T-cell acute lymphoblastic leukemia (T-ALL) and T-cell lymphoblastic lymphoma (T-LBL) are aggressive hematologic malignancies that are currently still treated by high-dose chemotherapy possibly followed by hematopoietic stem cell transplantation.1 Despite important progress in deciphering the genomic landscape of these diseases, the transition of novel targeted therapies towards clinical practice has remained largely unsuccessful.2 T-ALL and T-LBL, which the World Health Organization regards as one single disease entity, can be classified into different molecular genetic subtypes based on the aberrant expression of specific transcription factor oncogenes, such as LYL1, TLX1, TLX3, HOXA, NKX2-1, TAL1 or LMO2.2 Within these genetic subclasses, a variety of co-operative lesions converge towards activation of specific pathways, such as JAK-STAT or PI3K-AKT signaling.2 Interestingly, JAK-STAT pathway mutations seem to be more prevalent in LYL1+, TLX1+, TLX3+ and HOXA+ tumors, whereas mature TAL1+ leukemias/lymphomas more often display PI3K/AKT alterations.2
PIM1 is a highly conserved serine/threonine kinase involved in cell-cycle progression, transcription, apoptosis, drug resistance and cellular metabolism through phosphorylation of a myriad of known downstream targets.3 Formal proof of its oncogenic activity emerged from the analysis of Pim1 transgenic mice, which spontaneously developed T-cell lymphomas with a latency of several months.4 At the transcriptional level, PIM1 is a canonical JAK-STAT target gene that can be activated downstream of cytokine signaling. Recently, a number of studies have all shown that PIM1 might act as an attractive molecular target in human T-ALL.5–7 Indeed, recent work identified a case of adult T-ALL in which aberrant activation of PIM1 was driven by the T-cell receptor (TCR) translocation t(6;7)(p21;q34) (TCRβ-PIM1).7 In addition, PIM1 activation was also found to be more broadly implicated in T-ALL disease biology downstream of mutational activation of the JAK-STAT signaling pathway.6,7 Although these studies clearly point towards PIM1 as a novel therapeutic target for the treatment of T-ALL, initial drug evaluation experiments have largely been focused on human T-ALL and T-LBL cell lines, which often fail to provide an accurate representation of the primary disease.5,6 Therefore, additional in-depth pre-clinical in vivo evaluation of PIM inhibitors using patient-derived xenograft models of human T-ALL and T-LBL will be required to further facilitate the translation of these findings into clinical practice in the near future.
Here, we report the identification of a similar TCRβ-PIM1 translocation, t(6;7)(p21;q34), as previously described,7 in a case of pediatric T-LBL, suggesting that these PIM1 rearrangements are a rare but recurrent genetic abnormality in both pediatric and adult T-ALL and T-LBL. For this particular T-LBL case (see the Online Supplementary Methods for clinical information), initial FISH analysis revealed the presence of a TCRβ translocation in the major leukemic clone at diagnosis. Using Targeted Locus Amplification (TLA),8 with the TCRβ locus as viewpoint, we subsequently identified the genomic breakpoint of this rearrangement at 133 kb upstream of the PIM1 proto-oncogene (Online Supplementary Figure S1A and B). Quantitative RT-PCR (qPCR) confirmed that this T-LBL patient displayed strongly enhanced PIM1 expression as compared to normal CD34+ T-cell precursor cells (Online Supplementary Figure S1C). Moreover, analysis of a specific SNP (rs10507) in the 3’UTR of PIM1 confirmed skewed allelic expression (Online Supplementary Figure S1D). Furthermore, qPCR analysis revealed that this t(6;7)(p21;q34)-positive T-LBL was characterized by aberrant expression of the TLX1 oncogene (Online Supplementary Figure S1E), displayed trisomy 8, and contained focal deletions of the known tumor suppressor genes FHIT, LEF1, IKZF1 and CDKN2A (Online Supplementary Figure S2).2 Finally, TCRβ-PIM1+ T-LBL tumor cells also displayed an activating NOTCH1 mutation and a loss-of-function alteration targeting EP300 (Online Supplementary Table S1), but lacked any IL7R-JAK-STAT pathway mutations.
Using previously published microarray data of primary T-ALLs (n=64) and sorted subsets of normal human thymocyte precursors,9 we subsequently confirmed that high expression of PIM1 is largely restricted to LYL1+, TLX1+, TLX3+ and HOXA+ T-ALLs (Online Supplementary Figure S3A).6,7 In this cohort, PIM1 expression was also significantly correlated with different known JAK-STAT target genes, such as CISH (Pearson’s r = 0.845, P<10−5) and STAT4 (Pearson’s r=0.844, P<10−5) (Online Supplementary Table S2 and Online Supplementary Figure S3B). Furthermore, treating the TLX3+ IL7Rmut DND-41 cell line with the JAK inhibitor ruxolitinib led to reduced PIM1 mRNA levels (Online Supplementary Figure S3C) and we observed higher PIM1 mRNA levels in IL7Rmut/JAKmut patients versus IL7Rwt/JAKwt in another independent T-ALL cohort (n=117) (Online Supplementary Figure S3D),10 further confirming the tight association between PIM1 and JAK-STAT pathway activation.6,7 This second cohort also revealed that NKX2-1+ T-ALLs display low levels of PIM1 (Online Supplementary Figure S3E).2 Finally, similar ranges of PIM1 expression levels were also observed in two independent cohorts of primary T-LBL, as analyzed by qPCR (Online Supplementary Figure S3F and G). Thus, our data support the notion that PIM1 is activated in a significant fraction of human T-ALL and T-LBL patient samples, by rare TCR driven translocations or aberrant activation of the JAK-STAT signaling pathway.6,7
Next, in order to firmly validate PIM1 as a bona fide therapeutic target for the treatment of human T-ALL and T-LBL, we used two independent second-generation pan-PIM inhibitors (AZD120811 and TP-365412) to perform pre-clinical in vivo drug evaluation experiments. For this, we established a patient-derived xenograft (PDX) model from pleural effusion tumor material obtained from the TCRβ-PIM1+ TLX1+ NOTCH1mut T-LBL case. This PDX represents the primary disease as FISH analysis on PDX material confirmed the presence of the TCRβ-PIM1 translocation (Online Supplementary Figure S4A). Moreover, qPCR data showed similar PIM1 expression levels in primary pleural effusion cells and PDX cells (Online Supplementary Figure S4B). Treatment of this T-LBL xenograft was initiated when 1% of blasts were detected in the peripheral blood (PB) and consisted of 3 blocks (5 days on: 2 days off) either of AZD1208 (30 mg/kg, oral gavage), TP-3654 (125 mg/kg, oral gavage), or the respective controls (Figure 1A). Notably, both PIM inhibitors caused a significant delay in tumor development as evaluated by the percentage of hCD45+ tumor cells in PB or spleen weight (Figure 1B). Furthermore, similar patient-derived xenograft models were established from a TLX3+ PIM1high PHF6mut NOTCH1mut and a SIL-TAL1+ PIM1low FBXW7mut NOTCH1mut T-ALL patient sample (Online Supplementary Figure S4C and D). Notably, short-term in vivo PIM inhibition experiments (5 consecutive days; 125 mg/kg TP-3654, oral gavage) also delayed tumor development in the TLX3+ PIM1high T-ALL case to a similar extent as observed for the TCRβ-PIM1+ T-LBL xenograft (Figure 1C). However, and in contrast, no anti-leukemic effects were observed in the SIL-TAL1+ PIM1low patient-derived xenograft (Figure 1D). Given that we did not see any in vivo effect in the PIM1low case, and other PIM kinases (PIM2 and PIM3) are practially non-expressed in these patients (Online Supplementary Figure S5), we believe that the observed effects are largely due to specific PIM1 inhibition.
Figure 1.
In vivo PIM1 inhibition in PIM1high and PIM1low T-cell acute lymphoblastic leukemia (T-ALL)/T-cell lymphoblastic lymphoma (T-LBL) patients. (A) Schematic representation of the in vivo treatment schedule used for the PIM inhibitors TP-3654 and AZD1208; 4 mice per group were used. (B) Percentage of human CD45+ (%hCD45) cells in peripheral blood during treatment (left) and spleen weight of the different treatment groups after completion of treatment (right). *P<0.05; **P<0.01; ***P<0.005. (C) Delayed tumor development in TCRβ-PIM1+ T-LBL and TLX3+ PIM1high T-ALL xenografts after short-term in vivo PIM1 inhibition with TP-3654; 4 mice per group were used. (D) Short-term response of an SIL-TAL1+ PIM1low T-ALL to PIM1 inhibition using TP-3654; 4 mice per group were used.
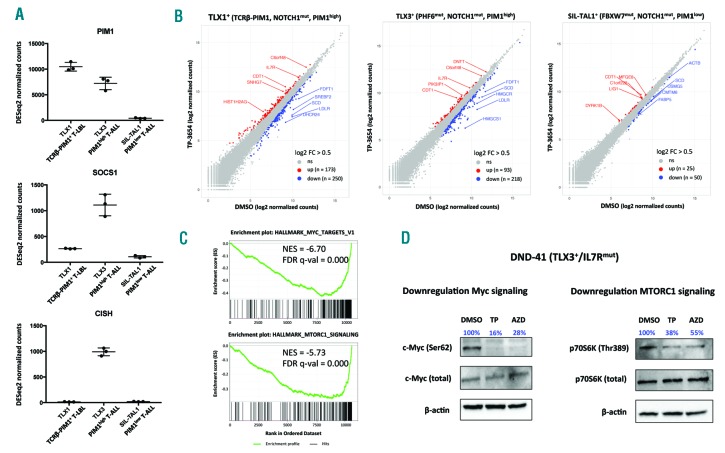
To further investigate the oncogenic role of PIM1 in primary T-ALL and T-LBL specimens, we subsequently performed RNA sequencing on biological triplicates of patient-derived xenograft cells obtained from the TCRβ-PIM1 PIM1high T-LBL, TLX3+ PIM1high T-ALL and SIL-TAL1+ PIM1low T-ALL cases treated with either DMSO or 1 mM TP-3654 ex vivo (24 hours) (Online Supplementary Figure S6). As expected, the transcriptional profiles of the DMSO control samples confirmed that high PIM1 expression occurred in the absence of JAK-STAT path way activation (as evidenced by low expression of the JAK-STAT target genes SOCS1 and CISH) in the TCRβ-PIM1+ T-LBL patient sample (Figure 2A), whereas high PIM1 levels coincided with aberrant activation of JAK-STAT signaling in the TLX3+ T-ALL case (Figure 2A). In line with the observed PIM1 levels and the therapeutic response described above, PIM1 inhibition induced transcriptional changes in the tumor cells from the TCRβ-PIM1+ PIM1high T-LBL (173 up and 250 down; |FC| >0.5, padj <0.05) and the TLX3+ PIM1high T-ALL patient sample (93 up and 218 down; |FC| >0.5, padj<0.05). In contrast, a limited transcriptional response was observed in the SIL-TAL1+ PIM1low T-ALL case (25 up, 75 down; |FC|>0.5, padj<0.05) (Figure 2B). Interestingly, Gene Set Enrichment Analysis (GSEA) revealed that genes significantly down-regulated upon PIM1 inhibition in both sensitive T-LBL and T-ALL samples were enriched for gene sets related to amino acid deprivation, MYC target genes and transcripts downstream of MTORC1 signaling (Figure 2C, Online Supplementary Table S3 and Online Supplementary Figure S7). These effects might, at least in part, be mediated by reduced phosphorylation at previously established PIM1 substrates, such as MYC at serine 62 and p70S6K at threonine 389, upon PIM inhibition, as shown for DND-41 (PIM1high/TLX3+/IL7Rmut) T-ALL cells (Figure 2D).
Figure 2.
Transcriptional effects of PIM1 inhibition in xenograft cells of human T-cell acute lymphoblastic leukemia (T-ALL) and T-cell lymphoblastic lymphoma (T-LBL). (A) DESeq2 normalized count values for PIM1, SOCS1 and CISH based on RNA sequencing data of DMSO control samples of xenograft spleen cells [collected after 24 hours (h) in culture] obtained from TCRβ-PIM1+ T-LBL, TLX3+ PIM1high T-ALL and SIL-TAL1+ PIM1low T-ALL patients, respectively. (B) Diagonal plots from RNA sequencing data of xenograft cells obtained from TCRβ-PIM1+ T-LBL, TLX3+ PIM1high T-ALL and SIL-TAL1+ PIM1low T-ALL patients with DMSO normalized counts on the x-axis versus TP-3654 normalized counts on the y-axis. Up-regulated genes are shown in red, down-regulated genes in blue (pad j < 0.05, |FC| > 0.5). (C) Gene Set Enrichment Analysis plots for down-regulated genesets upon PIM1 inhibition using TP-3654 in responsive TCRβ-PIM1+ T-LBL and TLX3+ PIM1high T-ALL patient samples. NES: Normalized Enrichment Score. (D) Western blot analysis of MYC (Ser62), total MYC, p70S6K (Thr389) and total 70S6K upon PIM inhibition by TP-3654 (6 h, 2.5 μM) or AZD1208 (6 h, 2.5 μM) in DND41 T-ALL cells.
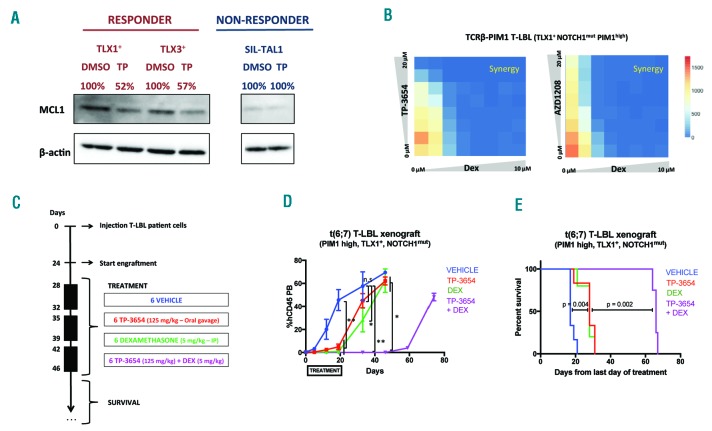
Finally, previous studies, for example in breast cancer, have shown that PIM1 inhibition can result in loss of anti-apoptotic MCL1 expression.13 In line with this, PIM1 inhibition by TP-3654 also resulted in decreased MCL1 expression in patient-derived xenograft cells from both responsive TCRβ-PIM1+ PIM1high T-LBL and TLX3+ PIM1high T-ALL patient samples (Figure 3A). In contrast, BCL2 protein levels were not affected by PIM inhibition (Online Supplementary Figure S8A).
Figure 3.
Synergistic effects of PIM1 inhibition and glucocorticoid therapy. (A) Western blot analysis of MCL1 protein levels in ex vivo treated xenograft spleen cells obtained from TCRβ-PIM1+ T-LBL, TLX3+ PIM1high T-cell acute lymphoblastic leukemia (T-ALL) and SIL-TAL1+ PIM1low T-ALL patients treated with PIM1 inhibitor TP-3654. (B) Combination treatment of TP-3654 and dexamethasone (72 hours) on an ex vivo co-culture system using tumor material from the TLX1+ TCRβ-PIM1+ T-cell lymphoblastic lymphoma (T-LBL) patient. Color code is based on the number of living cells per well. (C) Treatment schedule of in vivo combination treatment with TP-3654 and dexamethasone; 6 mice per group were used. (D) %hCD45 cells in peripheral blood during treatment for the different treatment groups. n.s.: not significant. *P<0.05; **P<0.01. (E) Kaplan-Meier curve representing survival during the in vivo combination drug treatment experiment.
Given that sustained MCL1 expression has been associated with glucocorticoid resistance in ALL,14 we wondered whether PIM1 targeting could also trigger sensitization towards glucocorticoid treatment in T-LBL and T-ALL. Therefore, we used a previously established ex vivo co-culture system15 to show that treatment of xenograft cells from the TCRβ-PIM1 PIM1high T-LBL case with TP-3654 or AZD1208 in combination with dexamethasone resulted in synergistic anti-leukemic activity (Figure 3B and Online Supplementary Figure S9). In addition, combination of the PIM inhibitors TP-3654 and AZD1208 with the BCL2 inhibitor ABT-199 on xenograft cells from TCRβ-PIM1+ PIM1high T-LBL patient also revealed strong synergism (CI<0.3) (Online Supplementary Figure S8B).
Finally, we used the TCRβ-PIM1+ T-LBL patient-derived xenograft to evaluate the PIM inhibition and dexamethasone combination therapy in vivo (Figure 3C). Notably, both TP-3654 as well as dexamethasone monotherapy caused a profound delay in tumor development as evaluated by the percentage of hCD45+ tumor cells in the peripheral blood (Figure 3D). However, combination of PIM inhibition and dexamethasone delayed leukemic blast recurrence in the periphery with approximately 40 days (Figure 3D). As expected, these effects coincided with a significant increase in survival for mice that received the combination therapy as compared to both mono-therapies (P=0.002) (Figure 3E).
Altogether, our study delivers strong pre-clinical in vivo evidence for the use of PIM inhibitors, potentially in combination with glucocorticoids, for the treatment of human T-ALL and T-LBL, and provides a rationale for including PIM1high T-ALL and T-LBL patients in clinical trials for second-generation pan-PIM inhibitors.
Supplementary Material
Acknowledgments
We want to thank all members of the Van Vlierberghe laboratory for critical review of the manuscript and their comments. In addition, we would like to thank Tolero Pharmaceuticals for providing in vivo quantities of TP-3654. Finally, we would like to acknowledge Cergentis for their assistance with TLA experiments.
Footnotes
Funding: this work was supported by the following funding agencies: Fund for Scientific Research Flanders (FWO), the Ghent University Special Research Fund (BOF), the European Hematology Association (EHA), Stand up to Cancer (the Flemish Cancer Society), the Belgian Foundation Against Cancer (STK), Kinderkankerfonds (a non-profit childhood cancer foundation under Belgain law), Associazione Italiana per la Ricerca sul Cancro (AIRC) (IG 19186 to GB) and Fondazione Umberto Veronesi (n. 2064 to VS). The computational resources and services used in this work were provided by the VSC (Flemish Supercomputer Center), funded FWO and the Flemish Government – department EWI.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Muffly L, Larson RA. Improving outcomes in childhood T-cell acute lymphoblastic leukemia: promising results from the Children’s Oncology Group incorporating nelarabine into front-line therapy. Transl Pediatr. 2012; 1(2):120–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017; 49(8):1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah N, Pang B, Yeoh KG, et al. Potential roles for the PIM1 kinase in human cancer - a molecular and therapeutic appraisal. Eur J Cancer. 2008; 44(15):2144–2151. [DOI] [PubMed] [Google Scholar]
- 4.van Lohuizen M, Verbeek S, Krimpenfort P, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989; 56(4):673–682. [DOI] [PubMed] [Google Scholar]
- 5.Lin YW, Beharry ZM, Hill EG, et al. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood. 2010; 115(4):824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padi SKR, Luevano LA, An N, et al. Targeting the PIM protein kinases for the treatment of a T-cell acute lymphoblastic leukemia subset. Oncotarget. 2017; 8(18):30199–30216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Starza R, Messina M, Gianfelici V, et al. High PIM1 expression is a biomarker of T-cell acute lymphoblastic leukemia with JAK/STAT activation or t(6;7)(p21; q34)/TRB@-PIM1 rearrangement. Leukemia. 2018. February 2 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.de Vree PJ, de Wit E, Yilmaz M, et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol. 2014; 32(10):1019–1025. [DOI] [PubMed] [Google Scholar]
- 9.Peirs S, Matthijssens F, Goossens S, et al. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood. 2014; 124(25):3738–3747. [DOI] [PubMed] [Google Scholar]
- 10.Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011; 19(4):484–497. [DOI] [PubMed] [Google Scholar]
- 11.Keeton EK, McEachern K, Dillman KS, et al. AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood. 2014; 123(6):905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foulks JM, Carpenter KJ, Luo B, et al. A Small-Molecule Inhibitor of PIM Kinases as a Potential Treatment for Urothelial Carcinomas. Neoplasia. 2014; 16(5):403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braso-Maristany F, Filosto S, Catchpole S, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016; 22(11):1303–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei G, Twomey D, Lamb J, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006; 10(4):331–342. [DOI] [PubMed] [Google Scholar]
- 15.Frismantas V, Dobay MP, Rinaldi A, et al. Ex vivo drug response profiling detects recurrent sensitivity patterns in drug-resistant acute lymphoblastic leukemia. Blood. 2017; 129(11):e26–e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.