Abstract
The communities of bacteria that reside in the intestinal tract are in constant competition within this dynamic and densely colonized environment. At homeostasis, the equilibrium that exists between these species and strains is shaped by their metabolism and also by pathways of active antagonism, which drive competition with related and unrelated strains. Importantly, these normal activities contribute to colonization resistance by the healthy microbiota, which includes the ability to prevent the expansion of potential pathogens. Disruption of the microbiota, resulting from, for example, inflammation or antibiotic use, can reduce colonization resistance. Pathogens that engraft following disruption of the microbiota are often adapted to expand into newly created niches and compete in an altered gut environment. In this review, we examine both the interbacterial mechanisms of colonization resistance and the strategies of pathogenic strains to exploit gaps in colonization resistance.
INTRODUCTION
The intestinal microbiota is composed of diverse communities of microorganisms that colonize different regions of the gastrointestinal tract. The composition of these communities varies along the length of the intestine, in addition to variation from the lumen to the mucous and enterocyte barrier.1 Dramatic transitions in the microbiota occur with age, and a major shift from the microbes present following initial colonization in the neonatal period to the mature state has been well documented.2–5Although the microbiota is highly complex, at higher taxonomic levels consistent patterns exist between healthy individuals.1 For example, the healthy colon is dominated by strict anaerobes in the Bacteroidetes and Firmicutes phyla.6,7 Furthermore, significant similarity between individuals is observed if diversity is assessed on the basis of the conservation of functional pathways present in the microbiota.6,8
Expansion of some opportunistic species including Enterococcus faecalis, Enterococcus faecium and members of the Enterobacteriaceae family that are typically present in low abundance to very high levels can have negative consequences for the host. For pathogenic taxa, such as Salmonella enterica serovar Typhimurium (Salmonella) or Clostridium difficile, expansion of these species in the gut triggers an inflammatory response and diarrhea (reviewed in refs.9,10). For other species, including Klebsiella pneumoniae, a member of the Enterobacteriaceae, and Enterococcus faecium, expansion in the gut does not trigger overt intestinal inflammation but is associated with an increased risk for patients to subsequently develop bacteremia.11,12 Increasingly, the strains that dominate the gut in clinical settings are resistant to one or more antibiotics. Indeed, in the United States, the Center for Disease Control has classified vancomycin-resistant Enterococcus (VRE), carbapenum-resistant Enterobacteriaceae, and extended spectrum β-lactamase producing Enterobacteriaceae (ESBLs) as serious or urgent threats.13
In mouse models and in patients, expansion of these potential pathogens can be triggered by disruption of the microbiota. The ability of the healthy microbiota to prevent expansion of these potential pathogens is termed colonization resistance, and is a critical function of the healthy microbiota under homeostatic conditions. The mechanisms of microbiota-mediated colonization resistance are diverse. In general, resistance mechanisms prevent expansion of pathogenic bacteria by either direct, bacteria-to-bacteria pathways, or by activating host immune defenses. Conversely, opportunistic bacterial species that dominate the gut use diverse strategies to exploit breakdowns in colonization resistance, leading to their expansion into newly created niches.
In this review, we examine direct interbacterial mechanisms of colonization resistance, as well as the strategies used by opportunistic species to compete and take advantage of the breakdown in colonization resistance that enables their expansion.
DIRECT MECHANISMS OF COLONIZATION RESISTANCE
The important function of the microbiota in providing colonization resistance has been recognized for decades.14,15 Indeed, work from Bohnhoff et al. in 1954 demonstrated that antibiotic-mediated disruption of the microbiota leads to a dramatic increase in susceptibility to Salmonella infection.15 These early pioneering studies and others generated interest in understanding the mechanisms of colonization resistance. Here we highlight some of the recent advances in our understanding of this critical function.
Metabolic
Nutrient limitation.
When food is consumed, host mechanical and chemical processes function to break down large complexes of protein, fat, and carbohydrates, thereby enabling absorption of nutrients. In addition to host-directed pathways, the microbiota plays an important role in digestion by expressing a complementary suite of catabolic pathways (Fig. 1). Microbiota-dependent digestion supports the growth of the bacterial community and generates byproducts that are utilized by the host, such as short-chain fatty acids (SCFAs). Competition for nutrients in the gut is intense, and evolution has shaped the cross-feeding patterns and substrate preferences of the microbiota to maximize utilization of the available substrates.16–18 Therefore, under homeostatic conditions, an exogenous species is unlikely to find an uncontested niche, and will be forced to compete with the established microbiota for nutrients.19,20 Interestingly, the odds of success for an exogenous species in establishing residence in the gut is increased if the microbiota of the gut harbors related taxa to the new species, suggesting that appropriate nutrient conditions exist.21
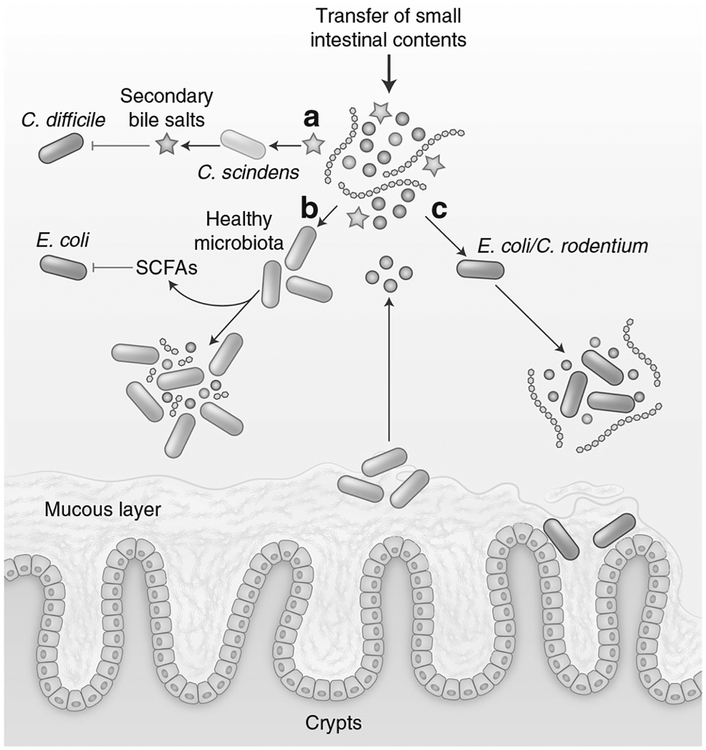
Fig. 1.
Microbiota metabolism contributes to colonization resistance. a Members of the microbiota, such as C. scindens, convert primary bile salts (blue stars) into secondary bile salts (red stars) that inhibit the vegetative growth of C. difficile. b The healthy microbiota ferments diet-derived simple sugars (purple/pink circles), complex polysaccharides (blue lines), and microbiota-liberated metabolites from the mucous layer (brown circles) to produce inhibitory SCFA. c In the absence of competing strains, E. coli and C. rodentium utilize the increased availability of simple sugars for replication
The abnormal expansion of Citrobacter rodentium (C. rodentium) or Escherichia coli (E. coli) in the gut is fueled by utilization of simple sugars.22–24 Kamada et al. found that C. rodentium competes for simple sugars with the endogenous microbiota.22Furthermore, shifting the diet to only simple sugars forced competition with strains like Bacteroides thetaiotaomicron that normally occupy different nutritional niches.22 E. coli is able to utilize a variety of sugars present in the mucous layer simultaneously, including galactose, fucose, mannose, N-acetyl glucosamine, arabinose, and ribose,23 and commensal strains of E. coli can provide colonization resistance against pathogenic E. coli O157 based on overlapping sugar utilization.25,26 Together, these results highlight the potential importance of carbon and energy source availability. However, additional studies are required to determine the contribution of nutrient limitation in the context of a normal diet and a complex, unperturbed microbiota in providing colonization resistance. In line with this, studies in both mice and humans have demonstrated that substrate availability and competition for those substrates along with subsequent micro-biota structure is strongly influenced by diet and the presence of appropriate substrate utilization pathways.27–29
In addition to competition for carbon and energy sources, invading bacteria compete with the microbiota for trace metals. For example, the probiotic Nissle 1917 E. coli strain protects mice from pathogenic Salmonella infection in manner that is dependent on iron uptake.30 Furthermore, zinc uptake pathways in Vibrio cholerae and Campylobacter jejuni are critical to the ability of these pathogens to compete with the densely colonizing microbiota.31The importance of this competition for trace metals is highlighted by observations demonstrating that immunization of mice to generate antibodies against iron-scavenging siderophores is sufficient to reduce the burden of Salmonella infection.32
Bile salts.
Bile salts are secreted into the small intestine by the host and function to increase fat solubilization and digestion.33 Bile salts are generated in the liver from enzymatic conversion of cholesterol, and the majority are conjugated to glycine or taurine through an amide linkage.34 The majority of secreted bile salts are reabsorbed in the small intestine; however, unabsorbed bile salts continue to the colon where they are metabolized by the microbiota into secondary bile acids. Microbiota-driven metabolism of bile salts begins with deconjugation from glycine or taurine, in a reaction catalyzed by bile salt hydrolases.35 These hydrolases are expressed by a large number of taxa within the normal microbiota.35 Further metabolism of bile salts is encoded in bile-acid-inducible (Bai) operons by a small subset of commensal species of the microbiota that includes Clostridium scindens and catalyze 7α/β-dehydroxylation35,36 (Fig. 1). Interestingly, secondary bile salts have been reported to induce the germination of C. difficile spores and inhibit the replication of vegetative cells in a concentration-dependent manner.37 Transfer of C. scindens restores colonization resistance against C. difficile in a bile-salt-dependent manner.38 In agreement, the kinetics of C. difficile germination and the initiation of pathology correlate with antibiotic-induced changes in bile salt levels and compositions,39 and varying antibiotic regimens leading to different bile salt levels correlated with susceptibility to C. difficile expansion.40 The relative resistance of various clinical C. difficile isolates to the secondary bile-salt lithocholic acid positively correlates with disease severity in murine infection models.41,42
SCFA-mediated inhibition.
The dominant phyla of the large intestine, Bacteroidetes and Firmicutes, are anaerobic bacteria that metabolize diverse substrates including complex polysaccharides, simple sugars, and glycans released from ingested fiber and also host mucus using fermentation pathways.43,44 This fermentation process leads to the generation of SCFAs (Fig. 1). The most abundant SCFA in the gut are acetate, propionate, and butyrate and the total concentration of SCFA in the proximal colon can reach concentrations of 70–140 mM.45,46 The majority of the SCFA are produced in the proximal colon and are absorbed by the host to support enterocyte metabolism.47 In the proximal colon, high levels of SCFA production contribute to acidification of the lumen.45 For example, using a wireless motility capsule approach in human subjects, Farmer et al. reported average pH values immediately distal to the ileal:caecal junction of 6.16 in healthy individuals.48 In vitro, numerous studies have demonstrated that SCFAs at an acidic pH are able to inhibit the replication of E. coli and Salmonella,49–51 and loss of SCFA production and acidity were correlated with in vivo susceptibility to Salmonella following streptomycin treatment.14 However, other work has argued against a significant role for direct SCFA-mediated inhibition in vivo.19,52 During homeostasis, the production of SCFAs by the microbiota also plays a critical role in maintaining the limited availability of oxygen and nitrates in the lumen of the gut.53,54 Recent work has demonstrated that butyrate production by Clostridia activates PPAR-γ in epithelial cells resulting in the repression of Nos2 expression, and activation of oxygen-consuming β-oxidation as opposed to anaerobic glycosis.54–56 Loss of butyrate production triggers increased oxygen and nitrate release into the lumen of the gut, which can be exploited by Enterobacteriaceae, as described below. In addition, several studies have demonstrated that the detection of SCFAs triggers a complex modulation of virulence factor expression in Salmonella.57–60
Therefore, the relative contribution of direct SCFA-mediated antagonism to colonization resistance compared with other mechanisms of colonization resistance or, indeed, other indirect effects of SCFAs remains unclear.
Active antagonism
Established members of the microbiota are also able to inhibit the expansion of new species through mechanisms of active antagonism. In the context of an exogenously introduced pathogen, these pathways contribute to colonization resistance. However, in many cases these mechanisms of antagonism can target the same or closely related species, suggesting that, in the absence of exogenous pathogenic bacteria, these pathways function to allow expressing strains to compete with other commensal strains capable of utilizing similar nutrients.
Bacteriocins.
Microbiota-derived antimicrobials have been identified that have activity against both Gram-positive and Gram-negative pathogens. The original natural variants of clinical antibiotics are produced through complex and diverse biosynthetic pathways, and include polyketides (erythromycin, tetracycline),61 non-ribosomally produced glycopeptides (vancomycin),61 aminoglycosides (neomycin),62 and lincosamides (clindamycin).63 Within the gut, the microbiota-produced antimicrobials are most frequently ribosomally synthesized and post-translationally modified peptides (RiPPs), also termed bacteriocins, which are processed and secreted from the cell64 (Fig. 2).
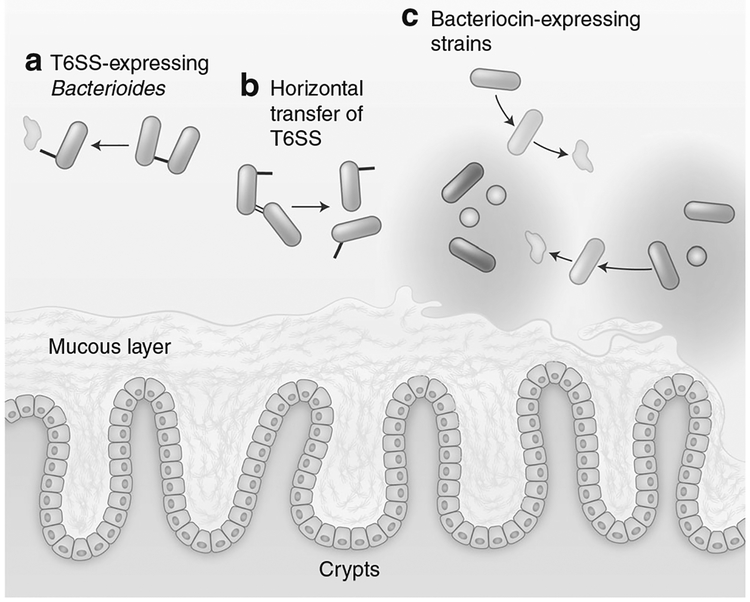
Fig. 2.
Mechanisms of active antagonism contribute to colonization resistance: a Constituents of the microbiota can express contact-dependent secretion systems that deliver antagonistic effector proteins. In Gram-negatives, such as Bacteroides, delivery is driven by the T6SS, while the Esx system has recently been identified as having an analogous role in Gram-positive bacteria. b In Bacteroides, the GAI-1 and GAI-2 variants of the T6SS are encoded on conjugative plasmids enabling horizontal gene transfer to other strains in the microbiota. c Short peptide antimicrobials, termed bacteriocins, are secreted by the microbiota and have activity against both related non-immune commensal strains and exogenous species
Some of the best-characterized bacteriocins are those produced by members of Lactobacillales order. Nisin is a prototypical member of the lantibiotic class of bacteriocin that is produced by Lactococcus lactis that inhibits a broad range of Gram-positive bacteria including Staphylococcus aureus, and Listeria monocyto-genes.65 Nisin functions by binding to Lipid II leading to pore formation and has bactericidal activity at nanomolar levels in Gram-positive bacteria where the peptidoglycan layer is exposed.66 As a result of its potency, Nisin has been used as a food preservative for decades to prevent the expansion of Gram-positive pathogens.65 Beyond Nisin, bacteriocin production in Lactobacillales is widespread. For example, Lactococcus salivarius UCC118 production of the bacteriocin Abp118 is able to protect mice from L. monocytogenes infection.67 In comparison to nisin, some bacteriocins target a narrower range of species.68 Thuricin CD is a bacteriocin produced by commensal Clostridium thuringiensis that has potent antimicrobial activity against C. difficile but does not target other members of the microbiota including Bifidobacteria or Lactobacillus.68
Although bacteriocins produced by Gram-positive bacteria can have high levels of activity against Gram-positive pathogens, most of these peptides lack significant activity against Gram-negative bacteria. However, recent work examining the bacteriocin NAI-107 found that this lantibiotic has activity against a small number of fastidious Gram-negative pathogens, and synergizes with polymyxin to inhibit a broader range of Gram-negative pathogens including Klebsiella pneuomoniae, and E. coli.69 Several groups have also attempted to artificially broaden the range of targets of bacteriocins.70,71 For example, Nisin that had been complexed to carbon and gold nanoparticles with a net positive charge had increased access to the periplasm of Gram-negative bacteria and antibacterial activity against E. coli and Pseudomonas aeruginosa.70 Similarly, fusion of Nisin to truncations of Gram-negative targeting antimicrobial peptides led to increased efficacy against E. coli.71
Using a bioinformatics approach, Donia et al. demonstrated that a large number of biosynthetic gene clusters are widely distributed among the different microbiota communities, and include pathways for the production of lantibiotics, thiazole/oxazole-modified microcins, and thiopeptides.72 In support of this bioinformatic data, the authors demonstrated that Lactococcus gasseri produces lactocillin, a thiopeptide with activity against a range of Gram-positive pathogens.72
Some strains of commensal Enterococcus express the bacteriocins AS-48, bac-31, and bac-21 from the pheromone-responsive conjugative plasmids pAD1, pYI17, and pPD1, respectively.73–75 Conjugative transfer of these plasmids is triggered by recognition of peptide pheromones secreted by recipient cells,76 which would allow for rapid horizontal propagation of the bacteriocin-producing phenotype. Interestingly, production of the pheromone cOB1 by commensal strains of Enterococcus can trigger cell death of VRE strains with cOB1 responsive mobile elements on the pTEF2spc plasmid and an IS-like elements in the main chromo-some.77 In vivo, expression of bac-21 increases the long-term colonization of commensal E. faecalis through increased competitiveness with other related strains.78 Furthermore, the high-density environment of the gut promotes conjugative transfer of bacteriocin-expressing plasmids from positive to negative strains.78 Finally, by engineering conjugation-defective pPD1 plasmids, Kommineni et al. were able to demonstrate that commensal E. faecalis carrying the engineered plasmids is able to de-colonize mice of pPD1-negative VRE.78 Potential probiotic Enterococcus strains have been identified that express bacteriocins targeting Gram-positive species including L. monocytogenes.79
The nasal microbiota provides colonization resistance against S. aureus, and Staphylococcus epidermidis or Streptococcus mitis have been negatively correlated with S. aureus abundance.80,81 To examine the mechanism of protection, Zipperer et al. screened nasal isolates for the ability to inhibit S. aureus and demonstrated that a commensal Staphylococus strain, Staphylococcus lugdunensis, is able to directly inhibit the growth of S. aureus through the production of a peptide antimicrobial, termed lugdunin.82 In addition to targeting MRSA, lugdunin also inhibits the growth of Listeria monocytogenes, VRE, and Streptococcus pneuomoniae.82
Members of the Gram-negative Enterobactericeae family can produce microcins, a class of short peptide bacteriocins, that have activity against closely related strains of bacteria.83 A large number of microcins have been identified in commensal strains of E. coli, including the probiotic Nissle 1917 strain.84 Recent work from Sassone-Corsi et al. demonstrated that probiotic E. coli Nissle 1917 could compete with both E. coli and Salmonella in a manner dependent on microcin expression.85 Interestingly, this successful competition occurred only in inflamed conditions but not in mice treated only with streptomycin. Under inflamed conditions the competition for iron becomes critical for Enterobacteriaceae expansion.30 By linking these observations, Sassone-Corsi et al. demonstrated that microcin expression is induced by iron deprivation, and that microcin-dependent competition in the gut depends on an iron-limited environment.85
Antimicrobial proteins.
In contrast to shorter peptides, some species of the microbiota also produce larger proteins with antimicrobial activities. For example, some members of the Bacteroidales, including Bacteroides fragilis and Bacteroides uniformis, can produce Bacteroidales-secreted antimicrobial proteins that inhibit related species and increase competitiveness in the gut.86,87 In addition, some B. fragilis strains can secrete a ubiquitin-like protein termed BfUbb that has antimicrobial activity against related strains.88 Similarly, some strains of Enterobacteriaceae carry plasmids with genes encoding colicins, a class of antimicrobial proteins that target related species lacking the appropriate immunity proteins.89 Unlike the best-characterized peptide antimicrobials which often target the cell wall, colicins must be transported by specific receptors on target Gram-negative cells into the periplasmic space and can have diverse activities inside the cell.89 Interestingly, work examining the dynamics of competition between colicin-resistant, colicin-sensitive, and colicin-producing E. coli strains has found that colicin production can actually promote diversity of related strains in the gut as equilibrium is reached.90
Type VI secretion systems.
Type VI secretion systems (T6SS) are complex structures that allow for export of effector molecules across the inner and outer membranes of Gram-negative bacteria and into adjacent bacteria or eukaryotic cells.91 Therefore, unlike antimicrobial compounds or toxic byproducts of metabolism, T6SS activity requires cell-to-cell contact for antagonism (Fig. 2). The best-characterized T6SS are those expressed by pathogenic Gammaproteobacteria.92 The delivered effectors antagonize neighboring cells through varied mechanisms of action, including degradation of NAD(P)+ and peptidoglycan hydrolysis.91,93–95 Critically, T6SS+ strains must encode an immunity protein that negates the activity of the effector to avoid self-targeting.93
In healthy adult individuals, Bacteroidetes are typically a major constituent of the anaerobic community in the large intestine. Interestingly, orthologs of the well-characterized Proteobacterial T6SS gene clusters are widespread in Bacteroidetes taxa,96 and can be horizontally transferred between strains97,98 (Fig. 2). T6SS in Bacteroidales are present in three forms termed GAI-1, GAI-2, and GAI-3.98 GAI-1 and GAI-2 are widely distributed among Bacterio-dales while GAI-3 is restricted to B. fragilis.98,99 In vivo, anindividual’s microbiota tends to harbor single variants of GAI-1/GAI-2 immunity/effector genes suggesting that a stable consortium of Bacteroidales in a gut share T6SS by horizontal gene transfer and subsequently resist colonization from new strains.99 On the basis of initial in silico identification of these orthologs, Russell et al. examined the expression of T6SS in vivo and found robust expression of T6SS components by both B. fragilis and Bacteroides eggerthii.96 In vitro, B. fragilis was able to inhibit the growth of a T6SS-strain of Bacteroides thetaiotaomicron in a T6SS-dependent manner.96 More recent reports have shown that B. fragilis T6SS are functional in vivo and mediate competition with related strains including T6SS-B. fragilis and Bacteroides vulgatus.100,101 Interestingly, both studies found that target strains with susceptibility to in vitro T6SS-antagonism are not necessarily susceptible to in vivo T6SS-antagonism, and species such as B. thetaiotaomicron can be unaffected by the presence of a T6SS-expressing competitor despite in vitro susceptibility.100,101 Importantly, T6SS-expressing B. fragilis can provide colonization resistance against pathogenic enterotoxigenic B. fragilis (ETBF).102
Recent work from Whitney et al. has demonstrated the existence of a cell-to-cell contact-dependent mechanism of antagonism in Gram-positive bacteria, including in the genomes of gut resident species.103 In this pathway, Gram-positive bacteria are able to secrete members of the LxG group of proteins, including toxins, through the Esx secretion system.103,104 For example, TelC is a cell wall targeting effector of the Esx secretion system,103 and putative NAD(P)+ hydrolyzing toxins with LXG domains have recently been identified.105 This suggests that, similar to Gram-negative T6SS, contact-dependent secretion systems in Gram-positive bacteria might also secrete a diverse group of effectors. However, it remains unclear if the Esx-type secretion system is capable of delivering toxins across the thick peptidoglycan layer of target Gram-positive cells in a manner analogous to intracellular delivery toxins by a T6SS.103
Consortia of bacteria can restore colonization resistance Under homeostatic conditions, the microbiota is a highly complex community that as a whole provides colonization resistance to exogenously introduced species. Clinically, fecal microbiota transplantation has been used successfully to treat recurrent infection with C. difficile.106,107 Therefore, transferring a complex community of bacteria can restore colonization resistance. As specific pathways of colonization resistance are identified, along with the microbiota constituents responsible for those pathways, it will likely be possible to design minimal consortia of bacteria that provide colonization resistance or clearance against specific clinically relevant species. In agreement, several papers have already identified small consortia that provide full or partial colonization resistance against VRE, L. monocytogenes, Salmonella, or C. difficile.38,108–112
STRATEGIES TO PATHOGENS TO EXPLOIT GAPS IN COLONIZATION RESISTANCE
As described above, under steady-state conditions, the healthy microbiota effectively excludes newly introduced potential pathogens through an array of mechanisms. In agreement, clinically relevant expansion of potential pathogens often occurs after the microbiota is perturbed with antibiotics (reviewed in ref. 113). Unfortunately, common nosocomial pathogens, including members of the Enterobacteriaceae, VRE, and C. difficile are particularly adept at dominating the gut following perturbation of the microbiota.
Perturbation of the healthy gut microbiota creates an opportunity for expansion, and drives intense competition between related strains to expand into those niches. The idea that species which expand into dysbiotic gut niches exhibit their own forms of colonization resistance and competition is demonstrated by the observation that initial colonization of the gut by a Salmonella strain prevents subsequent establishment of other isogenic wild-type strains.114 In this section, we examine the strategies utilized by these pathogens to exploit gaps in colonization resistance, compete for newly created niches, and subsequently maintain dominance in the dysbiotic gut.
Antagonism
Recent work has demonstrated that pathogens that expand into dysbiotic conditions utilize a variety of mechanisms to antagonize both the normal microbiota and other species or strains that are also seeking to rapidly expand. For example, as described above, the inflamed gut is an iron-limited environment. This environment can signal to initiate production of microcins in probiotic E. coli;85 however, pathogenic members of the Enterobacteriaceae such as Salmonella also use these environmental signals to upregulate expression of their own antimicrobial proteins, including Col1b.115 In line with this, pathogenic strains of E. coli causing urinary tract infections carry a greater number of microcin genes, and have higher prevalence of microcin H47 and colicin E1 compared to nonpathogenic E. coli strains.116 The gut microbiota has a high level of colonization resistance against Listeria expansion.109 Likely in response to this selective pressure, epidemic Listeria strains have virulence factors that target the microbiota.117,118 In the gut, Listeria expresses Listeriolysin S, which is able to inhibit the growth of members of the microbiota, including Lactococcus lactis, in order to promote infection.117,118
Many Gram-negative pathogens, including V. cholerae, P. aeruginosa, and S. typhimurium, have T6SS that are expressed during colonization and deliver a wide range of effectors to give a fitness advantage in a dysbiotic gut119–121 (Fig. 3). Strikingly, among Proteobacteria harboring a T6SS and an identifiable effector, 40% of strains had two effectors with different predicted activities, and 25% had three or more effectors.122 Work from LaCourse et al. found that P. aeruginosa secretes multiple effectors whose activities can be both synergistic and dependent on environmental conditions.122 In addition to its well-characterized Type III secretion systems SPI-1 and SPI-2, recent work has demonstrated that Salmonella also utilizes a T6SS to promote its expansion in the gut.121 The Salmonella T6SS is able to target commensal strains in vitro and ΔT6SS strains are deficient in in vivo colonization.121 Both Salmonella and Pseudomonas tightly regulate the activity of the T6SS and recognize cues present in the gut to activate the T6SS. These cues include both host-derived factors such as bile salts, as well as cell stress or kin cell lysis.120,121,123
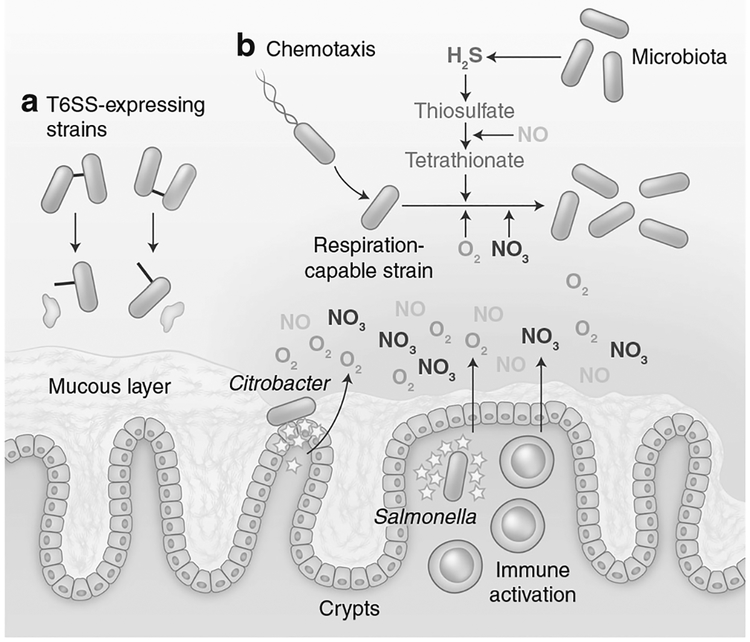
Fig. 3.
Expansion and engraftment strategies of invading species. a Some pathogens, such as Salmonella and P. aeruginosa, target either the microbiota or other opportunistic species through using T6SS’s and bacteriocin production. b Pathogens can drive host inflamma-tory responses (yellow stars) in order to trigger the release of nitrates or oxygen into the lumen of the gut (shaded gradient), and the formation of an oxidative environment. Together with novel metabolites formed by oxidation, such as tetrathionate, O2 and NO3 are used in anaerobic and aerobic respiration pathways by pathogens able to use these electron acceptors
Exploitation of nutrients available during inflammation and dysbiosis
Enterobacteriaceae can rapidly expand following initiation of either dysbiosis or inflammation. In 2007, two reports demonstrated that Salmonella and Citrobacter both drive host-driven inflammation in order to expand to high levels and compete with the micro-biota.124,125 Reducing inflammation using avirulent Salmonella or chemically inducing inflammation with DSS is sufficient to prevent or enable expansion in the gut, respectively.124,125 Subsequent work has found that the ability to utilize nutrients uniquely present in these conditions is essential for Enterobacteriaceae (Fig. 3). Importantly, and unlike the anaerobic fermenting bacteria of the microbiota, Enterobactericeae are able to perform both aerobic and anaerobic respiration, utilizing either O2 or other molecules as terminal electron acceptors in a membrane electron transport chain that generates proton motif force to produce ATP.
Two critical components of the inflammatory response to infection are the activation of inducible nitrate oxide synthase, iNOS, encoded by Nos2, and NADPH oxidase encoded in part by gp91phox. iNOS generates nitric oxide through the oxidation of L-arginine and nitric oxide,126 and NADPH oxidase generates reactive oxygen species.127 The production of these reactive oxygen and nitrogen species has long been recognized as two of the primary antibacterial effector mechanisms of the myeloid compartment.128 Importantly, the release of these reactive molecules also creates an oxidative environment, and reactive oxygen and nitrogen species can oxidize molecules present in the gut.
Tetrathionate was the first inflammation-associated terminal electron acceptor identified for Salmonella.129 Although tetrathionate is not present in the healthy gut, the microbiota produces hydrogen sulfide which is converted to thiosulfate by the host.130 iNOS-generated nitric oxide is capable of oxidizing thiosulfate to tetrathionate in the lumen.129 In this environment, Salmonella capable of respiring tetrathionate outcompete respiration-deficient mutants.129 The presence of tetrathionate as an anaerobic electron acceptor also enables Salmonella to utilize ethanolamine as a carbon source both in vivo and in vitro.131,132
Released nitric oxide can also react to form nitrate, and both NO and NO3 increase in the gut lumen during inflammation.133,134 E. coli are able to utilize nitrates, S-oxides, and N-oxides as terminal electron acceptors under anaerobic conditions through the production of molybdenum-containing reductases.135 As a result, E. coli with nitrate reductase activity have a competitive advantage over deficient strains in conditions of intestinal inflammation in DSS-treated or Il10fl/fl CD4-cre mice, as well as in mice with low-level inflammation following streptomycin treatment.54,133,136
The importance of anaerobic electron acceptors for Salmonella replication is highlighted by the observation that Salmonella expresses two chemotactic receptors Aer and Tsr that enable chemotaxis toward nitrate and tetrathionate in vitro and are critical for Salmonella to take advantage of the available electron acceptors in vivo.137,138 Furthermore, treatment with tungsten to inhibit the formation of the molybendum cofactor that is necessary for anaerobic reductases counters the competitive advantage gained by Enterobacteriaceae in the inflamed gut environment.139
The oxidative environment generated by iNOS also leads to the oxidation of carbon sources in the gut and generation of carbon sources that are absent under homeostatic conditions.140 In an in vitro assay, reactive nitrogen oxidizes galactose to galactarate and glucose to glucarate. In vivo, galactarate and glucarate increase in the lumen following streptomycin treatment, and this increase is Nos2 dependent.140 Both E. coli and S. Typhimurium are able to use glucarate and galactarate as carbon and energy sources, and mutants that are deficient in this pathway are outcompeted in the gut following antibiotic treatment.140
As described above, the normal microbiota plays an essential role in maintaining an anaerobic environment in the gut and depletion of Clostridia taxa leads to increased oxygen availability in the lumen of the gut.53,54 Under microaerophilic conditions, Enterobacteriaceae express cytochrome oxidase variants with increased oxygen affinity that enable them to utilize oxygen as a terminal electron acceptor at low oxygen tensions.141 Salmonella and E. coli with the ability to utilize oxygen in these conditions have a competitive advantage compared to deficient strains.53,54 Similarly, Citrobacter actively promotes oxygenation of the lumen by anchoring to enterocytes and promoting crypt hyperplasia using T3SS-delivered effectors.142 This functions to create an inflammatory environment where wild-type strains able to utilize oxygen outcompete deficient strains.142 Together, the presence of both oxygen and other anaerobic electron acceptors promotes Enterobacteriaceae expansion. Indeed, the loss of both pathways triggers a synergistic colonization defect in E. coli,54 and both pathways enable the usage of alternate carbon sources including ethanaloamine or 1,2-propanediol.131,143
Disruption of the microbiota can trigger transient increases in metabolites that are normally present but typically at low levels owing to utilization by the microbiota.144–147 For example, both Salmonella and C. difficile can utilize sialic acid that is cleaved from host mucus by microbiota-encoded sialidase enzymes during their enteric expansion.144,148 Alternatively, C. difficile, C. rodentium, and Salmonella can utilize microbiota-derived succinate during expansion.145,147,149 Work from Maier et al. found that Salmonella is able to utilize microbiota-generated H2 as an electron donor in the anaerobic gut to drive anaerobic respiration.150 In addition, Salmonella can also utilize fucose that is liberated in a microbiota-dependent manner.144 Fucose is a deoxyhexose sugar that is produced by the mammalian host, and, in the adult gut, glycans containing α(1–2)-linked fucose are exposed to the intestinal lumen.151 Members of the microbiota, such as Bacteroides acidifaciens or Bacteroides thetaiotaomicron, can cleave fucose from the conjugated proteins, thus enabling utilization by the microbiome.152,153 Following LPS stimulation of the immune system, fucosylation in the gut is increased in an fut2-dependent manner.152 Unexpectedly, increased availability of fucose increases tolerance of C. rodentium, without decreasing pathogen burden and correlating with reduced virulence gene expression.152 As described above, Citrobacter actively drives inflammatory responses partly in order to support its oxidative metabolism. Therefore, the release of fucose by fucosidase activity of the microbiota could provide substrates for Citrobacter, counteracting the importance of pathogen-driven intestinal inflammation. Overall, pathogens that expand in the gut are well adapted to take advantage of transient increases in microbiota-derived metabolites to fuel their expansion.
CONCLUSIONS
Under homeostatic conditions, the normal constituents of the gut microbiota exist in a constant state of competition for nutrients and niches. The intense competition that results has likely driven evolution of the bacteria present in this environment to both cooperate and compete. In most cases, the mechanisms of colonization resistance against pathogenic species described in this review are also active against commensal members of the microbiota and likely help to shape the equilibrium that exists between these species. Therefore, a newly introduced potential pathogen encounters a pre-existing web of interacting bacteria occupying all available niches and successfully competing with related strains. As a result, in most cases, the microbiota provides effective colonization resistance, and newly introduced pathogens are unable to gain a foothold and expand. If they are able to gain a foothold in the gut, often due to perturbation of the microbiota, pathogens that can expand in that environment use a similar collection of antagonistic pathways as the microbiota in addition to metabolic flexibility to successfully engraft.
In this review, we have focused on interbacterial mechanisms of colonization resistance; however, fungi, protozoa, helminths, and bacteriophage can also be present in the gut. Recent studies have highlighted the fact that non-bacterial gut residents including protists and helminths can influence mucosal immune responses or cell differentiation in the epithelial barrier,154–156 and it has been suggested that bacteriophage may play an important role in shaping the bacterial community.157,158 Therefore, it will be interesting to determine to what extent these non-bacterial gut residents contribute to colonization resistance through either direct or indirect mechanisms.
A potential layer of complexity in understanding colonization resistance is that the distribution of bacteria in the gut is not homogenous,159 and, therefore, the distribution of different effector mechanisms of colonization resistance is also likely to vary. To understand how potential pathogens try to establish themselves within the web of the microbiota, it will also be critical to understand the spatial organization of the gut microbiota and how the pathways of metabolic competition or exclusion and antagonism are distributed across the normal biogeography of the gut. Understanding the arrangement of both pathogens and the recovering microbiota following antibiotic treatment will likely be critical in predicting colonization outcomes. For example, our laboratory has demonstrated that Klebsiella and VRE reside in close proximity to the mucous layer, and that despite being physically close, K. pneuomoniae and VRE occupy different niches in the mouse gut.160 Recent work from Whitaker et al. demonstrated an elegant system for engineering Bacteroides species to express fluorescent proteins driven by promoters with varying activities to allow for the simultaneous imaging of multiple species in the gut.161 These engineered strains can be coupled with high-throughput imaging platforms designed for the microbiota.162 Expansion of these and similar techniques to other phyla will likely play a critical role in understanding colonization resistance in the complex spatial organization of the gut.
The multiple mechanisms of colonization resistance can function simultaneously in the gut. As a result, evaluating the relative importance of each individual pathway is difficult in this environment. While gene knockout strategies have been widely utilized in Gram-negative members of the microbiota, genetic manipulation of the Gram-positive taxa common in the gut remains more difficult. Progress has been made in improving the genetic tractability of Gram-positive members of the micro-biota.163–165 Alternatively, pathways and genes of interest from the microbiota can also be expressed in genetically tractable species.166 Recent work from Lim et al. introduced a system that allows tunable expression of a desired protein in Bacteroides in vivo.167 Overall, these advances in genetic manipulation of the microbiota will enable studies with microbiota compositions either lacking or overexpressing different components of colonization resistance.
ACKNOWLEDGEMENTS
M.T.S. is supported by a Canadian Institute of Health Research (CIHR) Fellowship. E.G. P. has received funding from NIH grants R01 AI042135, AI095706, U01 AI124275, and P30 CA008748.
Footnotes
ADDITIONAL INFORMATION
The online version of this article (https://doi.org/10.1038/s41385-018-0053-0) contains supplementary material, which is available to authorized users.
Competing interests: The authors declare no competing interests
REFERENCES
- 1.Sekirov I et al. Gut microbiota in health and disease. Physiol. Rev 90, 859–904 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Hasegawa M et al. Transitions in oral and intestinal microflora composition and innate immune receptor-dependent stimulation during mouse development. Infect. Immun 78, 639–50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer C et al. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YG et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature 486, 207–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ et al. A core gut microbiome in obese and lean twins. Nature 457, 480–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera-Chavez F & Baumler AJ The pyromaniac inside you: Salmonella metabolism in the host gut. Annu. Rev. Microbiol 69, 31–48 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Abt MC, McKenney PT & Pamer EG Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol 14, 609–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taur Y et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis 55, 905–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ubeda C et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest 120, 4332–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2013. https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf
- 14.Bohnhoff M, Miller CP & Martin WR Resistance of the mouse’s intestinal tract to experimental Salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J. Exp. Med 120, 805–16 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bohnhoff M, Drake BL & Miller CP Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med 86, 132–7 (1954). [DOI] [PubMed] [Google Scholar]
- 16.Rabiu BA & Gibson GR Carbohydrates: a limit on bacterial diversity within the colon. Biol. Rev. Camb. Philos. Soc 77, 443–53 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Friesen ML et al. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58, 245–60 (2004). [PubMed] [Google Scholar]
- 18.Crombach A & Hogeweg P Evolution of resource cycling in ecosystems and individuals. BMC Evol. Biol 9, 122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freter R et al. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect. Immun 39, 676–85 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira FC & Berry D Microbial nutrient niches in the gut. Environ. Microbiol 19, 1366–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stecher B et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6, e1000711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada N et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabich AJ et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun 76, 1143–52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang DE et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl Acad. Sci. USA 101, 7427–32 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maltby R et al. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS ONE 8, e53957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leatham MP et al. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect. Immun 77, 2876–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnenburg ED et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnenburg ED et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smits SA et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deriu E et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 14, 26–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gielda LM & DiRita VJ Zinc competition among the intestinal microbiota. MBio. 3, e00171–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sassone-Corsi M et al. Siderophore-based immunization strategy to inhibit growth of enteric pathogens. Proc. Natl Acad. Sci. USA 113, 13462–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Augustin O & Sanchez de Medina F Intestinal bile acid physiology and pathophysiology. World J. Gastroenterol 14, 5630–40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell DW The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem 72, 137–74 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Urdaneta V & Casadesus J Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. (Lausanne) 4, 163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang DJ et al. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta 1781, 16–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson KH Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol 18, 1017–9 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buffie CG et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koenigsknecht MJ et al. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect. Immun 83, 934–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theriot CM, Bowman AA & Young VB Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 1, 1–16 (2016). 10.1128/mSphere.00045-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis BB et al. Pathogenicity locus, core genome, and accessory gene contributions to Clostridium difficile virulence. MBio. 8, 1–15 (2017). 10.1128/mBio.00885-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BB, Carter RA & Pamer EG Bile acid sensitivity and in vivo virulence of clinical Clostridium difficile isolates. Anaerobe. 41, 32–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flint HJ et al. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc 74, 13–22 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Morrison DJ & Preston T Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 7, 189–200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Topping DL & Clifton PM Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev 81, 1031–64 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Macfarlane S & Macfarlane GT Regulation of short-chain fatty acid production. Proc. Nutr. Soc 62, 67–72 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Ruppin H et al. Absorption of short-chain fatty acids by the colon. Gastroenterology 78, 1500–7 (1980). [PubMed] [Google Scholar]
- 48.Farmer AD et al. Caecal pH is a biomarker of excessive colonic fermentation. World J. Gastroenterol 20, 5000–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cherrington CA et al. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J. Appl. Bacteriol 70, 161–5 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Roe AJ et al. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148(Pt 7), 2215–22 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Roe AJ et al. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol 180, 767–72 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olsan EE et al. Colonization resistance: the deconvolution of a complex trait. J. Biol. Chem 292, 8577–81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera-Chavez F et al. Depletion of Butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 19, 443–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Byndloss MX et al. Microbiota-activated PPAR-gamma signaling inhibits dys-biotic Enterobacteriaceae expansion. Science 357, 570–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alex S et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor gamma. Mol. Cell Biol 33, 1303–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly CJ et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 17, 662–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawhon SD et al. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol 46, 1451–64 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Hung CC et al. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol. Microbiol 87, 1045–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durant JA, Corrier DE & Ricke SC Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella typhimurium. J. Food Prot 63, 573–8 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Gantois I et al. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol 72, 946–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fischbach MA & Walsh CT Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev 106, 3468–96 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Park JW et al. Discovery of parallel pathways of kanamycin biosynthesis allows antibiotic manipulation. Nat. Chem. Biol 7, 843–52 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Janata J et al. Lincosamide synthetase—a unique condensation system combining elements of nonribosomal peptide synthetase and mycothiol metabolism. PLoS ONE 10, e0118850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arnison PG et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asaduzzaman SM & Sonomoto K Lantibiotics: diverse activities and unique modes of action. J. Biosci. Bioeng 107, 475–87 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Breukink E et al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–64 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Corr SC et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl Acad. Sci. USA 104, 7617–21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rea MC et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl Acad. Sci. USA 107, 9352–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brunati C et al. Expanding the potential of NAI-107 for treating serious ESKAPE pathogens: synergistic combinations against Gram-negatives and bactericidal activity against non-dividing cells. J. Antimicrob. Chemother 73, 414–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vukomanovic M et al. Nano-engineering the antimicrobial spectrum of lantibiotics: activity of Nisin against Gram negative bacteria. Sci. Rep 7, 4324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou L et al. Potentiating the Activity of Nisin against Escherichia coli. Front. Cell Dev. Biol 4, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donia MS et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomita H et al. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol 179, 7843–55 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martinez-Bueno M et al. A transferable plasmid associated with AS-48 production in Enterococcus faecalis. J. Bacteriol 172, 2817–18 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomita H et al. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J. Bacteriol 178, 3585–93 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clewell DB Bacterial sex pheromone-induced plasmid transfer. Cell 73, 9–12 (1993). [DOI] [PubMed] [Google Scholar]
- 77.Gilmore MS et al. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc. Natl Acad. Sci. USA 112, 7273–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kommineni S et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vimont A et al. Bacteriocin-producing Enterococcus faecium LCW 44: a high potential probiotic candidate from raw camel milk. Front. Microbiol 8, 865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frank DN et al. The human nasal microbiota and Staphylococcus aureus carriage. PLoS ONE 5, e10598 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bessesen MT et al. MRSA colonization and the nasal microbiome in adults at high risk of colonization and infection. J. Infect 71, 649–57 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Zipperer A et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–6 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Duquesne S et al. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep 24, 708–34 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Patzer SI et al. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149(Pt 9), 2557–70 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Sassone-Corsi M et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roelofs KG et al. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. MBio. 7, 1–10 (2016). 10.1128/mBio.01055-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chatzidaki-Livanis M, Coyne MJ & Comstock LE An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol. Microbiol 94, 1361–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chatzidaki-Livanis M et al. Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. MBio. 8, 1–12 (2017). 10.1128/mBio.01902-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cascales E et al. Colicin biology. Microbiol. Mol. Biol. Rev 71, 158–229 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirkup BC & Riley MA Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428, 412–4 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Silverman JM et al. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol 66, 453–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen L et al. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res 172, 19–25 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Russell AB et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Whitney JC et al. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Russell AB, Peterson SB & Mougous JD Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol 12, 137–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Russell AB et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 16, 227–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coyne MJ et al. Evidence of extensive DNA transfer between bacteroidales species within the human gut. MBio. 5, e01305–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coyne MJ, Roelofs KG & Comstock LE Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genom. 17, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Verster AJ et al. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe. 22, 411–19 e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chatzidaki-Livanis M, Geva-Zatorsky N & Comstock LE Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl Acad. Sci. USA 113, 3627–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wexler AG et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl Acad. Sci. USA 113, 3639–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hecht AL et al. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 17, 1281–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Whitney JC et al. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 6, 1–24 (2017). 10.7554/eLife.26938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang D et al. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang JY et al. Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J. Biol. Chem 293, 1504–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van Nood E et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med 368, 407–15 (2013). [DOI] [PubMed] [Google Scholar]
- 107.Kelly CR et al. Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: a randomized trial. Ann. Intern. Med 165, 609–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caballero S et al. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21, 592–602 e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Becattini S et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J. Exp. Med 214, 1973–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brugiroux S et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol 2, 16215 (2016). [DOI] [PubMed] [Google Scholar]
- 111.Ubeda C et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun 81, 965–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lawley TD et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim S, Covington A & Pamer EG The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev 279, 90–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lam LH & Monack DM Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PLoS Pathog. 10, e1004527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nedialkova LP et al. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS Pathog. 10, e1003844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smajs D et al. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol. 10, 288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Quereda JJ et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl Acad. Sci. USA 113, 5706–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Quereda JJ et al. Listeriolysin S is a streptolysin S-like virulence factor that targets exclusively prokaryotic cells in vivo. MBio 8, 1–15 (2017). 10.1128/mBio.00259-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fu Y, Waldor MK & Mekalanos JJ Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 14, 652–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.LeRoux M et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife 4, 1–25 (2015). 10.7554/eLife.05701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sana TG et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl Acad. Sci. USA 113, E5044–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.LaCourse KD et al. Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat. Microbiol 3, 440–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bachmann V et al. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl. Trop. Dis 9, e0004031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lupp C et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204 (2007). [DOI] [PubMed] [Google Scholar]
- 125.Stecher B et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Palmer RM, Ashton DS & Moncada S Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature 333, 664–6 (1988). [DOI] [PubMed] [Google Scholar]
- 127.Rigby KM & DeLeo FR Neutrophils in innate host defense against Staphylococcus aureus infections. Semin. Immunopathol 34, 237–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fang FC Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest 99, 2818–25 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winter SE et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Furne J et al. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem. Pharmacol 62, 255–9 (2001). [DOI] [PubMed] [Google Scholar]
- 131.Thiennimitr P et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl Acad. Sci. USA 108, 17480–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Price-Carter M et al. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol 183, 2463–75 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Winter SE et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lundberg JO et al. Greatly increased luminal nitric oxide in ulcerative colitis. Lancet 344, 1673–4 (1994). [DOI] [PubMed] [Google Scholar]
- 135.Iobbi-Nivol C & Leimkuhler S Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1827, 1086–1101 (2013). [DOI] [PubMed] [Google Scholar]
- 136.Spees AM et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 4, 1–10 (2013). 10.1128/mBio.00430-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rivera-Chavez F et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 9, e1003267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rivera-Chavez F et al. Energy taxis toward host-derived nitrate supports a Salmonella pathogenicity island 1-independent mechanism of invasion. MBio 7, 1–11 (2016). 10.1128/mBio.00960-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhu W et al. Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Faber F et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534, 697–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Borisov VB et al. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807, 1398–1413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lopez CA et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Faber F et al. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog. 13, e1006129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ng KM et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ferreyra JA et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 16, 770–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pham NT & Lawley TD Pathogens’ exploitation of the intestinal food web. Cell Host Microbe. 16, 703–5 (2014). [DOI] [PubMed] [Google Scholar]
- 147.Curtis MM et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 16, 759–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vimr ER et al. Diversity of microbial sialic acid metabolism. Microbiol Mol. Biol. Rev 68, 132–53 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Spiga L et al. An oxidative central metabolism enables Salmonella to utilize microbiota-derived succinate. Cell Host Microbe. 22, 291–301 e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maier L et al. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe. 14, 641–51 (2013). [DOI] [PubMed] [Google Scholar]
- 151.Becker DJ & Lowe JB Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R (2003). [DOI] [PubMed] [Google Scholar]
- 152.Pickard JM et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hooper LV et al. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl Acad. Sci. USA 96, 9833–8 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chudnovskiy A et al. Host−protozoan interactions protect from mucosal infections through activation of the inflammasome. Cell 167, 444–56 e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Howitt MR et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Escalante NK et al. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J. Exp. Med 213, 2841–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Carding SR, Davis N & Hoyles L Review article: the human intestinal virome in health and disease. Aliment Pharmacol. Ther 46, 800–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lusiak-Szelachowska M et al. Bacteriophages in the gastrointestinal tract and their implications. Gut Pathog. 9, 44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tropini C et al. The gut microbiome: connecting spatial organization to function. Cell Host Microbe. 21, 433–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Caballero S et al. Distinct but spatially overlapping intestinal niches for vancomycin-resistant Enterococcus faecium and Carbapenem-resistant Klebsiella pneumoniae. PLoS Pathog. 11, e1005132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Whitaker WR, Shepherd ES & Sonnenburg JL Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell 169, 538–46 e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Earle KA et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Heap JT et al. The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55 (2010). [DOI] [PubMed] [Google Scholar]
- 164.Heap JT et al. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol Methods 70, 452–64 (2007). [DOI] [PubMed] [Google Scholar]
- 165.Dodd D et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guo CJ et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell 168, 517–26 e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lim B et al. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell 169, 547–58 e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]