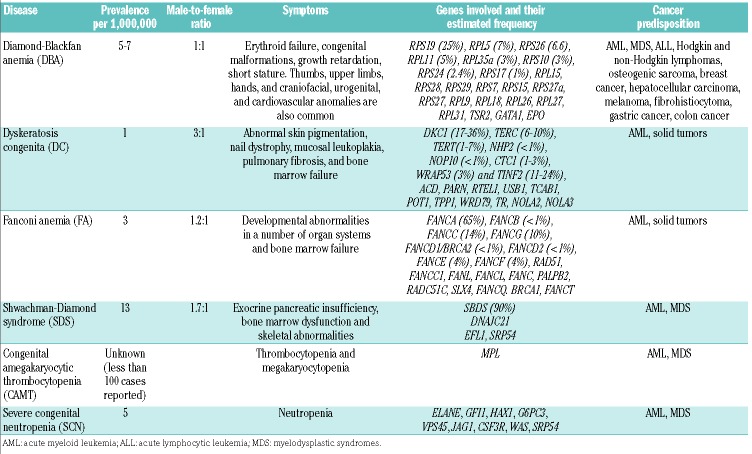
Table 1.
Inherited bone marrow failure syndromes.
Zebrafish model to study inherited bone marrow failure syndromes
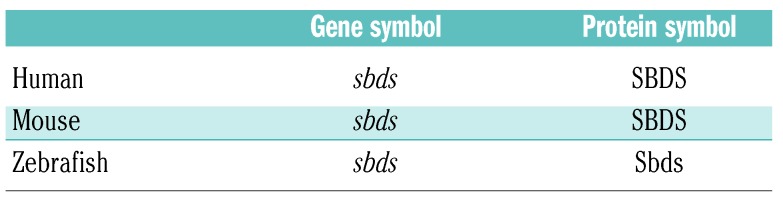
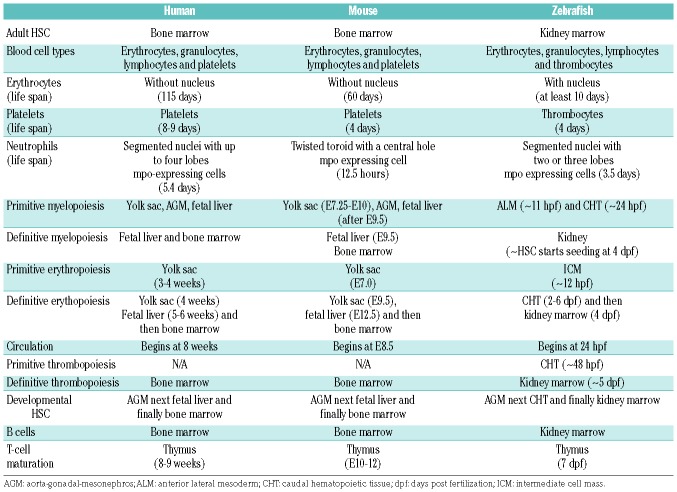
Zebrafish (Danio rerio) have gained popularity as a model organism for a number of reasons. Approximately 70% of all human genes have a zebrafish ortholog.10 Genes are orthologs if they evolved from a common gene, and orthologs typically share similar function. (The Human Genome Organization has adopted a nomenclature for gene and protein expression among different species, which we show using SDS as an example in Table 2.) In addition to lower maintenance and breeding costs, zebrafish provide major advantages to mice: their large clutch size of externally fertilized eggs, transparent embryos, quicker development (all major organs develop and begin functioning during the first 5 days), and short generational time to gamete formation.11 A high degree of genetic and morphological similarity in hematopoiesis between zebrafish and humans suggests that zebrafish can provide valuable insights into the pathogenesis of IBMFS. Developmental hematopoiesis in the zebrafish is comparable to that observed in mice or humans (Figure 1).12–15 One notable difference is that the site of definitive hematopoiesis lies in the zebrafish kidney perivascular space, not the bone marrow. Since the hematopoietic stem cell (HSC) niche provides protection and regulation of self-renewal and differentiation of HSC into blood cells, this difference may be important in non-cell autonomous processes.
Table 2.
Gene and protein nomenclature among species
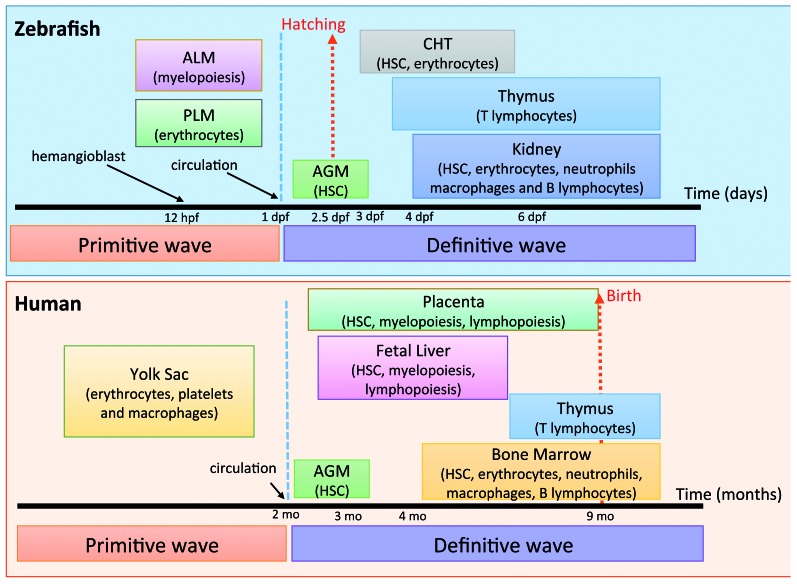
Figure 1.
Comparison of developmental hematopoiesis in humans and zebrafish. Primitive and definitive hematopoiesis occurs in both species. In human, hematopoietic stem cells (HSC) originate in aorta-gonad-mesonephros (AGM) and placenta, from where they colonize fetal liver and finally the bone marrow. In zebrafish, primitive hematopoiesis starts after hemangioblast formation around 12 hpf in the anterior lateral mesoderm (ALM) and posterior lateral mesoderm (PLM). Later, HSCs originate in the AGM and then mobilize to caudal hematopoietic tissue (CHT) prior to their final destination of the kidney (modified from Teittinen et al.12 and de Jong and Zon14).
Studies using zebrafish have facilitated our understanding of vertebrate hematopoiesis and aberrant hematopoiesis in diseases. Hematopoietic and non-hematopoietic lineage-specific transgenic reporter strains are available. They have been useful for the identification and characterization of genes for embryonic hematopoiesis, erythropoiesis, and modeling of human blood diseases (Table 3).16–19 In addition to a collection of zebrafish mutants induced by N-ethyl-N-nitrosourea or viral insertion,20,21 gene function can be studied by transgenic expression or genome editing by transcription activator-like effector nucleases (TALEN) or Cas nucleases acting on clustered, regularly interspaced, short palindromic repeats (CRISPR). Gene expression can be silenced temporarily and early during development by injection of morpholino antisense nucleotides (MO).
Table 3.
Comparison of human, mouse and zebrafish blood systems.
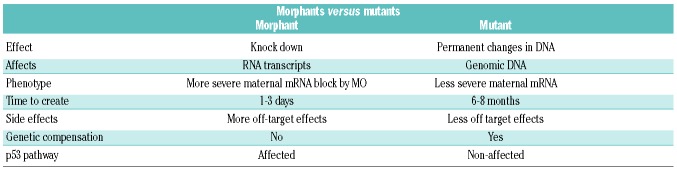
Zebrafish have provided a useful model organism for a quick validation and study of human disease candidate genes, including those involved in the pathophysiology of IBMFS (Table 4). MO-mediated knockdown was widely used to probe gene function, though this method has limitations. Phenotype of morphants (MO-injected animals) can differ and is often more severe than those of the corresponding mutants. There could be different reasons for this: 1) phenotypic rescue of zygotic mutants by maternal wild-type mRNA; 2) off-target effects of the MO; 3) hypomorphic nature of the mutant allele analyzed; or 4) genetic compensation in mutants but not in morphants (see Stainier et al.22). Moreover, injection of MO can cause Tp53 activation and cell death.23 In some instances, cell death can be prevented by simultaneous blocking of p53 by a second MO. This may lead to a misinterpretation of results, particularly in processes that depend on the Tp53 DNA damage response pathway (reviewed below). In some cases, results of MO knockdowns were not recapitulated with the genome editing techniques.24 Close examination of the differences in gene expression revealed a novel compensation mechanism that operates only after mutation but not after MO knockdown (Table 5).25
Table 4.
Comparison of mouse and zebrafish models for inherited bone marrow failure syndromes.
Table 5.
Diamond-Blackfan anemia
Diamond-Blackfan anemia is characterized by red cell hypoplasia, erythroid macrocytosis, and markedly reduced erythroid precursors in the bone marrow. Other hematopoietic lineages are usually normal at birth,26 but they may be affected later in childhood/adolescence.27 In addition to severe anemia, individuals with DBA may display physical anomalies that include thumb, upper limb, craniofacial, cardiovascular and kidney malformations, and short stature. DBA patients have a 25% higher risk of developing myelodysplastic syndromes (MDS), acute myeloid leukemia (AML), and osteosarcoma.
Diamond-Blackfan anemia is an autosomal dominant disorder with a disease incidence of 5-7 per million live births, equally distributed between genders.28 DBA patients have mutations in approximately 20 genes encoding ribosomal proteins; the most common (25%) is RPS19.29 Frameshift, splice defects, intragenic deletions and insertions, nonsense, as well as missense mutations have all been identified. Mutations involve other ribosomal genes: RPL5 (7%), RPL26 (6.6%), RPL11 (5%), RPS10 (3%), RPS26 (3%), RPL35A (3%), RPS24 (2.4%), RPS17 (1%), RPL15, RPS28, RPS29, RPS7, RPS15, RPS27a, RPS, RPL9, RPL18, RPL26, RPL27, RPL31.3 These findings support DBA as a disorder of ribosomal biogenesis and/or function. Mutations in three non-ribosomal proteins, GATA1, TSR2, and EPO, are also associated to DBA.3,29 It is hypothesized that DBA results from apoptosis due to aberrant activation of TP53 that induces cell cycle arrest or apoptosis in response to ribosomal stress.30 Some of the reports implicating TP53, as reviewed below, are based on MO-mediated effects.
In two different studies, rps19-deficient zebrafish were created using MO. Knockdown of rps19 in zebrafish recapitulates the hematopoietic and developmental phenotypes of DBA, including erythropoietic failure with severe anemia, with cell cycle arrest and increased apoptosis, with p53 upregulation. The rps19-deficient phenotype was rescued by injection of zebrafish rps19 mRNA.31–33 Moreover, these phenotypes were not rescued by expressing rps19 mRNAs with a missense or nonsense mutation found in DBA patients.32 Co-injection of MOs against rps19 and p53, showed a complete rescue of the morphological abnormalities, but did not rescue the hematologic defects. These results suggest that there is an erythroid specificity in Rps19 deficiency in zebrafish, independently of Tp53 activity. (See below for further discussion on Tp53 in DBA pathogenesis).34
Chakraborty et al. analyzed the effect of MO-mediated loss of rpl11 in zebrafish. Knockdown of rpl11 led to morphological defects in the developing brain, head, and eyes, and pericardial edema. These phenotypes appear specific as the investigators were able to suppress the morphant by co-injection of MO-resistant rpl11 mRNA. Similar to the loss of Rsp19 function, knockdown of rpl11 resulted in an upregulation of tp53 and mdm2. Moreover, co-injection of rpl11 and tp53 MO rescued the developmental defects and reduced apoptosis, suggesting that ribosomal dysfunction due to the loss of Rpl11 activates a Tp53-dependent response to prevent faulty embryonic development. An increase in tp53 expression and its target genes, cdkn1a and mdm2, was observed in rpl11 morphants. Genes involved in apoptosis (bik, bax, puma, and noxa) were also upregulated.35 Danilova et al. demonstrated that developmental and hematopoietic defects, and lower expression of α-E1 globin and hbae1.1 in Rps19-deficient fish were mediated by Tp53 upregulation. Upregulation of tp53 also occurred in zebrafish mutants for rps8, rps11 and rps18.31
Danilova et al. used a zebrafish rpl11 mutant to characterize the molecular pathways associated with ribosomal deficiency.36 This mutant showed anemia, decreased HSCs, and activation of the Tp53 pathway with altered expression in genes involved in cell cycle arrest (cdkn1a and ccng1) and apoptosis (bax and puma). Moreover, abnormal regulation of metabolic pathways with a shift from glycolysis to aerobic respiration, upregulation of genes involved in gluconeogenesis and insulin levels, decreased biosynthesis, and increased catabolism were observed. Nucleotide metabolism was affected by upregulation of adenosine deaminase (ada) and xanthine dehydrogenase/oxidase (xdh).33,37 They showed that treatment of mutant embryos with an exogenous supply of nucleosides resulted in downregulation of tp53 and its targets with normalization of ada and xdh levels. Interestingly, DBA patients show increased erythrocyte adenosine deaminase activity.38
Zhang et al. generated two zebrafish mutants using TALENs for rps19 and rps11. The knockout of both rps19 and rps11 resulted in the erythroid defects similar to DBA, such as lack of mature red blood cells (RBCs) and Tp53 activation. The mutants had significantly reduced production of globin proteins accompanied by either increased or unaffected level of mRNA transcripts. Furthermore, they observed decreased HSCs at 3 dpf in rps19 mutants and hemoglobin levels by 4 dpf. The authors concluded that this reduction in RBCs may be caused by a decreased cell survival and/or production of definitive HSCs.30 Similarly, Rowel et al. created a 5 bp deletion in rps19 zebrafish mutant using TALENs. Homozygous rps19 mutants showed developmental anomalies and anemia, and were dead by 5 days post fertilization (dpf). However, rps19 heterozygotes showed no difference to their wild-type siblings. Interestingly, exposure to cold stress during the first dpf resulted in a reduced number of RBCs.
To further investigate the biological functions of RPS7, Duan et al. used MO to knockdown rps7 in zebrafish.39 In rps7-deficient embryos, mdm2 and tp53 were activated, inducing the expression of downstream target genes involved in p53 pathway (bik, bax and puma, cdkn1a, and ccng1). rps7 morphants showed severe anemia with reduced expression of gata1 and the mature erythroid marker αe3 at 24 hours post fertilization (hpf). A marked suppression of hemoglobin at 48 hpf was observed, indicating that the deficiency of Rps7 might cause abnormal proliferation and/or differentiation of erythroid progenitors. There were also severe defects (short body length, tissue necrosis, and curved tail). Furthermore, simultaneous knockdown of the tp53 by co-injecting a tp53 MO resulted in partial rescue of morphological abnormalities. The lower levels of gata1 and α-E1 globin were partially rescued in the co-injected embryos, even though tp53, cdk1a, and mdm2 were still up-regulated.39
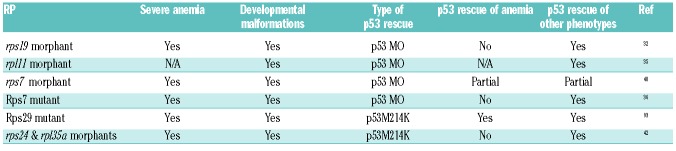
The contribution of tp53 to the pathological development of bone marrow failure syndromes may be tissue-and mutation-specific. Antunes et al. studied the effect of different rps7 and rpl11 mutations in zebrafish. rps7 mutant showed a stronger phenotype due to less maternal contribution of rps7 comparing to rpl11 mutant. Both mutants had severe anemia, morphological abnormalities, and increased apoptosis. Injection of p53 MOs rescued the apoptosis and the morphological phenotypes; however, it was unable to rescue anemia.40 Taylor et al. showed that rps29 mutants had defects in RBC development and increased apoptosis. Mutant embryos showed upregulation of tp53 and cdk1a expression. Mutation of tp53 in homozygous rps29 mutant embryos reversed the apoptotic and hematologic phenotypes. However, mutated tp53 did not fully rescue the embryonic lethality of rps29 mutants, suggesting that tp53-independent mechanisms were affected by rps29 knockdown.41 Yadav et al. knocked down five ribosomal protein genes (two DBA-associated, rpl35a and rps24, and three non-DBA-associated, rps3, rpl35 and rplp1), and analyzed these deficiencies on morphology and erythrocyte number in the presence and absence of p53 using MOs. They showed that any ribosomal protein deficiency led to anemia in zebrafish. Elimination of Tp53 function did not significantly affect the anemia, despite improving non-hematopoetic phenotypes.42 DBA zebrafish models have helped identify MDM2-ribosomal protein interactions, which may interfere with MDM2 inhibition to p53 function. p53 rescue of severe anemia in ribosomal protein deficiency zebrafish models varies (Table 6).31,35,43,44 Altogether, these findings suggest that there are p53-independent mechanisms involved in bone marrow failure. One p53-independent effect may be translational dysfunction. Zebrafish can provide a model organism to identify Tp53-independent pathways that contribute to marrow failure mice or humans.
Table 6.
RP deficiency and p53 rescue in zebrafish models.
Zebrafish may also be a valuable model organism for drug development for DBA treatment. Several groups have tested the hypothesis that L-leucine and L-arginine can stimulate translation via the mTOR pathway and rescue affected DBA fish. Treatment of rpl19 and rpl14 zebrafish morphants with L-leucine improved developmental defects and hemoglobin levels.45 Yadav et al. rescued the morphological defects of Rpl35a-deficient embryos and were able to improve erythroid cell number.42 They concluded that translation deficit, not Tp53 activation, is the primary defect perturbing erythropoiesis.42 While there have been anecdotal reports of leucine stimulation of erythropoiesis in DBA patients,46 definitive clinical trial results are still pending. Another study found that RAP-011, an activin receptor ligand trap, partially restored erythropoiesis in rpl11 morphants as well as rpl11 and rpl19 mutants.47 Zebrafish also provided an in vivo model for further drug development of SMER28, a small molecule inducer of ATG5-dependent autophagy.48 Given these results, we await clinical translation of SMER28 as a potential treatment for DBA.
Dyskeratosis congenita
Dyskeratosis congenita is associated with abnormal skin pigmentation, nail dystrophy, and oral leukoplakia. DC patients may have other organ involvement, including the pulmonary, gastrointestinal, skeletal, neurological, immunological, and ophthalmological systems. Eighty-five percent of DC patients experience bone marrow failure, which accounts for much of the DC-related mortality. Other causes of mortality include infections, pulmonary complications, and hematologic and non-hematologic malignancies.49–51
Dyskeratosis congenita is a genetically heterogeneous disorder, showing autosomal recessive, autosomal dominant, and X-linked inheritance. So far, at least 21 mutated genes have been identified that can cause DC: DKC1, TERC, TERT, NHP2, NOP10, CTC1, WRD79, TR, NOLA2, NOLA3, PARN, TPP1, POT1, CTC1, USB1, TCAB1, RTEL1, ACD, PARN, WRAP53 and TINF2 (http://telomerase.asu.edu/diseases.html).51–53 The X-linked DKC1 has a more severe phenotype compared with the autosomal dominant forms. Although there is a broad consensus that DC results from stem cell renewal failure due to defective telomere maintenance, some mutated genes (e.g. TERT, TERC, and DKC1) are required for pre-rRNA processing.2,49,50,54 How telomerase activity and impaired ribosomal biogenesis contribute to the pathophysiology of DC is still not known. Telomeres are complex DNA-protein structures at the end of chromosomes, and they shorten with each cell division. When telomeres become critically short, a DNA damage response is activated, causing cell cycle arrest or death. In humans, telomerase-based telomere elongation is the major mechanism that counteracts this process of continuous telomere shortening. In peripheral white blood cells, rapid telomere shortening occurs within the first year of life, followed by a more gradual decline over time.49 Genetic diseases that cause telomerase deficiency are associated with premature aging and cancer susceptibility. As in humans, zebrafish chromosomes possess telomeres that progressively decline with age, reaching lengths in old age comparable to those observed when telomerase is mutated.55 Several studies have helped to characterize its well-conserved molecular and cellular physiology. Different zebrafish mutants and morphants for telomere and telomerase research showed shorter lifespan, shorter telomeres, and different affected tissues (mainly brain, blood, gut and testes). These results make zebrafish an excellent model to unravel the connection between telomere shortening, tissue regeneration, aging and disease.55,56
Amsterdam et al. isolated the nop10hi2578 mutant allele where a viral insertion within the first intron resulted in nop10 decreased expression. This mutation is homozygously lethal by 5 dpf.21 nop10 encodes for a protein involved in 18S rRNA processing and is also part of the telomerase complex. Pereboom et al. observed that nop10 loss in this mutant line resulted in a failure of the 18S rRNA to be properly processed, which led to the instability of the 40S ribosomal subunit. Due to the loss of 18S RNA, ribosomal proteins cannot be incorporated into a ribosome subunit and interact with other proteins, including the E3 ubiquitin ligase Mdm2. Mdm2 regulates Tp53 by promoting its ubiquitination and degradation. By binding to Mdm2, Rps7 enhances the E3 ubiquitin ligase activity of Mdm2 that promotes the degradation of Rps7. Furthermore, they observed that an increase in Tp53-specific apoptosis is coupled to the increased binding of Mdm2 to the Rps7. They observed that nop10 mutants failed to form HSCs, a phenotype that is rescued by introducing a loss-of-function tp53 mutation. However, they detected no changes in telomere length in nop10 mutants.53 They concluded that the cytopenia(s) of DC could be the result of ribosome biogenesis defects. This would lead to Tp53-mediated apoptosis of HSCs during early development, caused partially by the association of Rps7 with Mdm2.53
Two different approaches were used by Zhang et al. to study DC in zebrafish. First, MO-mediated knockdown was used to study the mechanisms whereby dkc1 morphants result in HSC failure. Second, they performed retroviral-insertional mutagenesis of nola1. NOLA1 encodes for GAR1, involved in rRNA maturation, and is also a key component telomerase complex. No mutations in NOLA1 have been described in DC patients so far, but suspicion should be aroused in individuals with unexplained marrow failure or fibrosis. Both zebrafish models resulted in reduced hematopoiesis with reduction in runx1 and c-myb, increased tp53 expression, and defective ribosomal biogenesis without detectable changes in telomerase function. Their findings suggest that a telomerase-independent, Tp53-dependent mechanism contribute to hematopoietic failure in DC.50
Henriques et al. and Anchelin et al. studied the zebrafish telomerase reverse transcriptase tert mutant. These mutants develop normally for the first six months, but progressively develop tissue degeneration (gastrointestinal atrophy, loss of body mass, inflammation, a decrease in total blood cells and cell proliferation), and die prematurely. They also observed a Tp53-dependent response with increased transcripts of puma, cdkn1a, and ccng1a. Upregulation of cell cycle arrest inhibitors led to a G1 arrest and senescence. To study the effect of Tp53 in tert mutants, they created a double mutant tert−/−, tp53−/− and observed rescue of cell proliferation, which partially suppressed the degenerative phenotypes.55,57 In another study, Kishi et al. studied the effect of ablation of terfa; they found multiple malformations mainly in brain, spinal cord, and eye.58
Recently, 2 patients with a phenotype overlapping with DBA and DC (pure red cell aplasia, hypogammaglobulinemia, growth retardation, and microcephaly) harbored a de novo TP53 germline mutation that caused a C-terminal truncation in the last exon. This resulted in enhanced p53-mediated transcriptional activity. Using an MO that targets the 3’ splice site of intron 10, Toki et al. developed a zebrafish that displayed reduced number of erythrocytes, severe developmental defects, and died at 96 hpf.7
Fanconi anemia
Fanconi anemia is mostly an autosomal recessive condition characterized by congenital abnormalities, progressive bone marrow failure, chromosome fragility, and an early onset of cancers such as myelodysplastic syndromes (MDS)/acute myeloid leukemia (AML) and epithelial malignancies. FA is characterized by non-hematologic phenotype, including short stature, microcephaly, microphthalmia, hypogonadism, and infertility. The mechanisms by which FA leads to developmental anomalies in blood, skeleton, eyes, and gonads are poorly understood; however, genotoxic stress by chemicals, mutagens, and viruses may contribute.59,60
Mutations in at least 20 genes can cause FA. However, since some cases of FA cannot be assigned to any of these genes, additional genes still have to be identified.1,59 Proteins encoded by these genes constitute the FA pathway required for the efficient repair of damaged DNA. The FA core complex consists of at least 8 proteins: FANCA, FANCB, FANCC, FANCE, FANCF, FANCG FANCL, and FANCM. These proteins function as an E3 ligase and mediate the activation of the FANCD2 and FANCI (ID) complex. Once monoubiquitinated, the ID complex interacts with a third group of FANC proteins, including BRCA2 (FANCD1), FANCJ (BRIP1), FANCN (PALB2), FANCO (RAD51C), FANCP (SLX4), BRCA1, FAN1, histone H2AX, and RAD51, thereby contributing to DNA repair via homologous recombination.1,59,61,62 Until now, 20 genes have been associated with causing FA: FANCA, FANCB, FANCC, BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ, FANCL, FANCN, FANCP, FANCQ, RAD51, BRCA1, FANCT, FANCU, FANCV and FANCW. Information about all these genes is available on the public Fanconi Anemia Mutation Database (http://www.rockefeller.edu/fanconi/).
Although zebrafish contain the full complement of FA family members found in humans,63 loss-of-function models have been described for only a few. Liu et al. analyzed the zebrafish ortholog of the human FANCD2 gene using MO.64 They demonstrated developmental defects that arose during embryogenesis after fancd2 knockdown, phenocopying the reduction in body length, and smaller head and eyes, which are frequently observed among FA patients. This suggests that the FA pathway plays a similar role in zebrafish and humans. They showed that the defects in fancd2-deficient embryos were the result of inappropriate and selective activation of Tp53-mediated apoptotic pathways in highly proliferative cells.64
Titus et al. characterized the developmental and tissue-specific expression of FA pathway genes in zebrafish.63 They found maternal deposition of mRNA fanc genes can provide Fanc proteins to repair DNA damage encountered in rapid cleavage divisions. Zebrafish fancl mutants develop only as sterile males but without hematopoietic defects. The sex reversal was due to abnormal increase of germ cell apoptosis that compromises survival of developing oocytes and masculinizes the gonads. Interestingly, when the tp53 mutation was introduced, the sex reversal phenotype could be rescued.65 Botthoff et al. created a rad51 knockout zebrafish mutant. In this model, zebrafish lacking rad51 survived to adulthood, but they were all infertile males with fewer HSPCs in the kidney. In earlier stages (2 and 4 dpf), they found that rad51−/− embryos also had a lower number, increased apoptosis, and reduced proliferation of HSPCs compared with their wild-type siblings. To study the role of p53 in the rad51 mutants, they generated a zebrafish with mutations in both genes. After four months post fertilization, HSPCs were the same in wild-type and double mutants. The sex reversal was also corrected, but neither females nor male double mutants were fertile.66
Shwachman-Diamond syndrome
Shwachman-Diamond syndrome is an autosomal recessive disorder characterized by exocrine pancreatic insufficiency, bone marrow dysfunction, and skeletal abnormalities. Hematologic abnormalities are a major cause of morbidity and mortality, and include cytopenia(s), MDS, and AML. Neutropenia occurs in approximately 90% of patients and occurs as early as the neonatal period. Skeletal abnormalities, such as metaphyseal chondrodysplasia, thoracic dystrophy, and short stature are common in SDS. In 2003, mutations in the Shwachman–Bodian-Diamond syndrome (SBDS) gene were identified.70 In approximately 90% of cases, SDS is caused by two common mutations in exon 2 of SBDS: 183-184TA→CT introduces an in-frame stop codon (K62X) and 258+2T>C (C84Cfs) disrupts the donor splice site of intron 2, allowing a hypomorph to be produced.67 Fifty percent of cases are compound heterozygotes with respect to these two mutations. Boocock et al. found that both changes correspond to sequences that occur normally in the pseudogene. Both mutations can also occur in the same allele.67 Studies have identified additional changes in the coding sequence of SBDS that led to frameshift and missense mutations.
In 2007, Menne et al. characterized the function of the yeast SBDS ortholog Sdo1 in 60S maturation and translational activation of ribosomes.68 SBDS is a protein with a well-documented role in the later steps of ribosome biogenesis. SBDS interacts with the GTPase EFL1 to trigger release of eIF6 from the 60S ribosomal subunit. EIF6 is critical for biogenesis and nuclear export of pre-60S subunits and prevents ribosomal subunit association. Removal of eIF6 is a prerequisite for the association of the 60S with the 40S subunit, and thus for the formation of an actively functioning ribosome.4 Recently, mutations in DNAJC2169,70 and EFL171 have been identified in individuals with SDS-like conditions. All of the SDS-associated mutant genes affect ribosome maturation. These important discoveries advance the concept of SDS as a ribosomopathy, and beg the question as to how ribosomopathies like DBA, SDS, or del (5q) can result in different defects in hematopoietic and non-hematopoietic tissues.
There have been no reports of homozygosity for SBDS null alleles, suggesting that human SBDS is essential and that SDS patients carry at least one hypomorphic SBDS allele.67,72–75 This is consistent with the finding that mice homozygous for null alleles of sbds exhibit early embryonic lethality, indicating that SBDS function is an essential for life.30 While conditional knock-outs for sbds have been made, this approach is limited, costly, and time-consuming to generate. Thus, we and others have turned to the zebrafish also to study SDS. Venkatasubramani and Mayer used MO to knockdown sbds in zebrafish embryos, and study the effect in pancreas and myeloid development (Table 4). They observed an alteration in the spatial relationship between endocrine and exocrine pancreas. They also documented abnormal neutrophil distribution in the knockdown zebrafish model.76 In a subsequent study, also using MO, Provost et al. observed that their model fully recapitulated the spectrum of developmental abnormalities observed in SDS patients: loss of neutrophils, skeletal defects, and pancreatic hypoplasia, as well as changes in the ribosomal subunit ratio. In this case, loss of Tp53 did not rescue the developmental defects associated with loss of sbds in zebrafish morphants.77 Our recent work showed that sbds mutants obtained by CRISPR/Cas9 editing phenocopied SDS and displayed neutropenia, growth retardation, and atrophy of the pancreas.78
Congenital amegakaryocytic thrombocytopenia
Congenital amegakaryocytic thrombocytopenia is a rare autosomal recessive condition characterized by thrombocytopenia, absence of megakaryocytes, and occasional evolution to aplastic anemia or leukemia.79,80 Mutations in MPL have been described as the cause of CAMT.81 MPL gene encodes for myeloproliferative leukemia protein (CD110), the receptor for thrombopoietin. Mice with genetic ablation of Mpl showed normal development but a deficiency in megakaryocytes and severe thrombocytopenia.82 In zebrafish, disruption of mpl caused a severe reduction in thrombocytes (platelet equivalents), bleeding, and a decrease in HSCs. By phenocopying the human disease, affected zebrafish provide an accurate model to study this disease and for drug screening.83 Reduction in HSCs and repopulation defects in affected zebrafish demonstrate that c-Mpl function in hematopoiesis is highly conserved. Moreover, the partial rescue of thrombocyte number by IL-11 provides a model to finely dissect JAK/STAT signaling in thrombopoiesis.
Severe congenital neutropenia
Severe congenital neutropenia is a group of heterogeneous genetic disorders characterized by a maturation arrest at the promyelocyte stage of granulopoiesis and a high propensity to develop MDS/AML.84 Over the past eighteen years, the following mutations have been identified as causing SCN: ELANE, GFI1, HAX1, VPS45, JAGN, CSF3R, and WAS. ELANE is the most commonly mutated gene in SCN, but there is no zebrafish ortholog. However, zebrafish has proven to be a powerful model to validate and characterize the function of newly described gene candidates for SCN. Vacuolar Protein Sorting 45 Homolog (VPS45) encodes a protein associated with protein trafficking into distinct organelles. Biallelic mutations in this gene are the cause of SCN5. A zebrafish model of vps45 knockdown also showed a large decrease in neutrophils.85 Mutations in CSF3R cause SCN7.86 Pazhakh et al. mutated csf3r in zebrafish to study the effect on neutrophil production. They found that csf3r zebrafish mutants survive until adulthood with a 50% reduction in neutrophils and a substantial reduction in myeloid cells in the kidney marrow.87 Recently, SRP54 mutations have been identified as the second most common cause of SCN (with some features of SDS).88,89 Knockdown of SRP54 in zebrafish recapitulated the human phenotype of neutropenia, chemotaxis defect, and pancreatic exocrine insufficiency.88
Conclusions
Despite the identification of specific gene mutations and pathway involvement for the great majority of patients with IBMFS, little is known about how they result in single or multiple lineage cytopenias. Furthermore, very little is known about co-operating mutations that effect transformation to MDS, AML, or solid tumors. Patient-based studies are problematic owing to the rarity of these disorders and to the long latency before bone marrow failure or malignancy. Zebrafish provide a relatively inexpensive, rapidly developing, vertebrate model organism. Despite some differences in their respective hematopoietic organs, mutations or silencing of relevant zebrafish genes phenocopies human IBMFS. Studies on gene mutations or suppression in zebrafish have validated the role of ribosome biogenesis, and advanced the hypothesis that the TP53 pathway plays a major role in the pathophysiology of some of the IBMFS. Zebrafish modeling may also contribute to drug development, as suggested by studies on L-leucine and SMER28 for DBA.
Supplementary Material
Acknowledgments
SJC is supported by funding from NIH R01 HL128173, NIH R21 CA159203, Department of Defense Bone Marrow Failure Idea Development Award BM140102, Shwachman-Diamond Syndrome Foundation, Connor’s Heroes, and the CURE Childhood Cancer Foundation. Due to space restrictions, the authors deeply regret not being able to cite all of our colleagues’ publications.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/1/13
References
- 1.Rosenberg PS, Alter BP, Ebell W. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica. 2008;93(4):511–517. [DOI] [PubMed] [Google Scholar]
- 2.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Da Costa L, O’Donohue MF, van Dooijeweert B, et al. Molecular approaches to diagnose Diamond-Blackfan anemia: The EuroDBA experience. Eur J Med Genet. 2017. October 26 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Finch AJ, Hilcenko C, Basse N, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25(9):917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrillo J, Gonzalez A, Manguan-Garcia C, Pintado-Berninches L, Perona R. p53 path way activation by telomere attrition in X-DC primary fibroblasts occurs in the absence of ribosome biogenesis failure and as a consequence of DNA damage. Clin Transl Oncol. 2014;16(6):529–538. [DOI] [PubMed] [Google Scholar]
- 6.Ceccaldi R, Parmar K, Mouly E, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toki T, Yoshida K, Wang R, et al. De Novo Mutations Activating Germline TP53 in an Inherited Bone-Marrow-Failure Syndrome. Am J Hum Genet 2018;103(3):440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaubach T, Minella AC, Corey SJ. Cellular stress pathways in pediatric bone marrow failure syndromes: many roads lead to neutropenia. Pediatr Res. 2014;75(1-2):189–195. [DOI] [PubMed] [Google Scholar]
- 9.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. [DOI] [PubMed] [Google Scholar]
- 10.Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veldman MB, Lin S. Zebrafish as a Developmental Model Organism for Pediatric Research. Pediatr Res. 2008;64(5):470–476. [DOI] [PubMed] [Google Scholar]
- 12.Teittinen KJ, Grönroos T, Parikka M, Rämet M, Lohi O. The zebrafish as a tool in leukemia research. Leuk Res. 2012;36(9): 1082–1088. [DOI] [PubMed] [Google Scholar]
- 13.Davidson AJ, Zon LI. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 2004;23(43): 7233–7246. [DOI] [PubMed] [Google Scholar]
- 14.de Jong JLO, Zon LI. Use of the Zebrafish System to Study Primitive and Definitive Hematopoiesis. Annu Rev Genet. 2005;39(1):481–501. [DOI] [PubMed] [Google Scholar]
- 15.Palis J. Primitive and definitive erythropoiesis in mammals. Front Physiol. 2014;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood. 2008;111(7):3331–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen AT, Zon LI. Zebrafish blood stem cells. J Cell Biochem. 2009;108(1):35–42. [DOI] [PubMed] [Google Scholar]
- 18.Ellett F, Lieschke GJ. Zebrafish as a model for vertebrate hematopoiesis. Curr Opin Pharmacol. 2010;10(5):563–570. [DOI] [PubMed] [Google Scholar]
- 19.Bowman TV, Zon LI. Lessons from the Niche for Generation and Expansion of Hematopoietic Stem Cells. Drug Discov Today Ther Strateg 2009;6(4):135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solnica-Krezel L, Schier AF, Driever W. Efficient Recovery of Enu-Induced Mutations from the Zebrafish Germline. Genetics. 1994;136(4):1401–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amsterdam A, Burgess S, Golling G, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13(20):2713–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stainier DYR, Raz E, Lawson ND, et al. Guidelines for morpholino use in zebrafish. PLoS Genet. 2017;13(10):e1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robu ME, Larson JD, Nasevicius A, et al. p53 Activation by Knockdown Technologies. PLoS Genet. 2007;3(5):e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kok FO, Shin M, Ni CW, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi A, Kontarakis Z, Gerri C, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–233. [DOI] [PubMed] [Google Scholar]
- 26.Chiabrando D, Tolosano E. Diamond Blackfan Anemia at the crossroad between ribosome biogenesis and heme metabolism. Adv Hematol. 2010;2010:790632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giri N, Kang E, Tisdale JF, et al. Clinical and laboratory evidence for a trilineage haematopoietic defect in patients with refractory Diamond-Blackfan anaemia. Br J Haematol. 2000;108(1):167–175. [DOI] [PubMed] [Google Scholar]
- 28.Landowski M, O’Donohue MF, Buros C, et al. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum Genet. 2013;132(11):1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Da Costa L, Narla A, Mohandas N. An update on the pathogenesis and diagnosis of Diamond–Blackfan anemia. F1000Res. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Ear J, Yang Z, Morimoto K, Zhang B, Lin S. Defects of protein production in erythroid cells revealed in a zebrafish Diamond–Blackfan anemia model for mutation in RPS19. Cell Death Dis. 2014;5(7):e1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–5237. [DOI] [PubMed] [Google Scholar]
- 32.Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum Mol Genet. 2008;17(20):3204–3211. [DOI] [PubMed] [Google Scholar]
- 33.Danilova N, Bibikova E, Covey TM, et al. The role of the DNA damage response in zebrafish and cellular models of Diamond Blackfan anemia. Dis Model Mech. 2014;7(7):895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torihara H, Uechi T, Chakraborty A, Shinya M, Sakai N, Kenmochi N. Erythropoiesis failure due to RPS19 deficiency is independent of an activated Tp53 response in a zebrafish model of Diamond-Blackfan anaemia. Br J Haematol. 2011;152(5):648–654. [DOI] [PubMed] [Google Scholar]
- 35.Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of Ribosomal Protein L11 Affects Zebrafish Embryonic Development through a p53-Dependent Apoptotic Response. PLoS One. 2009;4(1):e4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101(35):12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danilova N, Sakamoto KM, Lin S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br J Haematol. 2011;152(2): 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danilova N, Gazda HT. Ribosomopathies: how a common root can cause a tree of pathologies. Dis Model Mech. 2015;8(9):1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan J, Ba Q, Wang Z, et al. Knockdown of ribosomal protein S7 causes developmental abnormalities via p53 dependent and independent pathways in zebrafish. Int J Biochem Cell Biol. 2011;43(8):1218–1227. [DOI] [PubMed] [Google Scholar]
- 40.Antunes AT, Goos YJ, Pereboom TC, et al. Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway. PLoS Genet. 2015;11(7):e1005326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, Zon LI. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol. 2012;40(3):228–237.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yadav GV, Chakraborty A, Uechi T, Kenmochi N. Ribosomal protein deficiency causes Tp53-independent erythropoiesis failure in zebrafish. Int J Biochem Cell Biol. 2014;49:1–7. [DOI] [PubMed] [Google Scholar]
- 43.Amsterdam A, Sadler KC, Lai K, et al. Many Ribosomal Protein Genes Are Cancer Genes in Zebrafish. PLoS Biol. 2004;2(5):e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16(5):369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Payne EM, Virgilio M, Narla A, et al. L-Leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood. 2012;120(11):2214–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pospisilova D, Cmejlova J, Hak J, Adam T, Cmejla R. Successful treatment of a Diamond-Blackfan anemia patient with amino acid leucine. Haematologica. 2007;92(5):e66–e67. [DOI] [PubMed] [Google Scholar]
- 47.Ear J, Huang H, Wilson T, et al. RAP-011 improves erythropoiesis in zebrafish model of Diamond-Blackfan anemia through antagonizing lefty1. Blood. 2015;126(7):880–890. [DOI] [PubMed] [Google Scholar]
- 48.Doulatov S, Vo LT, Macari ER, et al. Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med. 2017;9(376). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du HY, Pumbo E, Ivanovich J, et al. TERC and TERT gene mutations in patients with bone marrow failure and the significance of telomere length measurements. Blood. 2008;113(2):309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Morimoto K, Danilova N, Zhang B, Lin S. Zebrafish Models for Dyskeratosis Congenita Reveal Critical Roles of p53 Activation Contributing to Hematopoietic Defects through RNA Processing. PLoS One. 2012;7(1):e30188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruggero D, Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124(18):2784–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ballew BJ, Yeager M, Jacobs K, et al. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum Genet. 2013;132(4):473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereboom TC, van Weele LJ, Bondt A, MacInnes AW. A zebrafish model of dyskeratosis congenita reveals hematopoietic stem cell formation failure resulting from ribosomal protein-mediated p53 stabilization. Blood. 2011;118(20):5458–5465. [DOI] [PubMed] [Google Scholar]
- 54.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst. 2010;6(3): 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henriques CM, Carneiro MC, Tenente IM, Jacinto A, Ferreira MG. Telomerase Is Required for Zebrafish Lifespan. PLoS Genet. 2013;9(1):e1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carneiro MC, de Castro IP, Ferreira MG. Telomeres in aging and disease: lessons from zebrafish. Dis Model Mech. 2016;9(7):737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anchelin M, Alcaraz-Perez F, Martinez CM, Bernabe-Garcia M, Mulero V, Cayuela ML. Premature aging in telomerase-deficient zebrafish. Dis Model Mech. 2013;6(5):1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kishi S, Bayliss PE, Uchiyama J, et al. The Identification of Zebrafish Mutants Showing Alterations in Senescence-Associated Biomarkers. PLoS Genet. 2008;4(8):e1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodríguez-Marí A, Postlethwait JH. The Role of Fanconi Anemia/BRCA Genes in Zebrafish Sex Determination. The Zebrafish: Disease Models and Chemical Screens: Elsevier BV; 2011:461–490. [DOI] [PubMed] [Google Scholar]
- 60.Soulier J. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood. 2004;105(3):1329–1336. [DOI] [PubMed] [Google Scholar]
- 61.Geiselhart A, Lier A, Walter D, Milsom MD. Disrupted Signaling through the Fanconi Anemia Pathway Leads to Dysfunctional Hematopoietic Stem Cell Biology: Underlying Mechanisms and Potential Therapeutic Strategies. Anemia. 2012; 2012:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong H, Nebert DW, Bruford EA, Thompson DC, Joenje H, Vasiliou V. Update of the human and mouse Fanconi anemia genes. Hum Genomics. 2015;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Titus TA, Selvig DR, Qin B, et al. The Fanconi anemia gene network is conserved from zebrafish to human. Gene. 2006;371(2):211–223. [DOI] [PubMed] [Google Scholar]
- 64.Liu TX, Howlett NG, Deng M, et al. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell. 2003;5(6):903–914. [DOI] [PubMed] [Google Scholar]
- 65.Rodríguez-Marí A, Cañestro C, BreMiller RA, et al. Sex Reversal in Zebrafish fancl Mutants Is Caused by Tp53-Mediated Germ Cell Apoptosis. PLoS Genet. 2010;6(7): e1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Botthof JG, Bielczyk-Maczynska E, Ferreira L, Cvejic A. Loss of the homologous recombination gene rad51 leads to Fanconi anemia-like symptoms in zebrafish. Proc Natl Acad Sci USA. 2017;114(22):E4452–e4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boocock GR, Morrison JA, Popovic M, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97–101. [DOI] [PubMed] [Google Scholar]
- 68.Menne TF, Goyenechea B, Sánchez-Puig N, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nature Genet. 2007;39(4):486–495. [DOI] [PubMed] [Google Scholar]
- 69.Dhanraj S, Matveev A, Li H, et al. Biallelic mutations in DNAJC21 cause Shwachman-Diamond syndrome. Blood. 2017;129(11): 1557–1562. [DOI] [PubMed] [Google Scholar]
- 70.Tummala H, Walne AJ, Williams M, et al. DNAJC21 Mutations Link a Cancer-Prone Bone Marrow Failure Syndrome to Corruption in 60S Ribosome Subunit Maturation. Am J Hum Genet. 2016;99(1):115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stepensky P, Chacon-Flores M, Kim KH, et al. Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in aShwachman-Diamond like syndrome. J Med Genet. 2017;54(8):558–566. [DOI] [PubMed] [Google Scholar]
- 72.Erdos M, Alapi K, Balogh I, et al. Severe Shwachman-Diamond syndrome phenotype caused by compound heterozygous missense mutations in the SBDS gene. Exp Hematol. 2006;34(11):1517–1521. [DOI] [PubMed] [Google Scholar]
- 73.Kuijpers TW, Alders M, Tool AT, Mellink C, Roos D, Hennekam RC. Hematologic abnormalities in Shwachman Diamond syndrome: lack of genotype-phenotype relationship. Blood. 2005;106(1):356–361. [DOI] [PubMed] [Google Scholar]
- 74.Makitie O, Ellis L, Durie PR, et al. Skeletal phenotype in patients with Shwachman-Diamond syndrome and mutations in SBDS. Clin Genet. 2004;65(2):101–112. [DOI] [PubMed] [Google Scholar]
- 75.Dror Y, Donadieu J, Koglmeier J, et al. Draft consensus guidelines for diagnosis and treat ment of Shwachman-Diamond syndrome. Ann N Y Acad Sci. 2011;1242:40–55. [DOI] [PubMed] [Google Scholar]
- 76.Venkatasubramani N, Mayer AN. A Zebrafish Model for the Shwachman-Diamond Syndrome (SDS). Pediatr Res. 2008;63(4):348–352. [DOI] [PubMed] [Google Scholar]
- 77.Provost E, Wehner KA, Zhong X, et al. Ribosomal biogenesis genes play an essential and p53-independent role in zebrafish pancreas development. Development. 2012;139(17):3232–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oyarbide U, Kell MJ, Farinas J, Topczewski J, Corey S. Gene disruption of zebrafish Sbds phenocopies human Shwachman-Diamond Syndrome but suggests more global and lineage defects. Blood. 2016;128(22):336. [Google Scholar]
- 79.Ballmaier M. c-mpl mutations are the cause of congenital amegakaryocytic thrombocytopenia. Blood. 2001;97(1):139–146. [DOI] [PubMed] [Google Scholar]
- 80.Rose MJ, Nicol KK, Skeens MA, Gross TG, Kerlin BA. Congenital amegakaryocytic thrombocytopenia: the diagnostic importance of combining pathology with molecular genetics. Pediatr Blood Cancer. 2008;50(6):1263–1265. [DOI] [PubMed] [Google Scholar]
- 81.Ihara K, Ishii E, Eguchi M, et al. Identification of mutations in the c-mpl gene in congenital amegakaryocytic thrombocytopenia. Proc Natl Acad Sci USA. 1999;96(6):3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood. 1996;87(6):2162–2170. [PubMed] [Google Scholar]
- 83.Lin Q, Zhang Y, Zhou R, et al. Establishment of a congenital amegakaryocytic thrombocytopenia model and a thrombocyte-specific reporter line in zebrafish. Leukemia. 2017;31(5):1206–1216. [DOI] [PubMed] [Google Scholar]
- 84.Skokowa J, Germeshausen M, Zeidler C, Welte K. Severe congenital neutropenia: inheritance and pathophysiology. Curr Opin Hematol. 2007;14(1):22–28. [DOI] [PubMed] [Google Scholar]
- 85.Vilboux T, Lev A, Malicdan MCV, et al. A Congenital Neutrophil Defect Syndrome Associated with Mutations in VPS45. N Engl J Med. 2013;369(1):54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ward AC, van Aesch YM, Gits J, et al. Novel point mutation in the extracellular domain of the granulocyte colony-stimulating factor (G-CSF) receptor in a case of severe congenital neutropenia hyporesponsive to G-CSF treatment. J Exp Med. 1999;190(4):497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pazhakh V, Clark S, Keightley MC, Lieschke GJ. A GCSFR/CSF3R zebrafish mutant models the persistent basal neutrophil deficiency of severe congenital neutropenia. Sci Rep. 2017;7:44455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carapito R, Konantz M, Paillard C, et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J Clin Invest. 2017;127(11):4090–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bellanné-Chantelot C, Schmaltz-Panneau B, Marty C, et al. Mutations in SRP54 gene cause severe congenital neutropenia as well as Shwachman-Diamond-like syndrome. Blood. 2018;132(12):1318–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rowell J, Pietka G, Virgilio M, Pena O, Hockings C, Payne E. A Zebrafish Model of Diamond-Blackfan Anemia Results in Bone Marrow Failure and Demonstrates Defective Translation in Erythroid Cells By Ribosome Footprinting. Blood. 2017;130(Suppl 1):871. [Google Scholar]
- 91.McGowan KA, Mason PJ. Animal models of Diamond Blackfan Anemia. Semin Hematol. 2011;48(2):106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morgado-Palacin L, Varetti G, Llanos S, Gomez-Lopez G, Martinez D, Serrano M. Partial Loss of Rpl11 in Adult Mice Recapitulates Diamond-Blackfan Anemia and Promotes Lymphomagenesis. Cell Rep. 2015;13(4):712–722. [DOI] [PubMed] [Google Scholar]
- 93.Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, Zon LI. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol. 2012;40(3):228–237.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watkins-Chow DE, Cooke J, Pidsley R, et al. Mutation of the diamond-blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 2013;9(1):e1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uechi T, Nakajima Y, Nakao A, et al. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS One. 2006;1:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruggero D, Grisendi S, Piazza F, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299(5604):259–262. [DOI] [PubMed] [Google Scholar]
- 97.Gu BW, Bessler M, Mason PJ. A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc Natl Acad Sci USA. 2008;105(29):10173–10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jaskelioff M, Muller FL, Paik J-H, et al. Telomerase reactivation reverses tissue degeneration in aged telomerase deficient mice. Nature. 2011;469(7328):102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parmar K, D’Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668(1-2):133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rodriguez-Mari A, Postlethwait JH. The role of Fanconi anemia/BRCA genes in zebrafish sex determination. Methods Cell Biol. 2011;105:461–490. [DOI] [PubMed] [Google Scholar]
- 101.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16(12):7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang S, Shi M, Hui CC, Rommens JM. Loss of the mouse ortholog of the Shwachman-Diamond syndrome gene (Sbds) results in early embryonic lethality. Mol Cell Biol. 2006;26(17):6656–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lim KH, Chang YC, Chiang YH, et al. Expression of CALR mutants causes mpl-dependent thrombocytosis in zebrafish. Blood Cancer J. 2016;6(10):e481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu F, Kunter G, Krem MM, et al. Csf3r mutations in mice confer a strong clonal HSC advantage via activation of Stat5. J Clin Invest. 2008;118(3):946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.