Abstract
Background
Recently, nanomaterials have moved into biological and medicinal implementations like cancer therapy. Therefore, before clinical trials, their binding to plasma proteins like human serum albumin (HSA) and their cytotoxic effects against normal and cancer cell lines should be addressed.
Methods
Herein, the interaction of magnesium oxide nanoparticles (MgO NPs) with HSA was studied by means of fluorescence spectroscopy, circular dichroism (CD) spectroscopy, and docking studies. Afterwards, the cytotoxic impacts of MgO NPs on human leukemia cell line (K562) and peripheral blood mononucleated cells (PBMCs) were evaluated by MTT and flow cytometry assays to quantify reactive oxygen species (ROS) generation and apoptosis.
Results
It was demonstrated that MgO NPs spontaneously form a static complex with HSA molecules through hydrophobic interactions. Docking study based on the size of NPs demonstrated that different linkages can be established between MgO NPs and HSA. The CD investigation explored that MgO NPs did not alter the secondary structure of HSA. Cellular studies revealed that MgO NPs induced cytotoxicity against K562 cell lines, whereas no adverse effects were detected on PBMCs up to optimum applied concentration of MgO NPs. It was exhibited that ROS production mediated by IC50 concentrations of MgO NPs caused apoptosis-associated cell death. The pre-incubation of K562 with ROS scavenger (curcumin) inhibited the impact of MgO NPs -based apoptosis on cell fate, revealing the upstream effect of ROS in our system.
Conclusion
In summary, MgO NPs may exhibit strong plasma distribution and mediate apoptosis by ROS induction in the cancer cell lines. These data demonstrate a safe aspect of MgO NPs on the proteins and normal cells and their application as a distinctive therapeutic approach in the cancer treatment.
Keywords: spectroscopy, anticancer, magnesium oxide, nanoparticles, albumin, K562 cells
Introduction
Nanoparticles (NPs) and their nanomedicine application have been receiving great potentials in the recent years. Nanotechnology applications involve the fabrication of engineered organization of NP to develop unique and well-designed systems.1 Nano-scientists aim to fabricate prospective NPs with unique features, miniaturize large structures, and of course provide higher efficacy of NP-based systems.2 NPs, particularly owing to their high surface area, exhibit distinctive features that determine the engineering of biological interactions.3 In this regard, it is very critical to understand NP interaction at cellular and subcellular status. Also, NPs can be used as drugs to induce antiproliferative activity against cancerous cells.4,5 They can mediate reactive oxygen species (ROS) formation in the cancerous cells and switch on apoptotic, necrotic, and autophagic pathways.6 Indeed, NPs due to their high accessible area are prone to interact with cell and organelle membranes leading to membrane leakage and inducing cancer cell ablation.7 Therefore, scientists have decided to explore NPs demonstrating the most anticancer activity for therapeutic application, mostly for the development of NPs-derived anticancer drugs.1
To further explore the anticancer activity of NPs, it is required that experiments to be conducted in vitro as well as computational simulation models should be developed. Consequently, in vivo testing and clinical trials are performed to understand the fate of NPs in the biological systems. In vitro as well as computational simulation findings are often explored under in vivo conditions, since they simply guide in exploring the general impacts of a test on a living system. In NPs-based drugs discovery, for example, confirmation of NPs potency and their side effects in vitro are important, because in vitro experiments can sometimes provide potential outcomes with drug candidate agents that are relevant to in vivo conditions.8
To candidate NPs for anticancer activity, their side effects against healthy targets such as the protein structure9–12 and cell lines13–15 should be assessed. It has been well documented that plasma proteins like albumins are adsorbed on the NP surfaces as the protein corona in the biological system.16 Perturbing the native structure of the proteins may result in the NP-induced side effects against normal cells and tissues. Also, peripheral blood mononucleated cells (PBMCs) are in direct contact with NPs. Therefore, NP safety against the protein structure and normal cell lines should be studied in advance to provide important evidence of the side effects underlying the clinical application of nanomaterials.
The magnesium oxide NPs (MgO NPs) have been widely studied for antibacterial activity against food borne pathogens17 and antifungal effect against Candida albicans.18 Also, MgO NPs have been used as potential candidates in drug delivery,19 magnetic resonance imaging, and hyperthermia systems.20 However, their anticancer activity against human chronic myelogenous leukemia (K562) is not well explored.
The human serum albumin (HSA) is the main contributing protein in the formation of the protein corona on the NP surface. It contains an alpha helix structure with two well-defined subunits.21,22 A safe NP should induce marginal side effects on the protein structure.
In the present study, the interaction of MgO NPs with HSA will be studied by means of different spectroscopic and docking methods. Afterward, their anticancer activity against K562 cell line was explored. In addition, we tried to consider the anticancer efficacy of MgO NPs against K562 cell line and their safety toward HSA and PBMCs.
Materials and methods
Materials
MgO NPs (US3311) with purity 99.9%, size 10 nm, specific surface area (SSA) 85–120 m2/g, polyhedral, bulk density 0.68 g/cm3, and true density 3.58 g/m3 were purchased from US Nano Co (Houston, TX, USA). HSA (A9511) was obtained from Sigma-Aldrich Co. (St Louis, MO, USA). All other materials were of analytical grade.
Characterization of MgO NPs
MgO NPs were dissolved in double distilled water (DDW) as well as in 1% dimethyl sulfoxide (DMSO) and stirred for 30 minutes at room temperature. The size and hydrodynamic radius of MgO NPs were characterized by transmission electron microscopy (TEM, Zeiss electron microscope, EM10C – 100 kV; Carl Zeiss Meditec AG, Jena, Germany) and dynamic light scattering (DLS; Brookhaven Instrument, Holtsville, NY, USA) techniques, respectively. Also, the charge distribution of MgO NPs was determined using zeta potential analysis (Brookhaven Instrument).
Fluorescence emission spectra
The fluorescence spectroscopy was done using a Hitachi spectrofluorimeter (MPF-4 model; Hitachi, Tokyo, Japan) at 298K, 310K, and 315K. Excitation and emission slit widths were fixed at 5 nm and 10 nm, respectively. Excitation wavelength was set at 280 nm. HSA sample (2 µM, phosphate buffer, ionic strength of 10 mM, pH 7.4) was titrated with varying concentrations of MgO NPs ranging from 2 nM to 40 nM. Band intensity of HSA sample was corrected against buffer solution, NP solution, and inner filter effects.
Molecular docking
A (MgO)16 cluster with a size of 0.6 nm was used as the smaller MgO NP model. The Cartesian coordinates of atoms were directly obtained from Haertelt et al’s work.23 A cubic MgO cluster with a dimension of 1.5 nm was constructed by repetition of MgO unit cell and was considered as the larger NP model. A molecular docking analysis was performed using HEX 6.3 software.24
Circular dichroism experiment
Circular dichroism (CD) measurements were made on an Aviv model 215 spectropolarimeter (Lakewood, NJ, USA) using a 1.00 mm cuvette. CD spectrum was set in the range of 190–260 nm, and the outcomes were expressed as ellipticity (θ) in millidegrees. The HSA sample (5 µM, phosphate buffer [10 mM], pH 7.4) was titrated with different concentrations of MgO NPs ranging from 5 to 50 nM. CD signals of HSA were corrected against buffer and NP solutions.
Cell culture
K562 and PBMCs were used to examine the cell viability after incubating with MgO NPs. K562 cell line was purchased from Pasture Institute of Tehran, Tehran, Iran. Fresh PBMCs were obtained from National Institute of Genetic Engineering and Biotechnology, Tehran, Iran, under approval from the Ethical Committee of Pharmaceutical Sciences Branch, Islamic Azad University of Tehran. The K562 and PBMC cell lines were cultured in the RPMI-1640 cell culture medium supplemented with 10% FBS, 20 U/mL streptomycin, and 20 U/mL penicillin at 37°C in the incubator at 5% CO2.
Cell viability assay
The cytotoxicity of MgO NPs aganist K562 cells and PBMCs was assessed by MTT assay. Approximately, 5×103 cells/well were cultured in the 96-well plates and incubated with varying concentrations of MgO NPs for 24 hours. Afterward, the cells were incubated with 20 µL of MTT at 37°C for 4 hours followed by addition of 100 µL of DMSO at 37°C for 10 minutes. The absorbance of each sample was determined at 570 nm by a microplate reader (Expert 96; Asys Hitch, Ec Austria).
Measurement of ROS
Intracellular ROS was quantified by utilizing dichlorodihydro-fluorescein diacetate (DCFH-DA) staining. The DCFH-DA passively infiltrates the cell membrane where it interacts with ROS to produce a fluorophore dichlorofluorescein (DCF). Approximately, 3×105 cells/well were cultured in the 24-well plates and incubated with IC50 concentrations of MgO NPs for 24 hours, washed, stained with 20 µM DCFH-DA for 1 hour, washed, and the fluorescence signals were quantified by BD FACS Calibur (BD Biocsiences, San Jose, CA, USA) with excitation and emission wavelengths of 488 and 525 nm, respectively. To access the NP-induced ROS generation, an antioxidant (curcumin, 50 µM) was used. The cells were pretreated with single dose of 50 µM curcumin for 24 hours. Cells were then incubated with IC50 concentration of MgO NPs for additional 24 hours.
Quantification of apoptosis by Annexin V-FITC and propidium iodide
The K562 cells were cultured in a 96-well culture plate and incubated with IC50 concentration of MgO NPs for 24 hours. Normal, apoptotic, and necrotic cells were quantified using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide assay kit based on the manufacturer’s protocols. Thereafter, about 3.0×105 cells were washed, resuspended in the binding buffer, and dual stained with Annexin V-FITC and propidium iodide for 20 minutes at 4°C. To assess the NP-induced apoptosis by ROS, curcumin at a concentration of 50 µM was used. The cells were pretreated with a single dose of curcumin (50 µM) for 24 hours. Cells were then incubated with IC50 concentration of MgO NPs for additional 24 hours.
Statistical analysis
All tests were performed in triplicate. Data were reported as mean ± SD, and statistical significance was determined using one-way ANOVA followed by Dunnett’s post hoc test. The values of P<0.05, P<0.01, and P<0.001 were reported to be significant.
Results and discussion
Nanoparticle characterization
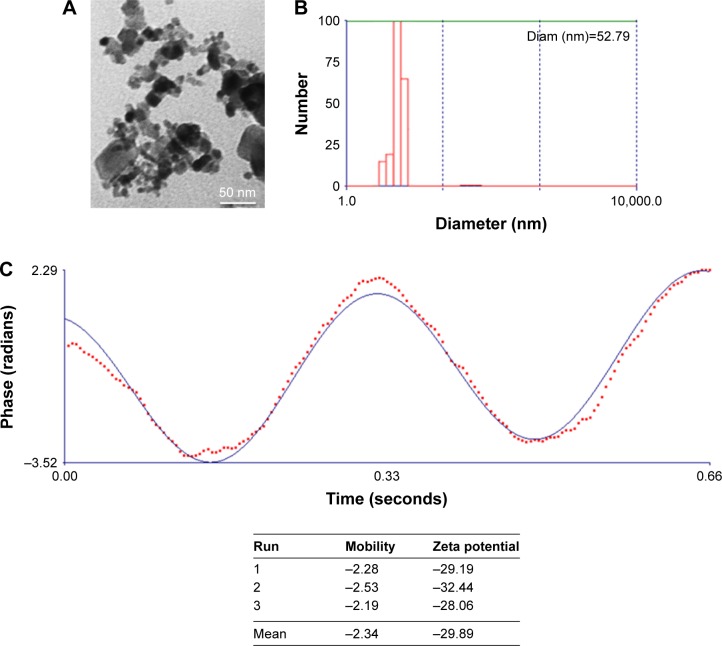
MgO NPs were characterized by means of TEM, DLS, and zeta potential investigations. TEM observation showed that the size of NPs in the dried state was about 10 nm (Figure 1A). DLS studies demonstrated that hydrodynamic radius of MgO NPs was 52.79 nm (Figure 1B). This difference between DLS and TEM outcomes is attributed to the presence of hydrated shell on the NP surface in the DLS investigations.25 To better determine the colloidal stability of MgO NPs, zeta potential measurement was performed. It was revealed that mean zeta potential of MgO NPs was −29.89 mV (Figure 1C). Because zeta potential of MgO NPs was around 30 mV, it may be concluded that MgO NPs exhibit a pronounced colloidal stability.25
Figure 1.
TEM observation (A), DLS data (B), and zeta potential measurement (C) of MgO NPs.
Abbreviations: TEM, transmission electron microscopy; DLS, dynamic light scattering; MgO NPs, magnesium oxide nanoparticles.
Fluorescence measurements
Steady-state fluorescence quenching studies
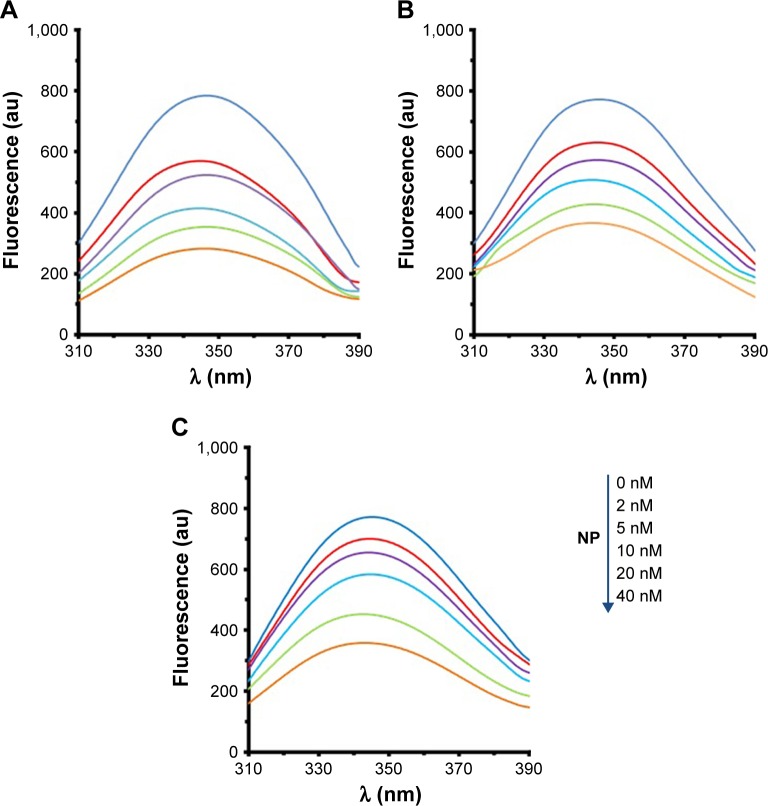
The fluorescence quenching investigation with MgO NPs was performed in 10 mM phosphate buffer, pH 7.4, at three different temperatures of 298K (Figure 2A), 310K (Figure 2B), and 315K (Figure 2C). In the investigations involving the HSA–MgO NP complex, the final concentration of MgO NP was 40 nM. In all the states, the excitation wavelength was 280 nm, so it can be concluded that the fluorescence emission detected was from tryptophan, tyrosine, and phenylalanine residues. Figure 2 shows that quenching mechanism occurs in the presence of MgO NPs in a concentration-dependent manner.
Figure 2.
The fluorescence quenching of HSA (2 µM) in the presence of varying concentrations of MgO NPs (2, 5, 10, 20, and 40 nM) at three different temperatures of 298K (A), 310K (B), and 315K (C).
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
A representative Stern–Volmer plot for the fluorescence quenching of the aromatic residues is depicted in Figure 3. The fluorescence quenching outcome was examined according to Equation 1.12
| (1) |
where, Fo, F, KSV, kq, and τo represent the fluorescence intensity in the absence of NPs, the fluorescence intensity in the presence of NPs, Stern–Volmer constant, the quenching rate constant of the HSA, and the fluorescence lifetime (10−8 seconds), respectively.
Figure 3.
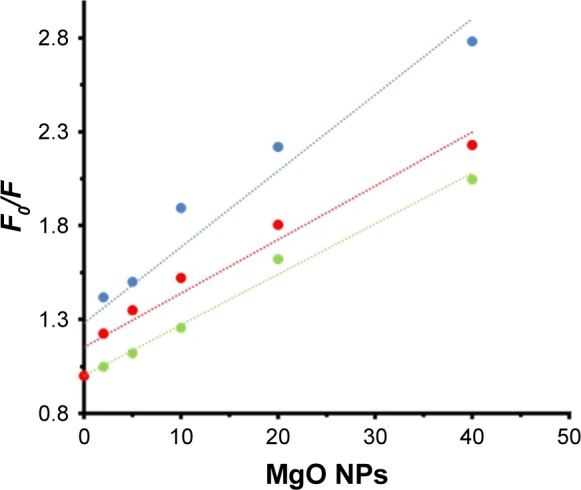
Stern–Volmer plot of HSA (2 µM) in the presence of varying concentrations of MgO NPs (2, 5, 10, 20, and 40 nM) at three different temperatures of 298K (●), 310K (●), and 315K (●).
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
The analysis was performed at three different temperatures: 298K, 310K, and 315K (Table 1).
Table 1.
Ksv and kq values of HSA in the presence of varying concentrations of MgO NPs at three different temperatures of 298K, 310K, and 315K
| T (K) | KSV(M−1) | kq(M−1s−1) | R2 |
|---|---|---|---|
| 298 | 4.05±0.59×107 | 4.05±0.59×1.015 | 0.93 |
| 310 | 2.85±0.24×107 | 2.85±0.24×1.015 | 0.96 |
| 315 | 2.71±0.21×107 | 2.71±0.21×1.015 | 0.99 |
Note: Data are reported as mean ± SD.
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
Judging by the calculated KSV values, the quenching mechanism can be described by a static model in which the HSA and MgO NPs form a complex. This outcome was based on the inverse relation between temperature and KSV values. Also, it was observed that HSA residues are more accessible to MgO NPs at lower temperature characterized by higher KSV value at 298K, while at higher temperature, HSA residues characterized with lower KSV values are less accessible to MgO NPs.
Also, the kq of the HSA for a static quenching mechanism should be an order of 1010 M−1⋅s−1. As it can be seen in Table 1, the kq value for the HSA–MgO NP complex is an order of 1015 M−1·s−1. Therefore, KSV and kq values indicated that quenching mechanism of HSA by MgO NPs was performed by a static system.
Binding constant and the number of binding sites
Figure 4 shows the representation of Hill’s equation:12
| (2) |
where Kb and n represent the binding constant and number of binding sites of the protein per NP, respectively. The variation of log (Fo − F/F) against the log ([MgO NPs]) for HSA at three different temperatures was estimated by Hill’s equation. It shows linear plots with distinctive binding constant at three different temperatures. Kb and n were estimated from the Y-interception and the slope of Hill’s plot, respectively (Table 2). The n values depicted in Table 2 indicating the existence of half to one independent binding site for MgO NPs in the HSA, at 298K–310K.
Figure 4.
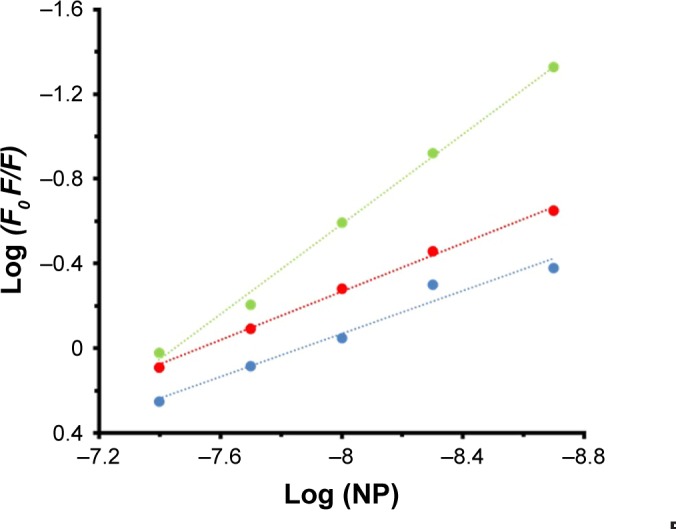
Hill’s plot of HSA (2 µM) in the presence of varying concentrations of MgO NPs (2, 5, 10, 20, and 40 nM) at three different temperatures of 298K (●), 310K (●), and 315K (●).
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
Table 2.
Log Kb and n values of HSA in the presence of varying concentrations of MgO NPs at three different temperatures of 298K, 310K, and 315K
| T (K) | Log Kb(M−1) | n | R2 |
|---|---|---|---|
| 298 | 4.01±0.44 | 0.50±0.06 | 0.97 |
| 310 | 4.31±0.47 | 0.57±0.07 | 0.99 |
| 315 | 7.92±0.79 | 1.06±0.12 | 0.99 |
Note: Data are expressed as mean ± SD.
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
Half-binding site reveals that two HSA molecules are adsorbed on the NP surfaces. However, as the temperatures increases, the n increases from 0.50 to 1.06. Also, the Kb values increased significantly with increasing temperature, indicating that marginal temperature-induced conformational changes of HSA may provide more favorite adsorption site of HSA on MgO NP surface.
Considering the Kb values, the distribution of a NP in plasma can be evaluated. A small Kb value reveals a weak binding affinity of NP to HSA and subsequent short life time, whereas a high Kb value indicates a strong binding affinity and longer life time.26
The strong binding of MgO NPs could be a consequence of their hydrophobic interactions with HSA. Therefore, it may be deduced that MgO NPs present enhanced capacity to enter into the tissues and can be applied as a potential candidate in the medical applications.26,27
Thermodynamic parameters
The driving interactions between a NP and the protein include hydrophilic and hydrophobic forces. The thermodynamic parameters can be calculated by van’t Hoff equation:28
| (3) |
where, ΔS° is entropy change, ΔH° exhibits enthalpy change, R is the universal gas constant and T is absolute temperature. The plot of ln Kb vs 1/T yields the calculation of ΔH° and ΔS° values (Figure 5, Table 3). The free energy change ΔG° was then calculated from the Gibbs–Helmholtz equation:28
| (4) |
Figure 5.
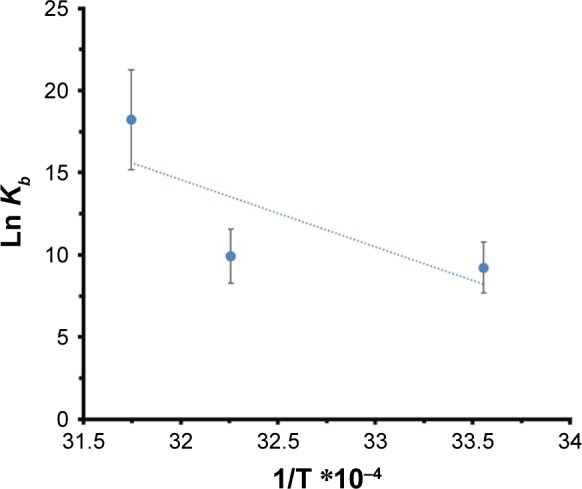
van’t Hoff plot of HSA/MgO NPs to calculate thermodynamic parameters.
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
Table 3.
Thermodynamic parameters of the HSA–MgO NPs complex at three different temperatures of 298K, 310K, and 315K
| T (K) | ∆G° (kJ/mol) | ∆H° (kJ/mol) | T∆S° (kJ/mol) |
|---|---|---|---|
| 298 | −22.81±2.27 | 363.01±55.03 | |
| 310 | −25.50±2.81 | −340.20±52.91 | 365.70±55.65 |
| 315 | −47.62±5.35 | 387.82±57.59 |
Note: Data are expressed as mean ± SD.
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
The values of ΔG°, ΔH°, and ΔS° are reported in Table 3.
The representative sign of both ΔH° and ΔS° are positive. So, the potential forces involved between MgO NPs and HSA were hydrophobic in nature. Negative ΔG° values verify the spontaneous binding process in the interactions.28
Thus, the data demonstrate the presence of only one binding site for MgO NPs in the HSA. Moreover, the Kb values for MgO NPs show that the interaction of albumin with MgO NPs is very strong.
In addition, at different temperatures, primary sites of MgO NPs seem to be located in different albumin regions where aromatic residues are located. Some hydrophilic subdomains which are more exposed to the hydrophilic environment can provide potential binding sites to MgO NPs. HSA has one tryptophan, 18 tyrosine, and 19 phenylalanine residues.22 It may be suggested that exposed aromatic residues are involved in the interaction of the HSA with MgO NPs.
However, the size of NPs cannot be easily controlled in the biological systems. Due to their high surface area and presence of counterpart ions, NPs may lose their colloidal stability and tend to form agglomerated species. Therefore, the size of NP can be dramatically changed and these different colloidal stabilities and corresponding different sizes of NPs may bind to different domains of the protein, although the chemical compound of NP is not changed. Indeed, it can be suggested that all hydrophilic and hydrophobic interactions are involved in the interaction of NPs with protein. However, the main contributing forces can be dictated by the size of NPs. For this purpose, molecular docking was done with MgO NPs of different sizes to reveal the role of the size of NPs on the kind of interactions between MgO NPs and HSA.
Molecular docking study
A (MgO)16 cluster with a size of 0.6 nm and a cubic MgO cluster with a size of 1.5 nm were used to explore the binding affinity of HSA to NPs with different cluster sizes.
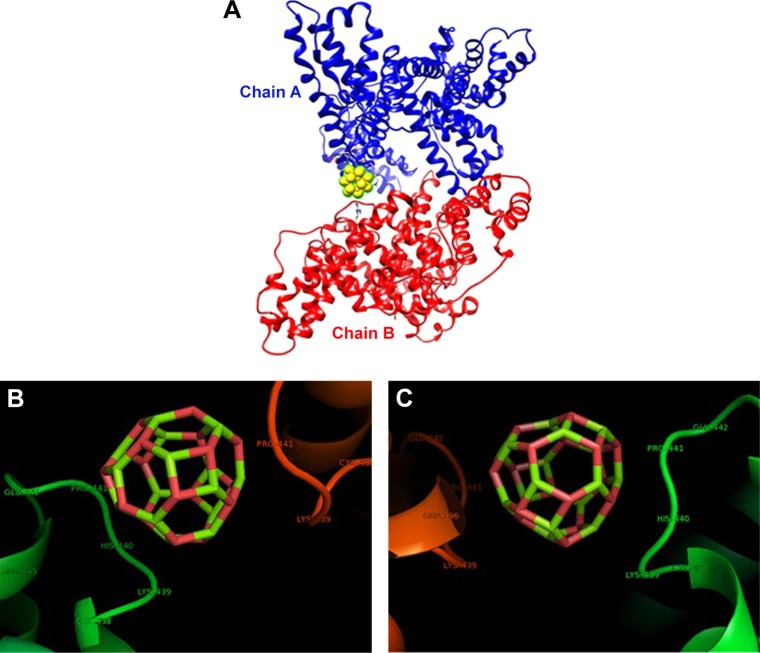
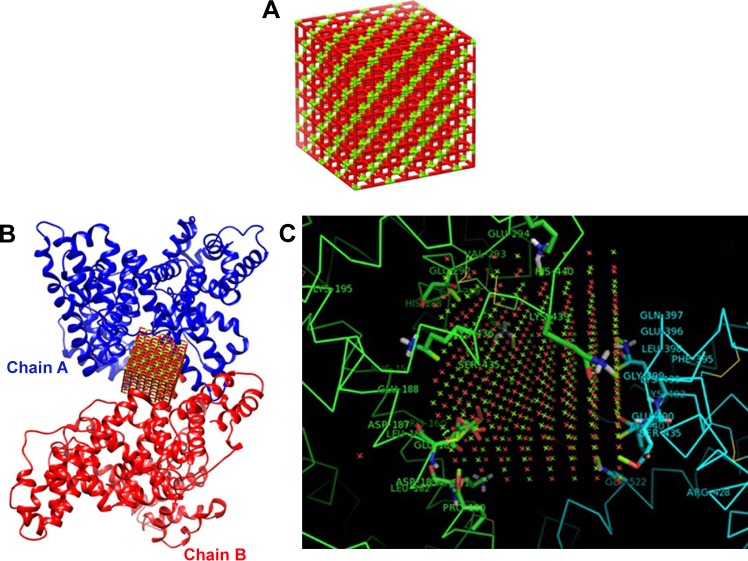
The crystallographic structure of HSA (PDB ID: 1AO6) was obtained from the Protein Data Bank (http://www.pdb.org). The molecular docking was performed with MgO NPs cluster of different sizes. The resulting binding energies for 0.6 nm (Figure 6A–C) and 1.5 nm MgO NP (Figure 7A–C) clusters were found to be −334.93 and −688.03 E-values, respectively. It was observed that the affinity of HSA to larger NPs is more than their smaller counterparts. Visualization of the docked site was performed using Chimera and PyMol graphical tools. The docked sites of MgO NPs with the sizes of 0.6 nm and 1.5 nm are demonstrated in Figures 6A and 7B, respectively. It was demonstrated that closest residues within 4 A° distance for MgO NP with the size of 0.6 nm are Glu-442.A, Pro-441.A, His-440.A, Lys-439.A, Cys-438.A, Arg-445.A, Pro-441.B, Cys-438.B, Lys-439.B, Glu-442.B, and Glu-396.B (Figure 6B and C). Therefore, it can be deduced that MgO NP with smaller size can interact with charged residues through electrostatic interactions. However, the binding site of MgO NPs having size of 1.5 nm within 4 A° and the spatial orientation in the binding site is depicted in Figure 7B. The nearest interacting residues are Gln-397.B, Glu-396.B, Leu-398.B, Gly-399.B, Glu-400.B, Phe-395.B, Pro-180.A, Val-293.A, Phe-156.A, Arg-160.A, Leu-182.A, Asp-183.A, Glu-184.A, Leu-186.A, and Phe-156.A (Figure 7C). It can be observed that larger NPs are prone to interact with hydrophobic residues by means of hydrophobic forces.
Figure 6.
(A) Docking site of interaction between MgO NPs (0.6 nm) and HSA; (B, C) the closest interacting residues with two rotational views.
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
Figure 7.
(A) The designed cluster of MgO NP (1.5 nm); (B) docking site of interaction between MgO NPs (1.5 nm) and HSA; and (C) the closest interacting residues.
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
There are different ways that NPs with different sizes can bind to the proteins. Every NP usually shows a particular binding site and NPs with different sizes can bind to different sites of protein. It was depicted that MgO NPs with different sizes can interact with different subdomains of HSA. Therefore, some important factors are involved in the mode of NP binding to proteins that allows the binding site to be characterized independent of the chemical composition of NPs.
Circular dichroism study
To explore the alteration in the secondary structure of the HSA upon addition of MgO NPs, CD spectroscopy was carried out at far UV-CD (190–260 nm) region. The CD band of HSA depicts two characteristic minima of α-helix at 208 and 222 nm, which is consistent with the native structure of HSA29,30 (Figure 8). The band of the HSA exhibits marginal alterations in the presence of different concentrations of MgO NPs. These data reveal that the folded structure of the HSA structure is not perturbed upon addition of MgO NPs.
Figure 8.
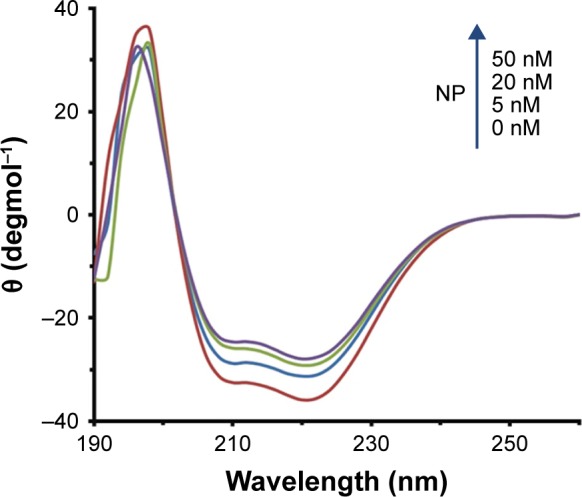
Ellipticity changes of HSA (5 µM) in the presence of varying concentrations of MgO NPs (5, 20, and 50 nM) in the far UV region.
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
This suggests that in the presence of 5, 20, and 50 nM of MgO NPs, HSA keeps the helical pattern similar to that determined for the native HSA (Table 4). Because, in the physiological conditions, NPs are effective at very low doses, our investigations with regard to the nanomolar dose of NPs should be sufficient to explore the details pertaining to NP binding.
Table 4.
The secondary structure of HSA in the presence of varying concentrations of MgO NPs was calculated by CDNN software
| [NP] nM | Helix% | Sheet% | Turn% | Random coil% |
|---|---|---|---|---|
| 0 | 63.7±3.72 | 8.4±0.22 | 11.2±0.12 | 16.7±1.09 |
| 5 | 63.3±3.71 | 8.5±0.23 | 11.2±0.12 | 17.0±1.10 |
| 10 | 62.9±3.69 | 8.1±0.21 | 11.5±0.13 | 17.5±1.11 |
| 50 | 62.7±3.67 | 8.2±0.21 | 11.6±0.13 | 17.5±1.11 |
Abbreviations: HSA, human serum albumin; MgO NPs, magnesium oxide nanoparticles.
Now, a question can be raised as to how it is possible that MgO NPs interacted with folded HSA molecules by means of hydrophobic interaction, while the secondary structure of HSA is intact? It may be suggested that MgO NPs induce partial structural changes and form hydrophobic forces with nonpolar residues close to the solvent area. Indeed, marginal NP-induced conformational changes result in maintaining the native structure of the protein; however, some hydrophobic patches can be displaced to the protein surface and provide an accessible site to be involved in the hydrophobic interactions.
To assess the safety of MgO NPs and compare the chemical composition, size, and functional groups of NPs on the HSA structure, literature survey was done. From the details summarized in Table 5, it can be deduced that MgO NPs do not induce significant structural changes in the HSA structure, whereas other studied NPs have shown remarkable conformational changes in the HSA structure.
Table 5.
Interaction of NPs with HSA based on the size, surface group, and types of NPs
| NPs | Size (nm) | Surface groups | Kind of interactions | Structural changes | Reference |
|---|---|---|---|---|---|
| MgO | 10 | – | Hydrophobic interaction | No changes | – |
| C60 | 50–110 | – | – | Alpha increase | 31 |
| CdTe-QDs | 5 | MPA | Electrostatic interaction | Alpha decrease | 32 |
| CdTe-QDs | 5 | N-Acetylcysteine | Electrostatic interaction | Alpha decrease | 32 |
| CdTe-QDs | 5 | Gluthatione | Electrostatic interaction | Alpha decrease | 32 |
| CdTe-QDs | 2–4.8 | MPA | Electrostatic interaction | Alpha decrease | 33 |
Abbreviations: HSA, human serum albumin; NPs, nanoparticles; CdTe-QDs, cadmium telluride-quantum dots; MPA, mercaptopropionic acid; C60, fullerene; MgO, magnesium oxide.
It can be concluded that size, chemical groups, and type of NPs can play important roles in the interaction of proteins with NPs. Indeed, MgO NPs show the strongest interaction (hydrophobic forces) and safest chemical composition among other NPs.
Therefore, it was demonstrated that MgO NPs can bind to HSA by means of hydrophobic interactions and induced partial structural changes to the HSA structure.
Now, the main application of MgO NPs in the cancer therapy should be considered. To address this objective, cytotoxicity of MgO NPs against cancerous cells and normal cells should be explored. Indeed, a NP is considered as a safe and potential candidate in cancer therapy when it exhibits selective cytotoxicity against tumor cells.
MTT assay
The data show that MgO NPs dramatically reduced the viability of K562 in a dose-dependent manner. At 1 µg/mL concentration, cell viability was markedly reduced to 78.5% in the K562 cells (P<0.05). However, MgO NPs did not mediate a remarkable decrease in the viability of normal cells (PBMCs) up to an optimum dose of 30 µg/mL, which was employed in this investigation (Figure 9). The IC50 concentration of MgO NPs for K562 cell line was 17.75 µg/mL. Therefore, it may be concluded that MgO NPs are more toxic against cancer cells as compared with normal cells. These data reveal selective cytotoxicity of MgO NPs against tumor cells.
Figure 9.
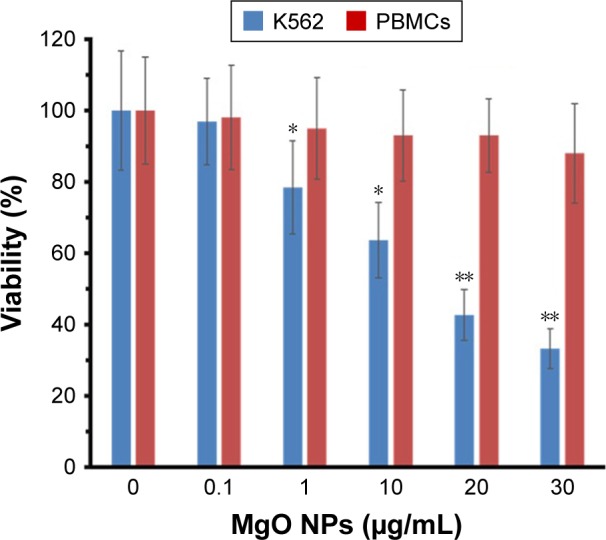
The effect of MgO NPs on the viability of K562 cells and PBMCs.
Notes: Data are reported as mean ± SD of three tests carried out in three replicates. *P<0.05 and **P<0.01, statistically significant differences relative to the control cells.
Abbreviations: MgO NPs, magnesium oxide nanoparticles; PBMCs, peripheral blood mononucleated cells.
ROS generation
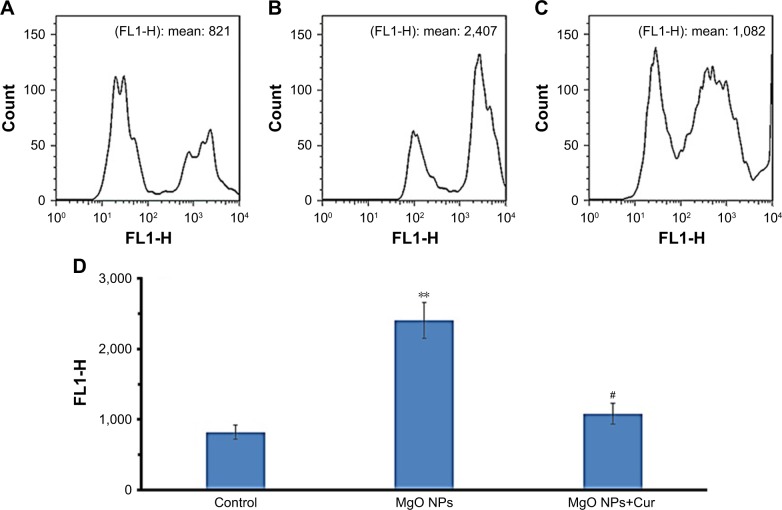
The intracellular ROS formation was determined following exposure to IC50 concentration of MgO NPs (17.75 µg/mL) using the H2DCF-DA compound in the K562 cell line (Figure 10A as control cells). The K562 cells treated with IC50 concentration of MgO NPs (17.75 µg/mL) for 24 hours (Figure 10B) demonstrated a significant increase in the mean ROS generation (FLI-1H: 2,407, P<0.01) as compared with control cells (FLI-1H: 821) (Figure 10A). However, pretreatment of cells with 50 µM curcumin for 24 hours resulted in the reduction of intracellular production of ROS (FLI-1H: 1,082, P<0.05) relative to NP-treated cells (Figure 10C). The histogram depicts the analysis of ROS generation by flow cytometry after incubation of K562 cells with MgO NPs for 24 hours (Figure 10D). Therefore, pretreatment of NP-treated cells with curcumin resulted in the reduction of ROS generation induced by MgO NPs.
Figure 10.
The intracellular ROS formation by IC50 concentration of MgO NPs.
Notes: Upper graphs show generation of ROS in the K562 cells using H2DCF-DA dye. Control cells (A); MgO NPs-treated cells (B); pretreatment of cells with curcumin (50 µM) followed by MgO NPs treatment (C). Cells were incubated with IC50 concentration of MgO NPs for 24 hours. Histogram analysis of ROS generation by flow cytometer after MgO NPs incubation with K562 cells for 24 hours (D). Values represent mean ± SD of three experiments. **P<0.01, compared to control, #P<0.05, compared to MgO NP-treated group.
Abbreviations: MgO NPs, magnesium oxide nanoparticles; ROS, reactive oxygen species; Cur, curcumin.
Flow cytometry analysis
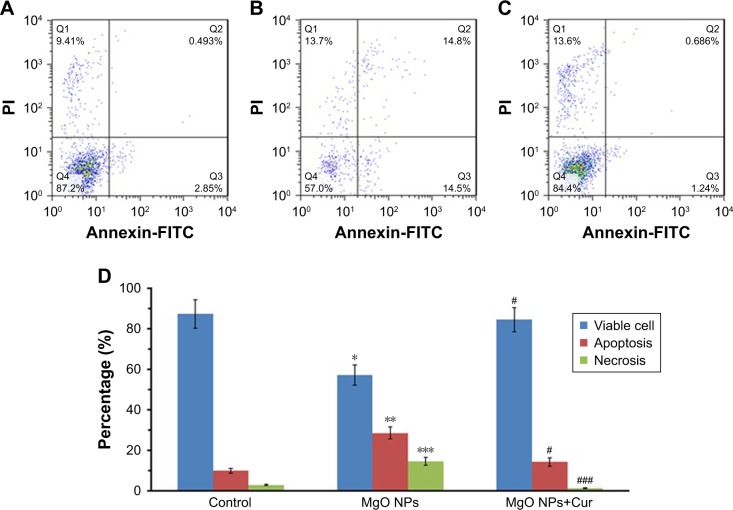
When K562 cells were treated with IC50 concentration of MgO NPs for 24 hours, the percentage of apoptotic cells significantly increased compared to control cells (Figure 11A). The exposure of K562 cells to MgO NPs caused induction of apoptosis (P<0.01) and necrosis (P<0.001) in the K562 cells (Figure 11B). However, pretreatment of the cells with curcumin for 24 hours (50 µM) resulted in the significant reduction of apoptosis (P<0.05) and necrosis (P<0.001) in MgO NPs-treated K562 cells compared to NP-treated cells with no pretreatment (Figure 11C). The histogram demonstrates the percentage of apoptosis and necrosis as determined by flow cytometer after 24 hours of incubation of K562 cells with IC50 concentration of MgO NPs (Figure 11D).
Figure 11.
Quantification of apoptosis by flow cytometry.
Notes: Control cells (A); MgO NPs-treated cells (B); pretreatment of cells with curcumin (50 µM) followed by MgO NPs treatment (C). Cells were incubated with IC50 concentration of MgO NPs for 24 hours. Histogram analysis of percentage of cells undergoing apoptosis and necrosis by flow cytometer after incubation of K562 cells with MgO NPs for 24 hours (D). Values represent mean ± SD of three experiments. *P<0.05, **P<0.01, ***P<0.001, compared to control; #P<0.05, ###P<0.001 compared to MgO NP-treated group.
Abbreviations: MgO NPs, magnesium oxide nanoparticles; FITC, fluorescein isothiocyanate; PI, propidium iodide; Cur, curcumin.
Human exposure to NPs is unpreventable as NPs become more extensively employed and as a result, nanotoxicology study is now growing up. However, while several NPs with different chemical compositions and implementation continue to expand, investigations to determine their interactions and their cytotoxic effect on the biological systems are few in the comparison.34 In the field of medicinal application in particular, NPs are being employed in the diagnostic and therapeutic devices to better diagnose and treat different illnesses. Exposure to NPs for medical applications is considered as an intentional exposure; accordingly, exploring the features of NPs and their impact on the human body is of high priority before considering their clinical application. Different outcomes have been reported for different NPs with regard to their adverse effects due to differences in the experimental conditions and varying colloidal stabilities of NPs.35,36 For NPs to be expanded in the clinical arena, it is crucial that nanotoxicology studies reveal and explore how chemical nature of NPs affect the adverse effects of NPs, in order to minimize their unenviable characteristics on the biological systems.
Cellular studies revealed that MgO NPs induced significant adverse effects on the cancer cell line; however, MgO NPs demonstrated marginal side effects on normal cell line up to an optimum concentration.
It was revealed that IC50 concentration of MgO NPs increased ROS levels to trigger apoptosis in the K562 cells. Preincubation of K562 cells with curcumin, an ROS scavenger, muted the effect of MgO NP-based apoptosis on cell fate, highlighting the upstream impact of ROS in our study. In summary, MgO NP provides a potential system of priming apoptosis by ROS production.
ROS are commonly produced in the biological systems. Accordingly, cells have developed enzymatic and nonenzymatic antioxidant defense systems that aid in inhibiting the ROS generation.37 Intracellular generation of ROS in response to NP exposure has been involved in upregulating transcription factors and inducing apoptosis and necrosis.38 It has been well documented that ROS trigger DNA sequence alterations in the form of DNA breakage and rearrangements.39 These changes may result in switching on apoptosis signaling pathways. The ROS-mediated upregulation of gene expression can be used as a potential approach for potential therapeutic appeal. Several antioxidant compounds have been demonstrated to limit ROS generation, oxidant-mediated DNA damage, and cell mortality.40 For example, polyphenols like curcumin show radical-scavenging ability and thereby inhibit apoptosis.41 In general, ROS generation and gene expression changes are involved in the apoptosis induction and the pathogenesis of cancer cells.38,39
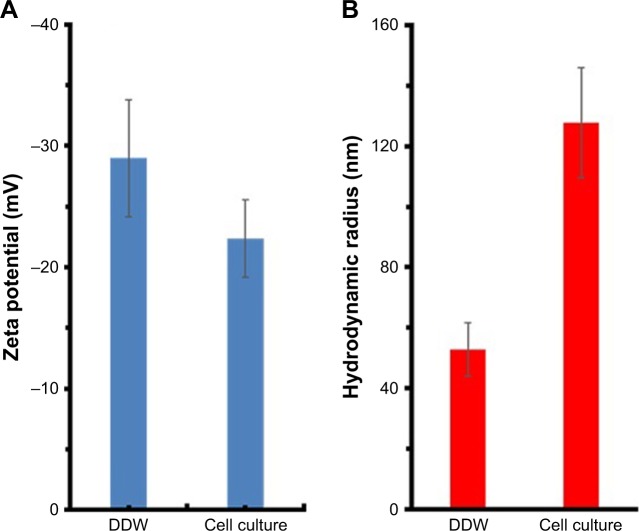
One of the crucial problems in cancer therapy lies in enhancing therapeutic efficiency of anticancer agents and inhibiting their side effects against normal tissues. In our MTT cytotoxicity test, MgO NPs exhibited a significant cytotoxicity on K562 cells; however, no induced cytotoxicity was observed against normal PBMC cells. One possible reason for the different toxicity of NPs against normal and cancer cells could be attributed to colloidal stability of NPs. It may be suggested that cancer cell membrane is more permeable to large NPs than normal cells.42 In other words, larger NPs are more prone to infiltrate the cancer cell lines as compared with normal cells.42 To address this hypothesis, DLS study in the cell culture medium with FBS (10%) was performed. It was observed that zeta potential of MgO NPs decreased from −28.89 mV in the DDW to −22.38 mV in the cell culture medium (Figure 12A). NPs having higher zeta potential values show high colloidal stability due to the strong repellent forces between NPs.43 For NPs with lower zeta potential, attractive interaction, for example, hydrophobic forces may overcome the repulsion between NPs.43 Therefore, NPs with low zeta potential are electrostatically destabilized, whereas NPs with high zeta potentials show a tendency to be dispersed. DLS study shows that the hydrodynamic radius of NPs increased from 52.79 nm in the DDW to 127.81 nm in the cell culture medium (Figure 12B). Therefore, NPs will tend to be agglomerated in the cell culture medium and lose their ability to enter the normal cells. However, cancer cells will be more vulnerable to the consequences of NP aggregation, finally enabling the penetration of larger NPs into the cell and inducing oxidative stress and apoptosis.
Figure 12.
(A) Zeta potential of MgO NPs in DDW and cell culture medium; (B) hydrodynamic radius of MgO NPs in the DDW and cell culture medium.
Abbreviations: MgO NPs, magnesium oxide nanoparticles; DDW, double distilled water.
Recently, selective toxicity of other NPs such as zinc oxide (ZnO)44 and iron oxide (Fe3O4) NPs45 on cancer cells have been studied. Premanathan et al explored that the ZnO NPs demonstrated a preferential capability to ablate human myeloblastic leukemia cells (HL60) as compared with PBMCs.44 Ahamed et al reported that Fe3O4 NPs show selective impact on cell mortality of cancer cells (HepG2 and A549), while inducing no side effect on normal (rat hepatocytes and IMR-90) cell lines.45 Shafaght et al claimed that copper oxide NPs triggered apoptosis in the human K562 cancer cell line via ROS generation, while no adverse effects were observed against PBMC cells.46
Recently, Moosavi et al demonstrated the concentration-dependent ability of photo-activated nitrogen-doped TiO2 NPs in triggering differentiation or cell mortality in the K562 leukemia cells.6
Conclusion
The MgO NPs demonstrated strong binding affinity through hydrophobic interactions with HSA molecules. It was revealed that MgO NPs induced marginal secondary structural changes in HSA molecule. The MgO NPs also exhibited selective cytotoxicity against K562 cell line and can be considered as a novel anticancer agent. The present study reveals that MgO NPs-mediated apoptosis is initiated through the ROS generation in the cancer cells. Further studies on other cancer types and normal cells should be conducted to explore whether anticancer effect of MgO NPs is equally applicable and productive against other cancer cell lines.
Acknowledgments
The financial support of Islamic Azad University, Pharmaceutical Science Branch, Tehran, Iran (IAUPS), is greatly acknowledged.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Fan W, Yung B, Huang P, Chen X. Nanotechnology for multimodal synergistic cancer therapy. Chem Rev. 2017;117(22):13566–13638. doi: 10.1021/acs.chemrev.7b00258. [DOI] [PubMed] [Google Scholar]
- 2.He Z, Alexandridis P. Ionic liquid and nanoparticle hybrid systems: Emerging applications. Adv Colloid Interface Sci. 2017;244:54–70. doi: 10.1016/j.cis.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Heydari H, Moosavifard SE, Elyasi S, Shahraki M. Nanoporous CuS nano-hollow spheres as advanced material for high-performance supercapacitors. Appl Surf Sci. 2017;394:425–430. [Google Scholar]
- 4.Bidkar AP, Sanpui P, Ghosh SS. Efficient induction of apoptosis in cancer cells by paclitaxel-loaded selenium nanoparticles. Nanomedicine. 2017;12(21):2641–2651. doi: 10.2217/nnm-2017-0189. [DOI] [PubMed] [Google Scholar]
- 5.Sengel-Turk CT, Hascicek C, Bakar F, Simsek E. Comparative evaluation of nimesulide-loaded nanoparticles for anticancer activity against breast cancer cells. AAPS PharmSciTech. 2017;18(2):393–403. doi: 10.1208/s12249-016-0514-2. [DOI] [PubMed] [Google Scholar]
- 6.Moosavi MA, Sharifi M, Ghafary SM, et al. Photodynamic N-TiO2 Nanoparticle Treatment Induces Controlled ROS-mediated Autophagy and Terminal Differentiation of Leukemia Cells. Sci Rep. 2016;6:34413. doi: 10.1038/srep34413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torchi A, Simonelli F, Ferrando R, Rossi G. Local Enhancement of lipid membrane permeability induced by irradiated gold nanoparticles. ACS Nano. 2017;11(12):12553–12561. doi: 10.1021/acsnano.7b06690. [DOI] [PubMed] [Google Scholar]
- 8.Fröhlich E, Salar-Behzadi S. Toxicological assessment of inhaled nanoparticles: role of in vivo, ex vivo, in vitro, and in silico studies. Int J Mol Sci. 2014;15(3):4795–4822. doi: 10.3390/ijms15034795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kermani ZR, Haghighi SS, Hajihosseinali S, et al. Aluminium oxide nanoparticles induce structural changes in tau and cytotoxicity of the neuroblastoma cell line. Int J Biol Macromol. 2018;120(Pt A):1140–1148. doi: 10.1016/j.ijbiomac.2018.08.182. [DOI] [PubMed] [Google Scholar]
- 10.Roshanfekrnahzomi Z, Badpa P, Esfandiari B, et al. Silica nanoparticles induce conformational changes of tau protein and oxidative stress and apoptosis in neuroblastoma cell line. Int J Biol Macromol. 2018 Sep 21; doi: 10.1016/j.ijbiomac.2018.09.118. Epub. [DOI] [PubMed] [Google Scholar]
- 11.Ajdary M, Moosavi MA, Rahmati M, et al. Health Concerns of Various Nanoparticles: A Review of Their in Vitro and in Vivo Toxicity. Nanomaterials. 2018;8(9):634. doi: 10.3390/nano8090634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeinabad HA, Kachooei E, Saboury AA, et al. Thermodynamic and conformational changes of protein toward interaction with nanoparticles: a spectroscopic overview. RSC Adv. 2016;6(107):105903–105919. [Google Scholar]
- 13.Bhamidipati M, Fabris L. Multiparametric assessment of gold nanoparticle cytotoxicity in cancerous and healthy cells: the role of size, shape, and surface chemistry. Bioconjug Chem. 2017;28(2):449–460. doi: 10.1021/acs.bioconjchem.6b00605. [DOI] [PubMed] [Google Scholar]
- 14.Mandani S, Majee P, Sharma B, et al. Carbon Dots as Nanodispersants for Multiwalled Carbon Nanotubes: Reduced Cytotoxicity and Metal Nanoparticle Functionalization. Langmuir. 2017;33(31):7622–7632. doi: 10.1021/acs.langmuir.7b00557. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Xu Y, Xi X, et al. Amino acid-modified chitosan nanoparticles for Cu2+ chelation to suppress CuO nanoparticle cytotoxicity. J Mater Chem B. 2017;5(19):3521–3530. doi: 10.1039/c7tb00344g. [DOI] [PubMed] [Google Scholar]
- 16.Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano. 2012;6(7):5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 17.Jin T, He Y. Antibacterial activities of magnesium oxide (MgO) nanoparticles against foodborne pathogens. J Nanopart Res. 2011;13(12):6877–6885. [Google Scholar]
- 18.Karimiyan A, Najafzadeh H, Ghorbanpour M, Hekmati-Moghaddam SH. Antifungal effect of magnesium oxide, zinc oxide, silicon oxide and copper oxide nanoparticles against Candida albicans. Zahedan J Res Med Sci. 2015;17(10):19–23. [Google Scholar]
- 19.Shen S, Chow PS, Chen F, Tan RB. Submicron particles of SBA-15 modified with MgO as carriers for controlled drug delivery. Chem Pharm Bull. 2007;55(7):985–991. doi: 10.1248/cpb.55.985. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Boubeta C, Balcells L, Cristòfol R. Self-assembled multifunctional Fe/MgO nanospheres for magnetic resonance imaging and hyperthermia. Nanomedicine: Nanotechnology. Biol Med. 2010;6(2):362–370. doi: 10.1016/j.nano.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Esfandfar P, Falahati M, Saboury A. Spectroscopic studies of interaction between CuO nanoparticles and bovine serum albumin. J Biomol Struct Dyn. 2016;34(9):1962–1968. doi: 10.1080/07391102.2015.1096213. [DOI] [PubMed] [Google Scholar]
- 22.Carter DC, He XM, Munson SH, et al. Three-dimensional structure of human serum albumin. Science. 1989;244(4909):1195–1198. doi: 10.1126/science.2727704. [DOI] [PubMed] [Google Scholar]
- 23.Haertelt M, Fielicke A, Meijer G, Kwapien K, Sierka M, Sauer J. Structure determination of neutral MgO clusters – hexagonal nanotubes and cages. Phys Chem Chem Phys. 2012;14(8):2849–2856. doi: 10.1039/c2cp23432g. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie DW, Venkatraman V. Ultra-fast FFT protein docking on graphics processors. Bioinformatics. 2010;26(19):2398–2405. doi: 10.1093/bioinformatics/btq444. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharjee S. DLS and zeta potential – What they are and what they are not? J Control Release. 2016;235:337–351. doi: 10.1016/j.jconrel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Baroli B, Ennas MG, Loffredo F, Isola M, Pinna R, López-Quintela MA. Penetration of metallic nanoparticles in human full-thickness skin. J Invest Dermatol. 2007;127(7):1701–1712. doi: 10.1038/sj.jid.5700733. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson GR. Plasma and tissue binding considerations in drug disposition. Drug Metab Rev. 1983;14(3):427–465. doi: 10.3109/03602538308991396. [DOI] [PubMed] [Google Scholar]
- 28.Das S, Ghosh P, Koley S, Singha Roy A, Roy AS. Binding of naringin and naringenin with hen egg white lysozyme: A spectroscopic investigation and molecular docking study. Spectrochim Acta A Mol Biomol Spectrosc. 2018;192:211–221. doi: 10.1016/j.saa.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Siddiqi M, Nusrat S, Alam P, et al. Investigating the site selective binding of busulfan to human serum albumin: Biophysical and molecular docking approaches. Int J Biol Macromol. 2018;107(Pt B):1414–1421. doi: 10.1016/j.ijbiomac.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Khatun S, Yasmeen S, Kumar A, Subbarao N. Calorimetric, spectroscopic and molecular modelling insight into the interaction of gallic acid with bovine serum albumin. J Chem Thermodyn. 2018;122:85–94. [Google Scholar]
- 31.Song M, Liu S, Yin J, Wang H. Interaction of human serum album and C60 aggregates in solution. Int J Mol Sci. 2011;12(8):4964–4974. doi: 10.3390/ijms12084964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai L, Lin C, Xu ZQ, et al. Spectroscopic studies on the interactions between CdTe quantum dots coated with different ligands and human serum albumin. Spectrochim Acta A Mol Biomol Spectrosc. 2012;97:366–376. doi: 10.1016/j.saa.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Xiao Q, Huang S, Su W, et al. Systematically investigations of conformation and thermodynamics of HSA adsorbed to different sizes of CdTe quantum dots. Colloids Surf B Biointerfaces. 2013;102:76–82. doi: 10.1016/j.colsurfb.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 34.Maity P, Bepari M, Pradhan A, Baral R, Roy S, Maiti Choudhury S. Synthesis and characterization of biogenic metal nanoparticles and its cytotoxicity and anti-neoplasticity through the induction of oxidative stress, mitochondrial dysfunction and apoptosis. Colloids Surf B Biointerfaces. 2018;161:111–120. doi: 10.1016/j.colsurfb.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 35.Fatemi M, Mollania N, Momeni-Moghaddam M, Sadeghifar F. Extracellular biosynthesis of magnetic iron oxide nanoparticles by Bacillus cereus strain HMH1: Characterization and in vitro cytotoxicity analysis on MCF-7 and 3T3 cell lines. J Biotechnol. 2018;270:1–11. doi: 10.1016/j.jbiotec.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 36.Izadiyan Z, Shameli K, Hara H, Mohd Taib SH. Cytotoxicity assay of biosynthesis gold nanoparticles mediated by walnut (Juglans regia) green husk extract. J Mol Struct. 2018;1151:97–105. [Google Scholar]
- 37.Manke A, Wang L, Rojanasakul Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. BioMed research international. 20132013 doi: 10.1155/2013/942916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandurangan M, Veerappan M, Kim DH. Cytotoxicity of zinc oxide nanoparticles on antioxidant enzyme activities and mRNA expression in the cocultured C2C12 and 3T3-L1 cells. Appl Biochem Biotechnol. 2015;175(3):1270–1280. doi: 10.1007/s12010-014-1351-y. [DOI] [PubMed] [Google Scholar]
- 39.van Aken B. Gene expression changes in plants and microorganisms exposed to nanomaterials. Curr Opin Biotechnol. 2015;33:206–219. doi: 10.1016/j.copbio.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 40.Phull AR, Abbas Q, Ali A, Raza H, Zia M, Haq IU. Antioxidant, cytotoxic and antimicrobial activities of green synthesized silver nanoparticles from crude extract of Bergenia ciliata. Future Journal of Pharmaceutical Sciences. 2016;2(1):31–36. [Google Scholar]
- 41.Dai C, Li D, Gong L, Xiao X, Tang S. Curcumin ameliorates furazolidone-induced DNA damage and apoptosis in human hepatocyte L02 cells by inhibiting ROS production and mitochondrial pathway. Molecules. 2016;21(8):1061–1068. doi: 10.3390/molecules21081061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honary S, Zahir F. Effect of zeta potential on the properties of nano-drug delivery systems – a review (Part 2) Trop J Pharm Res. 2013;12(2):265–273. [Google Scholar]
- 44.Premanathan M, Karthikeyan K, Jeyasubramanian K, Manivannan G. Selective toxicity of ZnO nanoparticles toward Gram-positive bacteria and cancer cells by apoptosis through lipid peroxidation. Nanomedicine. 2011;7(2):184–192. doi: 10.1016/j.nano.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Ahamed M, Alhadlaq HA, Khan MAM, Akhtar MJ. Selective killing of cancer cells by iron oxide nanoparticles mediated through reactive oxygen species via p53 pathway. J Nanopart Res. 2013;15(1):1225–1229. [Google Scholar]
- 46.Shafagh M, Rahmani F, Delirezh N. CuO nanoparticles induce cytotoxicity and apoptosis in human K562 cancer cell line via mitochondrial pathway, through reactive oxygen species and P53. Iran J Basic Med Sci. 2015;18(10):993–999. [PMC free article] [PubMed] [Google Scholar]