Abstract
Mutational processes occur in normal tissues from conception throughout life. Field cancerization describes the preconditioning of an area of epithelium to tumor growth. Pre-invasive lesions may arise in these fields, however only a minority of pre-invasive neoplasia progresses to overt malignancy. Within this review we discuss recent advances in our understanding of genomic instability processes in normal tissue, describe evolutionary dynamics in pre-invasive disease and highlight current evidence describing how increasing genomic instability may drive the transition from pre-invasive to invasive disease. Appreciation of the evolutionary rulebooks that operate in pre-invasive neoplasia may facilitate screening strategies, risk-stratification of pre-invasive lesions and precipitate novel preventative treatments in at-risk patient populations.
Keywords: Intratumor heterogeneity, genomic instability, cancer evolution, pre-invasive neoplasia, field cancerization
Introduction
Multicellular organisms repress individual cellular fitness to maintain ordered tissue homeostasis and consequently the fitness of the organism. The cancer phenotype represents the degradation of processes associated with this multicellular collectivism, whereby an individual cell achieves its own fitness at the expense of its host [1]. It was Peter Nowell in 1976 who first postulated a comprehensive framework describing that this process could be driven by evolutionary rules [2]. With the advent of large-scale sequencing technologies, this hypothesis has been confirmed [3–6]. In this review we summarise current evidence for genomic instability processes operating in normal tissues and discuss the dynamics underlying the pre-invasive stage of neoplasia that fuel the transition to invasive malignancy.
Genomic instability and clonal evolution in histologically normal tissues
Genomic instability refers to the higher rate of genetic aberrations observed in cancer and comprises the whole spectrum of genetic aberrations from point mutations through to chromosomal level aberrations. A recent deep-sequencing study of normal sun-exposed eyelid epidermis investigated whether genomic instability is detectable in normal tissue [7]. Through deep-sequencing of 74 cancer genes in 234 biopsies isolated from the eyelids of four different individuals, 3760 somatic mutations were identified, largely occurring in the ultraviolet mutational signature context. Evidence for ~140 driver gene mutations in a cm2 of epidermis was observed, with clonal expansions occupying up to several mm2 of skin area suggesting the presence of hundreds of evolving clonal populations [7]. Strong positive selection was observed within the skin samples, evidenced by an increased ratio of nonsynomous (protein-altering) mutations versus synonymous (background) mutations in six genes, including NOTCH1 and TP53. The sizes of clones containing mutations in these positively selected driver genes were compared to sizes of clones harboring synonymous mutations in non-driver genes (which would be selectively neutral). Clones containing driver mutations were significantly larger than clones containing neutral mutations for three of the six positively selected driver genes, however this difference in size was unexpectedly small. Martincorena et al. suggested that the observed limited clonal proliferation associated with driver gene events could represent a protection mechanism to guard against cancer development, potentially mediated by density dependent-growth constraints [7]. Simons re-analysed Martincorena et al.’s data and demonstrated that neutral growth dynamics were largely maintained in epidermal clones regardless of driver mutation status. It was noted that the observation of predominant neutral clonal growth created a paradox when coupled with evidence of positive selection of non-synonymous driver mutations [8]. In reply, Martincorena et al. discussed that the observations of neutral clonal growth dynamics and positive selection are not incompatible and could reflect initial clonal expansions falling beyond the detection limit of deep-sequencing. They suggested that initial exponential growth of a clonal population, due to acquisition of a driver mutation, could be followed by reversion to neutral drift due to physical growth constraint. [9]. This issue is yet to be fully resolved and it is likely that further studies will be necessary to ascertain if driver mutations do indeed produce a survival advantage in normal tissues [10].
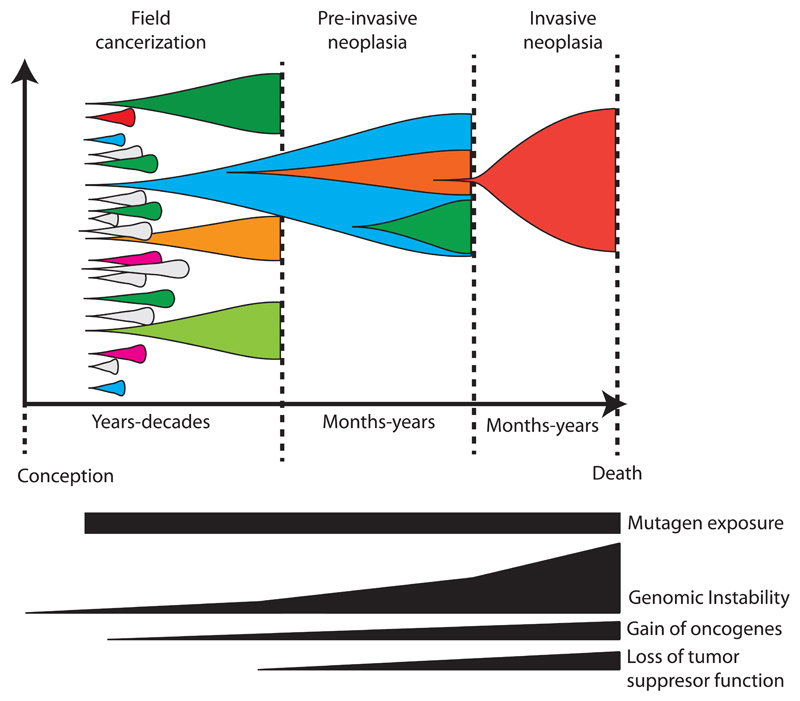
Mutational processes either from exogenous or endogenous origin, can potentially contribute to the development of pre-cancerous clones. These mutational processes are believed to have specific underlying mechanisms and can be characterized by their trinucleotide context, also known as mutational signatures [11]. Currently more than 30 different signatures have been identified [12]. There is strong evidence that several mutational processes are active from conception throughout life in normal tissue [13,14]. Observations of multifocal patches of clonal populations with cancer-related genetic aberrations are not restricted to the skin [7], but have amongst others also been found in lung epithelium [15–18], breast epithelium [19,20] and intestinal epithelium in patients with ileocolitis [21] (Figure 1). Collectively, literature points towards mixed mutational processes active in normal tissues from conception, varying in strength over space and time.
Figure 1. Evolutionary dynamics of ongoing mutational processes contributing to the development of neoplasia.
From conception, mutational processes contribute to the acquisition of passenger and driver mutations. This leads to the expansion of clonal populations within histologically normal-appearing tissue. Nevertheless, due to tumor-suppressive factors, most clonal populations are unable to progress toward genomic instability, occupying a relative “evolutionary cul-de-sac”. Through transcriptional reprogramming and loss of tumour suppressor genes, some clones do progress and acquire the capability to tolerate extensive levels of genomic instability. This increases the evolutionary tempo which eventually enables the acquisition of an invasive and metastatic phenotype. Please note that the timescale is not proportional.
Field cancerization predisposes normal tissue to the formation of neoplasia
The above studies provide evidence of a patchwork of clonal competition in epithelium and support Slaughter et al.’s hypothesis of field cancerization (or field defect) (Figure 1). They defined field cancerization as a patch of epithelium that has been preconditioned by carcinogen exposure, facilitating the process towards cancer formation and enabling the formation of multifocal malignant disease [22]. More recent studies have also implicated epigenetic dysregulation of cancer cells in the process of field cancerization in the colon [23,24] and esophagus [25,26]. Epigenetic changes induced in dermal fibroblasts by exposure to ultraviolet light may also contribute to field cancerization, through dysregulating local release of factors including cytokines and matrix-remodelling enzymes [27]. Collectively, these data led to the refined definition of field cancerization by Graham et al. as “the preconditioning of an area of epithelium to tumor growth, either as the result of a clonal proliferation of mutant cells through the epithelium without causing neoplasia or because of consistent changes to cells in the stromal compartment” [28].
In general, we can distinguish three types of fields, a normal field, carcinogen-exposed field and genetically-predisposed field. A normal field is the starting point in which a tissue has not yet acquired the molecular aberrations that predisposes to cancer development. In contrast, a carcinogen-exposed field has acquired (epi)genetic oncogenic aberrations either through exogenous factors (such as UV-exposure [29] and smoking [18]) or endogenous processes (such as age-related spontaneous deamination [13] or APOBEC-mutagenesis [30] and can underlie the emergence of a sporadic cancer. A patient with a cancer predisposing germline mutation, harbors a genetically predisposed field of epithelium. The germline mutation determines which type of epithelium is affected and is predisposed to field cancerization. For example, patients with familial adenomatous polyposis (FAP) have germline APC mutations, which causes a field defect in the colon even though the mutation is present in every cell. These patients will eventually develop hundreds to thousands of polyps which can eventually progress to a malignant outgrowth.
Evolutionary dynamics in pre-invasive lesions
Pre-invasive lesions such as colonic adenoma and Barrett’s Esophagus (BE) are characterised by abnormal histological features and may demonstrate cytological atypia consistent with dysplasia [31,32]. The majority of pre-invasive lesions do not progress to malignancy. The risk of transformation from colonic adenoma to carcinoma is estimated at only 0.25% a year [33] and less than five percent of patients with BE progress to esophageal adenocarcinoma (EAC) in their lifetimes [34,35]. In pre-invasive lesions that do progress to invasive neoplasia, the pre-invasive stage can last years to decades [36]. Within this section we use understanding gained through study of precursor lesions to gain insight into the evolutionary rules and dynamics that underlie the pre-invasive stage of carcinogenesis and drive the transition to invasive malignancy.
Barrett’s Esophagus
BE represents a precursor to EAC and is characterised by the replacement of normal squamous epithelium with glandular columnar epithelium within the lower portion of the esophagus [37]. Early work identified clonal heterogeneity within BE and suggested that clonal dynamics operating within BE may be subject to evolutionary rules [38]. BE epithelium is diverse, harboring multiple clonal populations and is highly mutated, demonstrating a somatic mutation rate higher than that of several other invasive carcinomas [39,40]. Martinez et al. conducted multi-color FISH on single-cells isolated from endoscopic brushings of non-dysplastic BE segments to map evolutionary dynamics occurring within BE over time. They observed that non-dysplastic BE is largely in a state of dynamic equilibrium with rare clonal contractions offset by clonal expansions leading to relative evolutionary stasis. Interestingly, clones containing CDKN2A-loss were observed to contract with time in this investigation. Since reflux control through acid suppression therapy was an inclusion criterion for this study, the authors hypothesised that CDKN2A-loss clones carry a survival advantage in an acid reflux environment and that following removal of this exogenous selection pressure genetic normalisation ensued [41]. Ross-Innes et al. performed targeted sequencing on 73 BE tissue samples taken from a single patient on multiple occasions over a three-year period. Six distinct clonal groups of neoplastic cells were identified and all six clones remained present within the Barrett’s Segment throughout follow-up. The only alteration in the clonal composition of the BE segment occurred following endoscopic treatment which resulted in the shrinkage of a single clone [40]. This again suggests that neoplastic clones largely exist in a state of equilibrium within BE and highlights the absence of clonal sweeps or fixation events within a three-year follow-up period.
BE is present in nearly all patients with EAC [42] and studies profiling matched BE epithelium with EAC from the same esophagectomy specimen show shared somatic mutations in the majority of cases, however the degree of molecular genetic overlap varies substantially [40,43,44]. Given that evidence for BE and EAC sharing a common ancestor exists, the question arises why do some Barrett’s clones progress to cause invasive cancer, whereas others remain as in situ precursor lesions? In BE it has been demonstrated that ecological measures of clonal diversity strongly predict progression to invasive EAC [45]. Expanding on this concept further studies suggest that genomic instability generates this diversity. For example, patients who do not progress to EAC have relatively stable somatic chromosomal aberrations including localised CDKN2A deletions, 9p loss and copy number neutral loss of heterozygosity. In contrast patients who do progress to EAC develop chromosome instability, genomic diversity and selection of somatic copy number alterations including amplifications and genome doublings events [46,47].
Insight into the genomic events associated with the acquisition of genomic instability and progression to EAC was provided by Stachler et al., who compared BE and EAC specimens identified in the same esophagectomy specimens. In eleven BE cases clonally unrelated to their paired EAC, representing BE populations that did not progress to EAC, only a single TP53 alteration was observed. Four unrelated BE segments contained homozygous CDKN2A deletions, whereas the EACs that developed in these patients lacked these alterations. In contrast, analysis of fourteen BE cases clonally related to their paired EAC, revealed that TP53 mutations were shared between BE and EAC segments in seven cases. In these cases, shared CDKN2A somatic alterations were not observed [43]. Given these findings, perhaps BE clones with early CDKN2A deletion are able to tolerate mutagenesis induced by an acidic environment through avoiding apoptosis. These clonal populations continue to proliferate but do not progress toward genomic instability, occupying a relative “evolutionary cul-de-sac” (Figure 1). On the other hand, clones that acquire early TP53 mutations, permissive for tolerance of DNA damage and ongoing genomic instability, undergo genome doubling and diversification acquiring further tumor suppressor gene inactivation and oncogene amplification events. This increases the evolutionary tempo operating within a BE segment, consequently leading to propensity for invasion [43,46]. In support of this theory, TP53 is recurrently mutated in dysplastic-BE [48], which is associated with occult EAC in up to 40% of cases [49], yet rarely mutated in non-dysplastic BE segments [48], which only progress to EAC in approximately 1 in 300 patients [50]. Furthermore, there is evidence to suggest TP53 mutations facilitate the transition toward genomic instability in EAC carcinogenesis. TP53 mutations were found to be pre-genome doubling events in 90% of 144 EACs analysed by whole exome sequencing [43] suggesting that they are acquired prior to the onset of chromosomal instability. We observed early ubiquitous TP53 disruption in eight multi-region sequenced EAC cases, all cases demonstrated evidence of genome doubling and in two cases chromothripsis was observed [51]. These findings suggest that TP53 loss might permit complex genomic rearrangements in BE, facilitating macro-evolutionary leaps toward EAC; fuelling a cancer dominated by large scale rearrangement events including chromothripsis and kataegsis [52].
Colonic adenoma
Colonic adenomas are thought to represent a precursor lesion to colonic adenocarcinoma on the basis of clinical and pathological evidence [53]. Adenomas are comprised of multiple genomically distinct populations of neoplastic cells [54,55]. DNA methylation analysis has revealed that crypts from a colonic adenoma are largely epigenetically diverse, regardless of spatial separation [56,57]. This suggests relative evolutionary stasis of clonal populations within adenoma crypts; since in a tumor comprising of clones undergoing continual expansions and contractions, spatially separated homogenous methylation patterns would be expected [58]. However, rare clonal expansions occurring within adenomas have been observed, suggesting that colonic adenoma, in a similar fashion to BE, may conform to a “punctuated” model of clonal evolution whereby expansion occurs rarely and stasis is the norm [56].
When adenomas are identified clinically they are removed, hence longitudinal monitoring of the adenoma to carcinoma transition is not feasible in the same fashion as that achieved in BE. However, evidence points toward the involvement of chromosomal instability processes. Colorectal carcinomas have been demonstrated to be more chromosomally instable than adenomas by whole genome single nucleotide polymorphism arrays [59] and separate analyses of genomic imbalances in the adenoma and carcinoma components present in “malignant polyps” (small adenocarcinomas arising in high grade adenomas) revealed considerably more chromosomal losses and gains within the carcinoma region as compared with the adenoma region suggesting increased chromosomal instability during tumor progression [60]. Interestingly gain of 20q has been associated with the adenoma to carcinoma transition in malignant polyps [60,61] and adenomas from patients with FAP with allelic imbalance in 20q have a significantly higher mutational rate than non 20q altered adenomas [55]. We noted 18q as the most frequently lost chromosomal region in aneuploid colorectal tumors [62]. Through analysis of colorectal adenomas and carcinomas in the same surgical specimens, we implicated this event in the adenoma to carcinoma transition. Three chromosomal instability suppressor genes, PIGN, RKHD2 and ZNF516, were identified on 18q. Silencing of these genes induced structural and numeric genomic instability through replication stress and chromosome missegregation events [62]. These findings suggest that punctuated events inducing genomic instability may increase the evolutionary tempo operating in pre-invasive disease, facilitating progression toward invasive carcinoma.
Cell-fate dynamics, transcriptional re-programming and the transition to invasive carcinoma
Further insight into the events that associate with the pre-invasive to invasive transition can be provided by animal models. Oncogenic events acquired during carcinogenesis can influence cell-fate dynamics pre-disposing to an invasive phenotype. High-grade esophageal dysplasia (HGD) represents the precursor lesion to esophageal squamous cell carcinoma (ESC). Frede et al. investigated a murine model of ESC development based on sorafenib- and diethylnitrosamine-initiated carcinogenesis. Genetic lineage tracing was performed, which demonstrated that HGD can arise from multiple cells, suggesting a polyclonal origin of this lesion. The transition of in situ HGD to invasive ESC could be initiated by induction of KRASG12D expression in esophageal epithelium. In HGD, a single proliferating population was observed, with a small propensity for producing daughter cells that continued to divide, whereas in KRASG12D induced ESC, a larger imbalance in cell-fate was observed in a subset of clones strongly biased towards production of dividing over non-dividing progeny. This study suggests that oncogenic events alter cell-fate dynamics influencing tumor growth and that this associates with invasive behaviour [63]. Tumor suppressor gene inactivation can also bias cell-fate toward proliferation, as TP53 mutant cells in the epidermis can produce an excess of proliferative over differentiated progeny in UV-exposed skin [64].
Deregulation of transcriptional networks has also been observed to associate with the development of invasive potential in animal-models. Transgenic expression in Zebrafish melanocytes of BRAFV600E, with concomitant deletion of TP53, leads to melanoma development, however only a small number of melanocytes develop into melanomas in this model [65]. Through use of an in vivo reporter of neural crest-progenitor (NCP) state it was demonstrated that clusters of melanocytes within a BRAFV600E mutant, TP53 deficient "cancer field”, regress to a NCP identity and it is these cells that display tumorigenic properties and progress to form invasive melanoma. SOX10, a transcription factor involved in melanocyte development, was identified as contributing to this reprogramming of melanocytes toward an embryonic NCP state [66]. Further studies support the concept that transcriptional reprogramming may underlie the generation of invasive potential (Figure 1). Interfollicular epidermis stem cells undergo reprogramming to a transcriptional state resembling embryonic hair follicle progenitors before progressing into invasive basal cell carcinoma, Wnt-/β-catenin signalling drives this cellular reprogramming in a SOX9 dependent manner [67,68]. Boumahdi et al. identified SOX2 as the most upregulated transcription factor in cancer stem cells derived from murine squamous cell carcinomas of the skin. In this study it was demonstrated that SOX2 has an essential role in squamous cell carcinoma initiation and that tumor epithelial cells expressing this transcription factor reactivated a gene network reminiscent of the embryonic epidermis [69].
Applying understanding of pre-invasive neoplasia to impact clinical care
Considerable effort is being directed towards the advancement of screening and diagnostic strategies to diagnose neoplasia at an early point in its development, when clinical intervention could be curative. For example, it is now feasible to identify cell-free DNA released from pre-invasive and minimally invasive lung neoplasia in plasma [70]; an approach that could be built upon for the early-detection of patients at risk of developing lung malignancy. As strategies such as these mature across tumor-types, the number of pre-invasive lesions diagnosed will increase. Only a minority of pre-invasive neoplasia transitions to invasive disease [33,35] and a proportion of pre-invasive neoplasia spontaneously regresses without treatment, e.g. in cases of cervical intraepithelial neoplasia [71] and pre-invasive bronchial lesions [72]. The challenge for the clinician will involve monitoring and risk-stratifying of identified pre-invasive lesions, to prevent morbidity associated with overtreatment. A thorough understanding of the evolutionary rules and genetic events governing the pre-invasive to invasive transition promises to inform these strategies by delineating low-risk and high-risk pre-invasive entities. There is already evidence to suggest that molecularly informed approaches to monitoring pre-invasive disease can show clinical utility. The Cytosponge is a non-endoscopic device designed to acquire BE samples, analysis of Cytosponge samples using a multi-biomarker panel including TP53 mutation status can identify patients at low-risk of progression to EAC [73]. In these patients invasive endoscopy could potentially be avoided. Additionally, an understanding of pre-invasive malignancy and the tissue microenvironment associated with malignant progression has the potential to precipitate novel, preventive treatment strategies. These include vaccination strategies and therapeutic modulation of inflammatory pathways within the tissue microenvironment that promote tumorigenesis [74]. Integration of data from experimental models of pre-invasive progression, large-scale sequencing initiatives and non-invasive monitoring of at-risk patients will be required to facilitate personalized approaches for early cancer treatment and prevention. To this end, there are already calls for a “big-data” efforts to improve understanding of pre-invasive disease [75].
Conclusion
Genomic instability processes occur throughout life from conception, this generates diversity and acts as a substrate for selection. Within this review we discuss early mutational processes occurring within normal-appearing tissue that lead to gain of oncogenic events and loss of tumor suppressor gene activity, a process known as field cancerization or field defect. This predisposes tissue fields to the development of pre-invasive histological atypia such as BE. Studies of BE reveal an epithelium consisting of multiple clones that are largely evolutionary static. Some clones may be born to be bad and develop genomic instability resulting in rapid acquisition of oncogene events and tumor suppressor gene loss, facilitating the transition to invasive malignancy. Studies in animal models demonstrate that transcriptional reprogramming and oncogene-induced disruption of cell-fate may also contribute to the transition from pre-invasive to invasive disease. We now enter an exciting stage in cancer medicine, where large-scale genomic analyses are providing a previously unachievable resolution by which the evolutionary dynamics underlying carcinogenesis can be appreciated in detail. Although work still needs to be done to understand the contribution of the stromal compartment to the development of invasive neoplasia, we may reach a point at which the evolutionary rules that dictate the development of cancer can be appreciated. This understanding would lead to more meaningful, rational monitoring of pre-invasive neoplasia and targeted intervention to intercept cancer formation at the earliest stage, significantly reducing cancer-related morbidity.
Highlights.
Discussion of evidence regarding clonal evolution in normal tissue.
An overview of field cancerization.
Evolutionary dynamics operating in the pre-invasive to invasive transition.
Clinical applications of this knowledge.
Acknowledgements
C.S. is Royal Society Napier Research Professor. This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001169), the UK Medical Research Council (FC001169), and the Wellcome Trust (FC001169 ); by the UK Medical Research Council (grant reference MR/FC001169 /1); CS is funded by Cancer Research UK (TRACERx), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rosetrees Trust, NovoNordisk Foundation (ID 16584), the Prostate Cancer Foundation, the Breast Cancer Research Foundation, the European Research Council (THESEUS) and Support was provided to CS by the National Institute for Health Research, the University College London Hospitals Biomedical Research Centre, and the Cancer Research UK University College London Experimental Cancer Medicine Centre.
Abbreviations
- BE
Barrett’s Esophagus
- EAC
esophageal adenocarcinoma
- HGD
High-grade esophageal dysplasia
- ESC
esophageal squamous cell carcinoma
- FAP
Familial Adenomatous Polyposis
References
- 1.Chen H, Lin F, Xing K, He X. The reverse evolution from multicellularity to unicellularity during carcinogenesis. Nat Commun. 2015;6 doi: 10.1038/ncomms7367. [DOI] [PubMed] [Google Scholar]
- 2.Nowell P. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England journal of medicine. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, Massie CE, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47(4):367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, Li Y, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DIS, Zairis S, Abate F, Liu Z, Elliott O, Shin Y-J, Lee J-K, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348(6237):880–886. doi: 10.1126/science.aaa6806. [•• This work provides an in-depth analysis of ongoing clonal competition in histologically normal-appearing eyelid epidermis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simons BD. Deep sequencing as a probe of normal stem cell fate and preneoplasia in human epidermis. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(1):128–133. doi: 10.1073/pnas.1516123113. [•• This work is a re-analysis of [••7] and largely observes neutral competition of clones regardless of their mutational status.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martincorena I, Jones PH, Campbell PJ. Constrained positive selection on cancer mutations in normal skin. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(9):E1128–1129. doi: 10.1073/pnas.1600910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons BD. Reply to martincorena et al.: Evidence for constrained positive selection of cancer mutations in normal skin is lacking. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(9):E1130–1131. doi: 10.1073/pnas.1601045113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petljak M, Alexandrov LB. Understanding mutagenesis through delineation of mutational signatures in human cancer. Carcinogenesis. 2016;37(6):531–540. doi: 10.1093/carcin/bgw055. [DOI] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, Stratton MR. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47(12):1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang ML, Kinde I, Tomasetti C, McMahon KW, Rosenquist TA, Grollman AP, Kinzler KW, Vogelstein B, Papadopoulos N. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(35):9846–9851. doi: 10.1073/pnas.1607794113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazdar AF, Minna JD. Multifocal lung cancers--clonality vs field cancerization and does it matter? Journal of the National Cancer Institute. 2009;101(8):541–543. doi: 10.1093/jnci/djp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung J, Kishimoto Y, Sugio K, Virmani A, McIntire DD, Minna JD, Gazdar AF. Allele-specific chromosome 3p deletions occur at an early stage in the pathogenesis of lung carcinoma. JAMA. 1995;273(7):558–563. [PubMed] [Google Scholar]
- 17.Tang X, Shigematsu H, Bekele BN, Roth JA, Minna JD, Hong WK, Gazdar AF, Wistuba II. Egfr tyrosine kinase domain mutations are detected in histologically normal respiratory epithelium in lung cancer patients. Cancer Res. 2005;65(17):7568–7572. doi: 10.1158/0008-5472.CAN-05-1705. [DOI] [PubMed] [Google Scholar]
- 18.Jakubek Y, Lang W, Vattathil S, Garcia M, Xu L, Huang L, Yoo SY, Shen L, Lu W, Chow CW, Weber Z, et al. Genomic landscape established by allelic imbalance in the cancerization field of a normal appearing airway. Cancer Res. 2016;76(13):3676–3683. doi: 10.1158/0008-5472.CAN-15-3064. [• The authors characterize the copy number landscape of normal-appearing airway of patients with early-stage non-small cell lung cancer with the tumor.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsberg LA, Rasi C, Pekar G, Davies H, Piotrowski A, Absher D, Razzaghian HR, Ambicka A, Halaszka K, Przewoznik M, Kruczak A, et al. Signatures of post-zygotic structural genetic aberrations in the cells of histologically normal breast tissue that can predispose to sporadic breast cancer. Genome Res. 2015;25(10):1521–1535. doi: 10.1101/gr.187823.114. [• The authors characterize the copy number landscape of histologically normal-appearing breast cells in 282 females with sporadic breast cancer.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274(5295):2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 21.Galandiuk S, Rodriguez-Justo M, Jeffery R, Nicholson AM, Cheng Y, Oukrif D, Elia G, Leedham SJ, McDonald SA, Wright NA, Graham TA. Field cancerization in the intestinal epithelium of patients with crohn's ileocolitis. Gastroenterology. 2012;142(4):855–864 e858. doi: 10.1053/j.gastro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6(5):963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 23.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, Buckmeier J, et al. Mgmt promoter methylation and field defect in sporadic colorectal cancer. Journal of the National Cancer Institute. 2005;97(18):1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 24.Alonso S, Dai Y, Yamashita K, Horiuchi S, Dai T, Matsunaga A, Sanchez-Munoz R, Bilbao-Sieyro C, Diaz-Chico JC, Chernov AV, Strongin AY, et al. Methylation of mgmt and adamts14 in normal colon mucosa: Biomarkers of a field defect for cancerization preferentially targeting elder african-americans. Oncotarget. 2015;6(5):3420–3431. doi: 10.18632/oncotarget.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, Laird PW, et al. Fields of aberrant cpg island hypermethylation in barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60(18):5021–5026. [PubMed] [Google Scholar]
- 26.Lee YC, Wang HP, Wang CP, Ko JY, Lee JM, Chiu HM, Lin JT, Yamashita S, Oka D, Watanabe N, Matsuda Y, et al. Revisit of field cancerization in squamous cell carcinoma of upper aerodigestive tract: Better risk assessment with epigenetic markers. Cancer Prev Res (Phila) 2011;4(12):1982–1992. doi: 10.1158/1940-6207.CAPR-11-0096. [DOI] [PubMed] [Google Scholar]
- 27.Hu B, Castillo E, Harewood L, Ostano P, Reymond A, Dummer R, Raffoul W, Hoetzenecker W, Hofbauer GF, Dotto GP. Multifocal epithelial tumors and field cancerization from loss of mesenchymal csl signaling. Cell. 2012;149(6):1207–1220. doi: 10.1016/j.cell.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham TA, McDonald SA, Wright NA. Field cancerization in the gi tract. Future Oncol. 2011;7(8):981–993. doi: 10.2217/fon.11.70. [DOI] [PubMed] [Google Scholar]
- 29.Dotto GP. Multifocal epithelial tumors and field cancerization: Stroma as a primary determinant. J Clin Invest. 2014;124(4):1446–1453. doi: 10.1172/JCI72589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middlebrooks CD, Banday AR, Matsuda K, Udquim KI, Onabajo OO, Paquin A, Figueroa JD, Zhu B, Koutros S, Kubo M, Shuin T, et al. Association of germline variants in the apobec3 region with cancer risk and enrichment with apobec-signature mutations in tumors. Nat Genet. 2016 doi: 10.1038/ng.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fléjou J-F. Barrett’s oesophagus: From metaplasia to dysplasia and cancer. Gut. 2005;54(suppl 1):i6–i12. doi: 10.1136/gut.2004.041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jass JR, Sobin L. Histological typing of intestinal tumours. Springer Science & Business Media; 2012. [Google Scholar]
- 33.Eide TJ. Risk of colorectal cancer in adenoma-bearing individuals within a defined population. Int J Cancer. 1986;38(2):173–176. doi: 10.1002/ijc.2910380205. [DOI] [PubMed] [Google Scholar]
- 34.Qiao Y, Hyder A, Bae SJ, Zarin W, O'Neill TJ, Marcon NE, Stein L, Thein H-H. Surveillance in patients with barrett/'s esophagus for early detection of esophageal adenocarcinoma: A systematic review and meta-analysis. Clin Trans Gastroenterol. 2015;6:e131. doi: 10.1038/ctg.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conio M, Blanchi S, Lapertosa G, Ferraris R, Sablich R, Marchi S, D'Onofrio V, Lacchin T, Iaquinto G, Missale G, Ravelli P, et al. Long-term endoscopic surveillance of patients with barrett's esophagus. Incidence of dysplasia and adenocarcinoma: A prospective study. The American journal of gastroenterology. 2003;98(9):1931–1939. doi: 10.1111/j.1572-0241.2003.07666.x. [DOI] [PubMed] [Google Scholar]
- 36.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, Kinzler KW, et al. Comparative lesion sequencing provides insights into tumor evolution. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(11):4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang KK, Sampliner RE. Updated guidelines 2008 for the diagnosis, surveillance and therapy of barrett's esophagus. Am J Gastroenterol. 2008;103(3):788–797. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 38.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, Rabinovitch PS, Reid BJ. Evolution of neoplastic cell lineages in barrett oesophagus. Nat Genet. 1999;22(1):106–109. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leedham SJ, Preston SL, McDonald SA, Elia G, Bhandari P, Poller D, Harrison R, Novelli MR, Jankowski JA, Wright NA. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human barrett's oesophagus. Gut. 2008;57(8):1041–1048. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O'Donovan M, Malhotra S, di Pietro M, Ivakhno S, He M, Weaver JMJ, et al. Whole-genome sequencing provides new insights into the clonal architecture of barrett's esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47(9):1038–1046. doi: 10.1038/ng.3357. [•• This work provides novel insights into how competing clones within Barrett's esophagus might develop into esophageal adenocarcinoma.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez P, Timmer MR, Lau CT, Calpe S, del Carmen Sancho-Serra M, Straub D, Baker A-M, Meijer SL, Ten Kate FJ, Mallant-Hent RC. Dynamic clonal equilibrium and predetermined cancer risk in barrett’s oesophagus. Nature communications. 2016;7 doi: 10.1038/ncomms12158. 12158. [• The authors perform a longitudinal study of clonal dynamics in barrett's esophagus through multi-color in situ fluorescence hybridization and provide estimates of in vivo rates of clonal expansion and contraction. They provide further evidence that baseline genetic diversity in BE predicts progression] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theisen J, Stein HJ, Dittler HJ, Feith M, Moebius C, Kauer WKH, Werner M, Siewert JR. Preoperative chemotherapy unmasks underlying barrett’s mucosa in patients with adenocarcinoma of the distal esophagus. Surgical Endoscopy. 2002;16(4):671–673. doi: 10.1007/s00464-001-8307-3. [DOI] [PubMed] [Google Scholar]
- 43.Stachler MD, Taylor-Weiner A, Peng S, McKenna A, Agoston AT, Odze RD, Davison JM, Nason KS, Loda M, Leshchiner I, Stewart C, et al. Paired exome analysis of barrett’s esophagus and adenocarcinoma. Nature genetics. 2015;47(9):1047–1055. doi: 10.1038/ng.3343. [•• The authors compare Barrett's esophagus and esophageal adenocarcinoma specimens. They implicate genome doubling and aneuploidy to cancer progression within this disease.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, Wang L-D, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discovery. 2012;2(10):899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merlo LMF, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC. A comprehensive survey of clonal diversity measures in barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer prevention research (Philadelphia, Pa) 2010;3(11):1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Galipeau PC, Paulson TG, Sanchez CA, Arnaudo J, Liu K, Sather CL, Kostadinov RL, Odze RD, Kuhner MK, Maley CC, et al. Temporal and spatial evolution of somatic chromosomal alterations: A case-cohort study of barrett's esophagus. Cancer Prevention Research. 2014;7(1):114–127. doi: 10.1158/1940-6207.CAPR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu J, Ajani J, Hawk E, Ye Y, Lee JH, Bhutani MS, Hofstetter WL, Swisher SG, Wang K, Wu X. Genome-wide catalogue of chromosomal aberrations in barrett’s esophagus and esophageal adenocarcinoma: A high-density snp array analysis. Cancer prevention research (Philadelphia, Pa) 2010;3(9):1176–1186. doi: 10.1158/1940-6207.CAPR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weaver JMJ, Ross-Innes CS, Shannon N, Lynch AG, Forshew T, Barbera M, Murtaza M, Ong C-AJ, Lao-Sirieix P, Dunning MJ, Smith L, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014;46(8):837–843. doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heitmiller RF, Redmond M, Hamilton SR. Barrett's esophagus with high-grade dysplasia. An indication for prophylactic esophagectomy. Annals of Surgery. 1996;224(1):66–71. doi: 10.1097/00000658-199607000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desai TK, Krishnan K, Samala N, Singh J, Cluley J, Perla S, Howden CW. The incidence of oesophageal adenocarcinoma in non-dysplastic barrett's oesophagus: A meta-analysis. Gut. 2012;61(7):970–976. doi: 10.1136/gutjnl-2011-300730. [DOI] [PubMed] [Google Scholar]
- 51.Murugaesu N, Wilson GA, Birkbak NJ, Watkins T, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, Rowan A, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discovery. 2015 doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, MacRae S, Grehan N, O'Donovan M, Miremadi A, Yang T-P, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48(10):1131–1141. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leslie A, Carey FA, Pratt NR, Steele RJC. The colorectal adenoma–carcinoma sequence. British Journal of Surgery. 2002;89(7):845–860. doi: 10.1046/j.1365-2168.2002.02120.x. [DOI] [PubMed] [Google Scholar]
- 54.Thirlwell C, Will OCC, Domingo E, Graham TA, McDonald SAC, Oukrif D, Jeffrey R, Gorman M, Rodriguez–Justo M, Chin–Aleong J, Clark SK, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138(4):1441–1454.e1447. doi: 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 55.Borras E, San Lucas FA, Chang K, Zhou R, Masand G, Fowler J, Mork ME, You YN, Taggart MW, McAllister F, Jones DA, et al. Genomic landscape of colorectal mucosa and adenomas. Cancer Prev Res (Phila) 2016;9(6):417–427. doi: 10.1158/1940-6207.CAPR-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphries A, Cereser B, Gay LJ, Miller DSJ, Das B, Gutteridge A, Elia G, Nye E, Jeffery R, Poulsom R, Novelli MR, et al. Lineage tracing reveals multipotent stem cells maintain human adenomas and the pattern of clonal expansion in tumor evolution. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(27):E2490–E2499. doi: 10.1073/pnas.1220353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang H, Salomon MP, Sottoriva A, Zhao J, Toy M, Press MF, Curtis C, Marjoram P, Siegmund K, Shibata D. Many private mutations originate from the first few divisions of a human colorectal adenoma. The Journal of Pathology. 2015;237(3):355–362. doi: 10.1002/path.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegmund KD, Marjoram P, Woo Y-J, Tavaré S, Shibata D. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proceedings of the National Academy of Sciences. 2009;106(12):4828–4833. doi: 10.1073/pnas.0810276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, Curtis C. A big bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [•• The authors molecularly profile colorectal tumors and find evidence for a neutral model of growth.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirsch D, Camps J, Varma S, Kemmerling R, Stapleton M, Ried T, Gaiser T. A new whole genome amplification method for studying clonal evolution patterns in malignant colorectal polyps. Genes, chromosomes & cancer. 2012;51(5):490–500. doi: 10.1002/gcc.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthäi A, Cuesta MA, Terhaar sive Droste JS, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58(1):79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 62.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, Chew SK, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494(7438):492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frede J, Greulich P, Nagy T, Simons BD, Jones PH. A single dividing cell population with imbalanced fate drives oesophageal tumour growth. Nat Cell Biol. 2016;18(9):967–978. doi: 10.1038/ncb3400. [• This work describes a mouse model of esophageal tumors and suggests that within esophageal tumors there may be a bias towards producing dividing over non-dividing daughter cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klein AM, Brash DE, Jones PH, Simons BD. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by uv b during preneoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(1):270–275. doi: 10.1073/pnas.0909738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, Aster JC, et al. Braf mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Current biology : CB. 2005;15(3):249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 66.Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, Tan JL, Fogley RD, van Rooijen E, Hagedorn EJ, Ciarlo C, et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351(6272) doi: 10.1126/science.aad2197. [•• This work demonstrates that in addition to oncogene and tumor supressor gene alterations transcriptional re-programming, leading to the emergence of progenitor identity, may be a rate limiting step in the development of melanoma.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larsimont JC, Youssef KK, Sanchez-Danes A, Sukumaran V, Defrance M, Delatte B, Liagre M, Baatsen P, Marine JC, Lippens S, Guerin C, et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell stem cell. 2015;17(1):60–73. doi: 10.1016/j.stem.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 68.Youssef KK, Lapouge G, Bouvree K, Rorive S, Brohee S, Appelstein O, Larsimont JC, Sukumaran V, Van de Sande B, Pucci D, Dekoninck S, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nature cell biology. 2012;14(12):1282–1294. doi: 10.1038/ncb2628. [DOI] [PubMed] [Google Scholar]
- 69.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, Brohee S, et al. Sox2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511(7508):246–250. doi: 10.1038/nature13305. [•• The authors demonstrate that SOX2 begins to be expressed in pre-invasive skin lesions and is expressed heterogenously in human squamous cell carcinoma. They demonstrate that SOX2 is an important regulator of of skin tumor initiation cells and cancer stem cells.] [DOI] [PubMed] [Google Scholar]
- 70.Izumchenko E, Chang X, Brait M, Fertig E, Kagohara LT, Bedi A, Marchionni L, Agrawal N, Ravi R, Jones S, Hoque MO, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nature communications. 2015;6 doi: 10.1038/ncomms9258. 8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trimble CL, Piantadosi S, Gravitt P, Ronnett B, Pizer E, Elko A, Wilgus B, Yutzy W, Daniel R, Shah K, Peng S, et al. Spontaneous regression of high-grade cervical dysplasia: Effects of human papillomavirus type and hla phenotype. Clinical Cancer Research. 2005;11(13):4717–4723. doi: 10.1158/1078-0432.CCR-04-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bota S, Auliac J-B, Paris C, Métayer J, Sesboue R, Nouvet G, Thiberville L. Follow-up of bronchial precancerous lesions and carcinoma in situ using fluorescence endoscopy. American journal of respiratory and critical care medicine. 2001;164(9):1688–1693. doi: 10.1164/ajrccm.164.9.2012147. [DOI] [PubMed] [Google Scholar]
- 73.Ross-Innes CS, Chettouh H, Achilleos A, Galeano-Dalmau N, Debiram-Beecham I, MacRae S, Fessas P, Walker E, Varghese S, Evan T, Lao-Sirieix PS, et al. Risk stratification of barrett's oesophagus using a non-endoscopic sampling method coupled with a biomarker panel: A cohort study. The Lancet Gastroenterology & Hepatology. 2017;2(1):23–31. doi: 10.1016/S2468-1253(16)30118-2. [DOI] [PubMed] [Google Scholar]
- 74.Spira A, Disis ML, Schiller JT, Vilar E, Rebbeck TR, Bejar R, Ideker T, Arts J, Yurgelun MB, Mesirov JP, Rao A, et al. Leveraging premalignant biology for immune-based cancer prevention. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(39):10750–10758. doi: 10.1073/pnas.1608077113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campbell JD, Mazzilli SA, Reid ME, Dhillon SS, Platero S, Beane J, Spira AE. The case for a pre-cancer genome atlas (pcga) Cancer Prev Res (Phila) 2016;9(2):119–124. doi: 10.1158/1940-6207.CAPR-16-0024. [DOI] [PubMed] [Google Scholar]