Abstract
Large-scale genome sequencing studies have identified a wealth of mutations in human tumors and have dramatically advanced the field of cancer genetics. However, the functional consequences of an altered gene in tumor progression cannot always be inferred from mutation status alone. This underscores the critical need for complementary methods to assign functional significance to mutated genes in cancer. Transposons are mobile genetic elements that serve as powerful tools for insertional mutagenesis. Over the last decade, investigators have employed mouse models with ondemand transposon-mediated mutagenesis to perform unbiased genetic screens to identify clinically relevant genes that participate in the pathogenesis of human cancer. Two distinct DNA transposon mutagenesis systems, Sleeping Beauty (SB) and PiggyBac (PB), have been applied extensively in vivo and more recently, in ex vivo settings. These studies have informed our understanding of the genes and pathways that drive cancer initiation, progression, and metastasis. This review highlights the latest progress on cancer gene identification for specific cancer subtypes, as well as new technological advances and incorporation of the CRISPR/Cas9 toolbox into transposon-mediated functional genetic studies.
Introduction
Insertional mutagenesis screens in worms, flies, and mice have yielded fundamental discoveries in biology and led to the identification of critical signaling pathway components [1,2]. The molecular reconstruction of a Tcl/mariner DNA transposon from fish revolutionized the field and generated new opportunities for in vivo genome engineering. This re-awakened element, called Sleeping Beauty (SB), was the first synthetic transposon to be mobilized in mammalian cells [3●●,4]. Successful applications include germline and somatic mutagenesis in mice [5–9]. PiggyBac (PB), another DNA transposon originating from the cabbage looper moth, was subsequently shown to be active in mammalian cells and developed for in vivo somatic mutagenesis [10,11].
The SB and PB systems utilize two components, an engineered transposon that harbors a mutagenic gene trap element, and an enzyme termed the transposase. When both components are present in the same cell, the transposase binds the inverted repeat/direct repeat sequences and catalyzes mobilization of the transposon. For in vivo studies, independent transgenic mouse lines are bred to induce transposon mobilization. Insertion of the transposon harboring the gene trap into a new genomic location disrupts gene function by introducing gain-of-function or loss-of-function mutations depending on the orientation and location of the element (Figure 1). Bioinformatic analysis identifies common insertion sites (CISs) representing genomic windows with more transposon insertions than predicted by chance [12–15], revealing genes that accelerate tumorigenesis when their functions are altered.
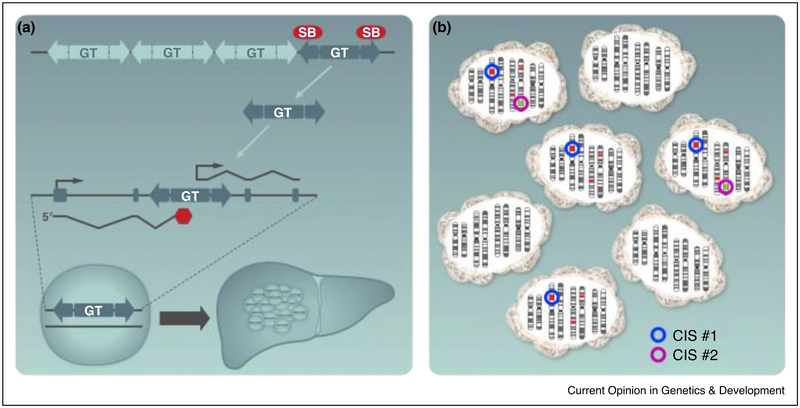
Figure 1. Transposon mobilization disrupts gene function by introducing gain-of-function or loss-of-function mutations.
(a) The SB and PB systems utilize two transgenic mouse lines, one harboring a concatemerized mutagenic gene trap (GT) that can disrupt gene function, and a second line carrying a ubiquitous or tissue-specific transposase that binds the transposon ends and catalyzes mobilization to new genomic sites. SB mutagenesis is depicted in liver cells, leading to the formation of liver tumors in vivo. The gene trap can alter gene function in two ways. In one or both orientations, a splice acceptor is followed by a polyadenylation (pA) signal. When the transposon inserts into a gene, the gene trap may splice to the transcript and the pA signal will prematurely truncate the mRNA, thereby disrupting expression of a candidate tumor suppressor. Additionally, a strong promoter followed by a splice donor (SD) is usually present in only one orientation. Transposon insertions that utilize this promoter/SD may drive overexpression of candidate oncogenes. (b) Bioinformatic analysis identifies common insertion sites (CISs) that represent genomic windows with more transposon insertions than predicted by chance. CISs 1 and 2 (represented as blue and purple circles) are found in independent tumors. Different methods of CIS identification are used to uncover genes that accelerate tumorigenesis, including Monte Carlo-based and Poisson distribution methods, Gaussian Kernel Convolution methods, and gene-centric common insertion site (gCIS) analysis.
Remarkably, tumors that arise from transposon mutagenesis in mice accurately model the anatomical and histological features of human cancers. Early whole-body mutagenesis screens using a ubiquitous SB transposase knocked into the Rosa26 locus identified genes that promote hematopoietic cancers and sarcomas [5,7]. This was followed by screens in the liver and gastrointestinal system using tissue-specific Cre mouse lines that direct expression of the SB transposase to individual tissues [16,17]. In recent years, screens have been performed for many other tumor types including melanoma, neurofibroma, medulloblastoma, breast, prostate, and thyroid cancers (Table 1). Transposon mobilization has been shown to promote tumorigenesis alone or in cooperation with sensitized genetic backgrounds.
Table 1.
Transposon mutagenesis screens published after 2014
| Tissue/Tumor type | Transposon system | Sensitized background/System | Reference |
|---|---|---|---|
| Invasive lobular breast carcinoma | Rosa26Lox66SBLox71;T2/Onc | Cdh1 mutant | [20] |
| Triple negative breast cancer | Rosa26-LSL SBase; T2/Onc2 and T2/Onc3 | Pten mutant | [23] |
| Triple negative breast cancer | Rosa26-LSL SBase; T2/Onc2 | Truncated beta-catenin | [24] |
| Breast cancer | Rosa26-LSL SBase; T2/Onc2 | Trp53+/− | [25] |
| Chronic liver injury | Rosa26-LSL SBase; T2/Onc3 | CCl4 treatment | [32] |
| Steatosis-associate hepatic tumors | Rosa26-LSL SBase; T2/Onc | High fat diet | [33] |
| Pancreatic cancer | Rosa26LSL-PB; ATP1-S2 | KrasLSL-G12D | [39] |
| Metastatic prostate cancer | Rosa26Lox66SBLox71; T2/Onc3 | Pten mutant | [48] |
| Osteosarcoma | Rosa26-LSL SBase; T2/Onc | LSL-Trp53R270H | [49] |
| Medulloblastoma recurrence | Math1-SB11; T2/Onc and T2/Onc2 | Ptch+/− | [53] |
| Lung cancer | Rosa26-LSL SBase; T2/Onc and T2/Onc2 | LSL-Trp53R270H; p19ARF−/−; Pten mutant | [66] |
| B-cell acute lymphoblastic leukemia | Rosa26-LSL SBase; T2/Onc | Stat5b-CA | [67] |
| Melanoma | Rosa26-LSL SBase; T2/Onc2 and T2/Onc3 | BrafV600E | [68] |
| Thyroid cancer | Rosa26-LSL SBase; T2/Onc2 | HrasG12V | [69] |
| Multiple intestinal neoplasia | Rosa26-LSL SBase; T2/Onc2 | Tgfbr2fl/fl | [70] |
| Gastric cancer | Rosa26-LSL SBase; T2/Onc3 | Smad4KO/+ | [71] |
| Gastrointestinal tract | Rosa26Lox66SBLox71; T2/Onc2 | Apcmin; KrasG12D; Smad4KO; Tp53R172H | [72] |
| Neurofibroma | Rosa26-LSL SBase; T2/Onc | Nf1fl/fl | [73] |
| New technologies | |||
| Quantitative insertion site sequencing (QiSeq) | Rosa26PB; Rosa26LSL-PB; ATP1–3 | KrasLSL-G12D | [38] |
| Single-cell transposon insertion sequencing (SBCapSeq) | Rosa26-LSL SBase; T2/Onc2 | Tp53+/−; Tp53LSL−R172H/+ | [45] |
| RNA sequencing of SB-induced tumors | Rosa26-LSL SBase; T2/Onc | LSL-Trp53R270H | [46] |
| Single-copy SB mutagenesis | Rosa26-LSL SBase; Inactivating transposon (ITP2m) | PtenSBm2/+; Blmm3/m3 | [47] |
| Ex vivo screens | |||
| Epithelial-mesenchymal transition in HCC | Rosa26-LSL SBase; T2/Onc2 | Pten mutant | [56] |
| Growth factor independence and B-cell leukemogenesis | SB11; SB100; T2/Onc | Murine Ba/F3 cells | [57] |
| Colorectal cancer (recellularized human colon model) | SB100; modified T2/Onc | Human colonic epithelial cells (hCEC-APCshRNA) | [58] |
| Drug resistance | |||
| Fludarabine resistance in chronic lymphocytic leukemia (CLL) | PB transposon; HyPBase transposase | Human CLL cells | [50] |
| Braf inhibitor resistance in melanoma | Rosa26-LSL SBase; T2/Onc | BrafV618E | [51] |
| Transposon screens incorporating CRISPR/Cas9 | |||
| CRISPR/Cas9 somatic mutliplex mutagenesis | CRISPR-SB (sgRNA and Cas9); hSB5 | KrasLSL-G12D | [61] |
| PiggyBac in vivo CRISPR library screening | PB-CRISPRM1; PB-CRISPR-M2 | CD-1 | [62] |
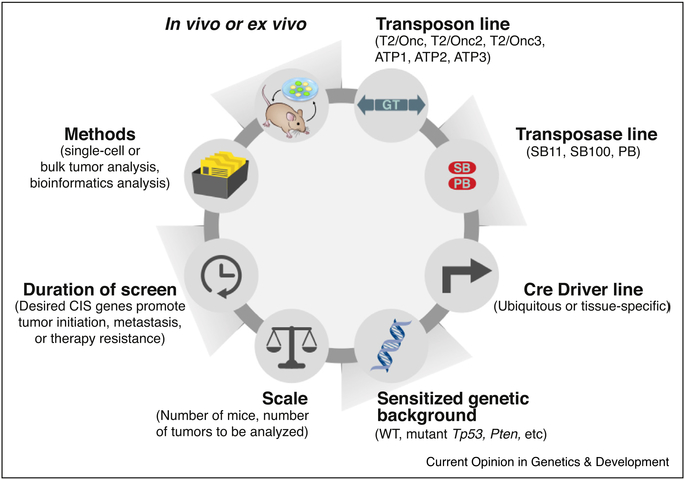
Several important factors must be considered for any transposon screen, including the choice of the transposon gene trap and transposase, the selection of the Cre driver line used to induce tissue-specific expression of the transposase, the scale and duration of the genetic screen, and the specific methods used for the identification and analysis of transposon insertions (Figure 2). A number of excellent studies have examined important statistical considerations for transposon mutagenesis screens and will not be discussed in detail here [12,18,19].
Figure 2. Important considerations for performing transposon screens.
Critical considerations for performing transposon screens include: 1) the selection of the transposon gene trap vector (SB lines = T2/Onc, T2/Onc2, T2/Onc3; PB lines = ATP1, ATP2, ATP3 that differ based on the transposon copy number within the concatemer and site of integration on different chromosomes), 2) the transposase line, 3) the selection of the Cre driver line used to induce expression of the transposase, 4) whether transposon mutagenesis occurs alone or in combination with a sensitized genetic background, 5) the scale of the screen (depending on the number of mice and tumors that are desired for analysis), 6) the duration of the screen (depending on what types of CIS genes are desired at early or late stages of tumorigenesis), 7) the specific methods used for the identification and analysis of transposon insertions, and 8) whether in vivo or ex vivo mutagenesis is desired.
Breast cancer
Significant effort has focused on the identification of genes that contribute to breast cancer pathogenesis. Several key advantages of insertional mutagenesis over other functional approaches can be highlighted using recent Sleeping Beauty studies as examples. Invasive lobular carcinoma (ILC), the second most common breast cancer subtype, is characterized by loss of E-cadherin (CDH1). Cdh1 null mice do not develop mammary tumors, suggesting that additional mutations are required for ILC development. One SB screen performed in mammary-specific Cdh1 deficient mice uncovered recurrent and mutually exclusive insertions in genes implicated in actin cytoskeleton regulation in ILC [20●●]. This revealed driver mutations that were not readily identified in human datasets. For example, heterozygous inactivating insertions were identified in Nonmuscle myosin Ha heavy chain 9 (Myh9), resulting in dosage reduction of MYH9. Yet, driver mutations in MYH9 were not readily detected in human ILC because the gene is rarely mutated or exhibits shallow deletions, which are putative heterozygous deletions. Analysis of human ILCs revealed that MYH9 was commonly altered by heterozygous copy number loss, which correlated with reduced expression of MYH9 mRNA. Experimental validation confirmed that Myh9 haploinsufficiency induced ILC formation in vivo. Consistent with this, it has been proposed that haploinsufficiency of candidate genes in commonly deleted regions may have detectable tumor suppressing activity only in the context of cooperating genetic events [21,22]. Given the heterogeneity of transposon-induced tumors which mimics the complexity of human cancers, similar screens may uncover additional context-dependent alterations in other tumor types.
Another advantage provided by a recent SB transposon mutagenesis study was the ability to pinpoint two candidate cancer genes (Transformation related protein 53 binding protein 2 (Trp53bp2) and Protein Phosphatase 1 Regulatory Subunit 12B (Ppp1r12b), encoding Protein Phosphatase PP1-targeting subunits) with orthologues that are present in a large region on human chromosome 1q, which is known to be frequently amplified in human breast cancer [20●●]. The large size of this amplicon made it particularly difficult to identify critical driver gene(s). Subsequent validation studies showed that SB insertions in these two genes caused truncation of the PP1-targeting subunits, and that expression of the truncated subunits induced tumor formation in genetically engineered mice. This demonstrates the power of integrating CIS gene lists with available copy number data to prioritize driver genes that are present in large windows of amplifications or deletions in human cancer.
SB screens on different mutant backgrounds identified additional breast cancer susceptibility genes. Classification of SB-induced tumors in Pten mutant mice identified a collection of different breast cancer subtypes, including basal-like (triple negative), luminal A, and HER2 positive tumors [23]. Functional validation studies identified eight tumor suppressor genes, including Transcriptional Repressor GATA Binding 1 (Trps 1) as a metastasis tumor suppressor in triple negative breast cancer (TNBC). Interestingly, multiple independent breast cancer (BC) screens used the K5-Cre transgenic mouse line to drive expression of the SB transposase [23–25]. The K5 Cre drives expression in all mammary epithelium cell populations, including basal cells and luminal cells, that likely contributed to the development of different breast cancer subtypes in these studies.
Chen and colleagues performed SB mutagenesis in breast epithelial cells alone or in combination with stabilized N-terminally truncated β-catenin [24]. Integration of this screening approach with survival prediction analysis led to the identification of six gene pairs with prognostic value that could stratify breast cancer subtypes. This demonstrates the utility of incorporating functional mutagenesis screens with expression and survival data to identify novel subtyping biomarkers.
Liver cancer
One of the most frequently modeled tumors using the SB system is hepatocellular carcinoma (HCC). This is likely due to the availability of excellent mouse models, prevalence of the disease, and accessibility of tumor tissues for genomic and histopathologic analysis. The first liver-specific screen for HCC, performed on a mutant Tp53 background, identified known driver genes (Epidermal growth factor receptor, Egfr, and Tyrosine-protein kinase Met) and new potential therapeutic targets including Ubiquitin conjugating enzyme E2H (Ube2h) [16]. Retrotransposon-like 1 (Rtl1), initially identified in a T2/Onc3-induced screen [26], was subsequently validated as a novel gene that promotes HCC development [27]. Another study identified genes that cooperate with MYC to accelerate liver tumorigenesis, revealing a tumor suppressor role for Steroid Receptor Coactivator 2/Nuclear Receptor Coactivator 2 (Src-2/Ncoa2) [28]. Interestingly, SRC-2 promotes survival and metastasis in other tumor types, suggesting a tissue-specific and context-dependent role for SRC-2 in tumorigenesis [29]. Suresh and colleagues recently illuminated the mechanisms of tumor suppression by SRC-2 in liver and provided evidence that SRC-2 may exhibit oncogenic or tumor suppressor activity depending on the target genes and nuclear receptors that are expressed in distinct tissues [30].
Chronic infection with hepatitis B virus (HBV) is the most common risk factor for developing HCC. To identify genes that cooperate with HBV-induced liver inflammation in HCC development, Jenkins and colleagues performed a SB mutagenesis screen using transgenic mice expressing the HBV surface antigen (HBsAg) [31]. This near-saturating screen identified early-stage and late-stage drivers of tumorigenesis, including many CIS genes involved in cellular metabolic processes. Recently, two studies have been performed in the context of chronic liver damage and hepatic steatosis, recapitulating additional settings in which HCC frequently develops [32,33]. Interestingly, chronic liver injury enhanced tumor penetrance and significantly altered SB insertion profiles, reflecting distinct selective pressures exhibited by this tumor type.
PiggyBac mutagenesis
PiggyBac serves as a complementary mutagenesis system to SB and has several important distinctions. First, SB integrates into ‘TA’ dinucleotides whereas PB requires ‘TTAA’ motifs that occur less frequently in mammalian genomes. Second, SB has no preference for insertion into genes [34], while PB more frequently integrates into active transcription units [35–37]. Third, PB transposase activity is more efficient in mammalian genomes and has the capability to mobilize larger payloads. Fourth, unlike SB, PB mobilizes without creating footprint mutations. This means that an excision event restores the DNA sequence that existed before the insertion occurred. While this may cause less genomic damage during mutagenesis, it may also preclude the identification of some PB insertions. Finally, SB exhibits more local hopping, whereby the transposon favors mobilization to sites in proximity to the donor concatemer in transgenic mice. This allows for a higher mutational coverage across the genome and may be exploited for targeted regional mutagenesis. However, Rad and colleagues have suggested that PB may be superior for regional mutagenesis because it produces fewer nonspecific insertions around the donor locus [38●●].
Although fewer PB screens have been performed to date, a versatile PB mutagenesis platform was generated by developing a series of transgenic mouse lines that carry different transposon constructs. Most transposon lines carry PB and SB inverted terminal repeats, allowing mobilization with either transposase [11,38●●]. Transgenic lines with three distinct promoter/enhancer elements were generated, each with the transposon concatemer present in a broad range of copies and on different chromosomes. Interestingly, the selection of the promoter used in the transposon line skews the tumor spectrum in whole body screens, with the murine stem cell virus (MSCV) 5’LTR inducing primarily hematopoietic cancers, the CMV early enhancer/chicken beta actin (CAG) promoter producing more solid tumors, and the phosphoglycerate kinase (PGK) promoter generating a mix of both tumor types. This phenomenon has also been observed in SB screens. Early SB screens using the T2/Onc or T2/Onc2 mouse strains, which utilized the MSCV promoter, exhibited a tendency to develop hematopoietic tumors. Replacing the MSCV promoter with the CAG or PGK promoters increased the incidence of solid tumors [26]. This modularity provides investigators great flexibility by offering the ability to alter the tumor type and incidence when performing whole body or tissue-specific SB and PB screens.
A conditional PB mouse model identified novel oncogenic networks including FOXP1 as an oncogenic transcription factor in pancreatic cancer [39●●]. This screen also helped elucidate the genetic basis of different histologic subtypes of pancreatic cancer. Insertions in Fidgetin (Fign), which encodes a member of the AAA-ATPase superfamily, were significantly enriched in hepatoid tumors, a rare pancreatic cancer subtype. These results implicate altered regulation of Fign in the development of hepatoid pancreatic cancer, however direct evidence for this awaits validation. Interestingly, hepatoid cancers have not been described in pancreas-specific SB screens [40,41], and the identified CISs from these SB screens only partially overlapped with candidate cancer genes found in the conditional PB pancreatic cancer screen. This is likely due to different integration preferences, highlighting the complementarity of these two systems.
In addition to SB and PB, other transposable elements have been mobilized in mouse cells including Tol2, Minos, and a codon-optimized mouse LINE-1 retrotransposon, ORFeus [6,42]. Application of these systems in future studies may further expand the repertoire of available insertional mutagenesis platforms for cancer gene discovery.
In contrast to transposon mutagenesis, chemical mutagenesis is another complementary method with unique features, most notably the capability to induce point mutations that are typically not modeled by insertional mutagenesis approaches [43]. Although next-generation sequencing allows genome-wide detection of chemically-induced mutations, existing limitations include prohibitive costs and high background mutation rates.
Technological advances and ex vivo screens
Since the advent of transposon mutagenesis, numerous technological advances in the recovery of insertions and identification of fusion transcripts have been made. Most transposon screens utilize ligation-mediated PCR (LM-PCR) to recover transposon insertions from tumor DNA. However, the use of restriction enzymes in the LM-PCR reaction generates bias due to uneven distribution of restriction sites across the genome. Shearing of genomic DNA before LM-PCR increases the insertion recovery and allows ‘truncal’ insertions that occur early in tumor development to be distinguished from ‘branch’ insertions that occur later in tumor development [44]. Recently, a semi-quantitative transposon insertion site sequencing method using acoustic DNA shearing (QiSeq) has been described [38●●]. Further refinement of sequencing methods has uncovered transposon insertions in single tumor cells (SBCapSeq) [45●]. This powerful method detected clonal insertion events in a myeloid leukemia mouse model and identified cooperating events in individual tumor cells. RNA sequencing of SB-induced tumors has been particularly useful for identifying fusion transcripts between the transposon elements and endogenous transcripts in order to determine whether a specific SB insertion generates a gain-of-function or loss-of-function mutation [46].
Another clever screening approach combined targeted gene inactivation to single-copy transposon mobilization to identify novel genes that cooperate with Pten in suppressing prostate, breast, and skin tumorigenesis [47●●]. There are several unique benefits to using this model. First, a single-copy SB transposon limited the number of insertions to one per cell, which may reduce the number of passenger insertions. Second, transposition occurred simultaneously in the same cells that underwent Pten inactivation, potentially enhancing the sensitivity of the screen. Finally, unlike most other SB screens, the gene trap promoted only inactivating mutations but not activating mutations. Although this precluded the identification of putative oncogenes, this greatly simplified the interpretation of the roles of uncharacterized CIS genes as putative tumor suppressors. Additional studies utilizing this approach are warranted to gain a better understanding of how altering transposon gene-trap choice and copy number impacts the presence of passenger mutations and degree of tumor heterogeneity.
SB and PB screens have been employed to identify genes that drive metastasis [39●●,48,49] and therapy resistance [50–52]. Recently, Morrissy and colleagues developed a SB-driven mouse model of metastatic medulloblastoma that recapitulated post-treatment tumor recurrence. The investigators performed a mutagenesis screen in mice to identify genes that drive primary medulloblastoma, then resected tumors and employed image-guided radiotherapy, which is the standard therapy for children with this disease [53●]. Sequencing of SB-induced tumors revealed a poor degree of overlap between the SB insertions in primary mouse tumors and the insertions in tumors that recurred after treatment. Consistent with this, whole genome sequencing of human tumors at the time of diagnosis and at recurrence revealed significant genetic divergence. Striking results from the mouse and human data showed that the dominant clone at recurrence arose in part through clonal selection of a minor clone that was present at the time of diagnosis. This suggests that surgery and radiation generate evolutionary pressure, thereby allowing divergent clones to become resistant to therapies. The authors proposed that therapeutic strategies targeting truncal mutations in the primary tumor are destined to fail if the mutation is lacking after recurrence. They also advocated that sequencing of biopsies at recurrence should guide future clinical trials and treatment decisions. In the future, transposon-based mutagenesis systems may be further refined to screen for better drug targets at recurrence for different tumor types.
Although in vivo transposon screens have proven to be effective for cancer gene discovery, they are also time and resource intensive. This prompted several groups to develop ex vivo based systems wherein human or mouse cells growing in culture are mutagenized and screened for the acquisition of specific phenotypes in vitro or in vivo. This approach relies upon stable or transient expression of the transposase and is easily modified for conditional or dose-dependent expression of the transposon or transposase in different settings. This also avoids local hopping, inherent to SB and PB screens. Mutagenesis of human bone explant mesenchymal cells utilizing a hybrid lentiviral and SB mutagenesis system generated myxofibrosarcomas when transplanted into mice [54]. Other cell-based screens have identified genes involved in transformation of neural stem cells into glioma-initiating cells [55] and genes driving epithelial-mesenchymal transition in immortalized mouse hepatoblasts [56]. One of the largest ex vivo SB screens performed to date identified genes that promote growth factor independence and transformation of Ba/F3 cells, an IL-3-dependent murine pro-B cell line. Recurrent insertions were identified in JAK/STAT and MAPK pathway genes in addition to a large number of genes that are mutated or associate with survival of leukemia patients but had not previously been linked to these pathways [57●]. Enforced expression of individual CIS genes promoted growth factor independence and tumori-genesis in vivo, validating this approach. Finally, an ex vivo recellularized human colon model identified genes that drive colorectal cancer progression [58●]. This advance contributed a unique tissue-engineering method and enabled the implementation of forward genetic screening in human tissues under physiologic conditions.
Incorporation of the CRISPR/Cas9 mutagenesis toolbox into transposon-mediated screens
The clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) mutagenesis system has emerged as a revolutionary tool for interrogating phenotypes and pathways in unbiased high-throughput screens [59,60]. Several groups have incorporated the CRISPR toolbox into transposon studies by using PB and SB to deliver individual single guide RNAs (sgRNAs) to mice. For example, Weber and colleagues generated a CRISPR-SB vector with sgRNA and Cas9 cassettes flanked by SB inverted repeats, generating a system for multiplexed mutagenesis of large gene sets in adult mice [61]. When the CRISPR-SB and SB transposase vectors were delivered via tail vein injection, animals developed HCC and intrahepatic cholangiocarcinoma (ICC). PB was also identified as an efficient delivery vehicle for in vivo CRISPR library screening in mice [62].
One major difference between CRISPR screening and transposon screening is that Cas9 derivatives are engineered for the identification of gain-of-function (GOF) or loss-of-function (LOF) mutations, but not at the same time. In contrast, transposon screening allows for simultaneous identification of GOF and LOF mutations in the same mutagenized tumor or clone, which may more accurately reflect the complexity of human tumors. Furthermore, transposon-mediated delivery provides a non-viral alternative for efficient delivery of CRISPR libraries in mice and for cell-based screens.
Concluding remarks
One understudied advantage provided by transposon mutagenesis screens over cDNA overexpression or RNA interference screens is that important non-coding and regulatory regions of the genome may be detected in an unbiased manner. To date, the vast majority of transposon screens have focused on validating protein-coding genes. Two notable exceptions include the identification of competing endogenous RNAs (ceRNAs) that suppress Pten in a BrafV600E-induced mouse model of melanoma [63] and the identification of a Cdkn2a cis-regulatory region in a recent conditional PB pancreatic cancer screen [39●●]. In future studies, transposon screens should be harnessed to identify and rigorously validate non-coding RNAs and critical regulatory regions that drive cancer progression. However, this will depend on the extent to which mouse and human non-coding RNAs exhibit significant conservation at the primary sequence level.
In summary, transposon mutagenesis studies have provided important insights into the functional consequences of mutated genes in human tumors. As new technologies continue to emerge, these studies will undoubtedly improve our understanding of clonal evolution in primary tumors and metastases, the genetic basis of histologic subtypes for different tumors, and mechanisms underlying drug resistance. However, several key questions remain. For example, can we continue to improve and refine the SB and PB systems? One current limitation of transposon mutagenesis is that it does not faithfully reproduce the full spectrum of mutations seen in human cancer, namely point mutations and reciprocal translocations. In the future, innovative modifications to these systems may be developed for this purpose. Also, what is the best way to integrate the CIS gene identification with human genomic data? One possibility is to link transposon insertion data and associated databases [64,65] with existing Cancer Genome Atlas datasets (TCGA) and oncogenomic databases including cBioPortal, the Catalog of Somatic Mutations in Cancer (COSMIC), Oncomine, and the International Cancer Genome Consortium (ICGC). Direct integration of available functional data with human clinical datasets will accelerate the development of novel diagnostic and therapeutic strategies for human malignancies.
Acknowledgements
I thank Barrett Updegraff, Nicole Novaresi, Mahesh Padanad, and Joshua Mendell for critical reading of the manuscript and Jose Cabrera for assistance with figures. KAO is a CPRIT Scholar in Cancer Research and a Kimmel Scholar. This work was supported by the National Cancer Institute (R01CA207763 to KAO), the Cancer Prevention Research Institute of Texas (CPRIT, RP150676 to KAO), the Welch Foundation (I-1881 to KAO), the Sidney Kimmel Foundation (SKF-15-067 to KAO), and the LUNGevity Foundation (2015-03 to KAO). I apologize to colleagues whose important contributions could not be cited due to space limitations.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
● of special interest
●● of outstanding interest
- 1.St Johnston D: The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet 2002, 3: 176–188. [DOI] [PubMed] [Google Scholar]
- 2.Uren AG, Kool J, Berns A, van Lohuizen M: Retroviral insertional mutagenesis: past, present and future. Oncogene 2005, 24: 7656–7672. [DOI] [PubMed] [Google Scholar]
- 3.●●.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z: Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 1997,91: 501–510. [DOI] [PubMed] [Google Scholar]; This study describes the re-awakening of the Tcl/mariner DNA transposon from fish, which revolutionized the field and generated new opportunities for in vivo genome engineering.
- 4.Luo G, Ivics Z, Izsvak Z, Bradley A: Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci USA 1998, 95: 10769–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier LS, Carlson CM, Ravimohan S, DupuyAJ, Largaespada DA: Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 2005, 436: 272–276. [DOI] [PubMed] [Google Scholar]
- 6.Copeland NG, Jenkins NA: Harnessing transposons for cancer gene discovery. Nat Rev Cancer 2010, 10: 696–706. [DOI] [PubMed] [Google Scholar]
- 7.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA: Mammalian mutagenesis using a highly mobile somatic sleeping beauty transposon system. Nature 2005,436: 221–226. [DOI] [PubMed] [Google Scholar]
- 8.Dupuy AJ, Fritz S, Largaespada DA: Transposition and gene disruption in the male germline of the mouse. Genesis 2001, 30: 82–88. [DOI] [PubMed] [Google Scholar]
- 9.Horie K, Kuroiwa A, Ikawa M, Okabe M, Kondoh G, Matsuda Y, Takeda J: Efficient chromosomal transposition of a Tc1/mariner-like transposon sleeping beauty in mice. Proc Natl Acad Sci USA 2001, 98: 9191–9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T: Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 2005, 122: 473–483. [DOI] [PubMed] [Google Scholar]
- 11.Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice S, Campos LS, Yusa K, Banerjee R, Li MA, de la Rosa J et al. : PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 2010, 330: 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brett BT, Berquam-Vrieze KE, Nannapaneni K, Huang J, Scheetz TE, Dupuy AJ: Novel molecular and computational methods improve the accuracy of insertion site analysis in sleeping beauty-induced tumors. PLoS One 2011, 6: e24668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ridder J, Uren A, Kool J, Reinders M, Wessels L: Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol 2006, 2: e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A: High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 2002, 32: 153–159. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG: New genes involved in cancer identified by retroviral tagging. Nat Genet 2002, 32: 166–174. [DOI] [PubMed] [Google Scholar]
- 16.Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, Silverstein KA, Sarver A, Starr TK, Akagi K, Tessarollo L, et al. : A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnot 2009, 27: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O’Sullivan MG, Matise I, Dupuy AJ, Collier LS et al. : A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009, 323: 1747–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeNicola GM, Karreth FA, Adams DJ, Wong CC: The utility of transposon mutagenesis for cancer studies in the era of genome editing. Genome Biot 2015, 16:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann MB, Jenkins NA, Copeland NG, Mann KM: Sleeping beauty mutagenesis: exploiting forward genetic screens for cancer gene discovery. Curr Opin Genet Dev 2014, 24: 16–22. [DOI] [PubMed] [Google Scholar]
- 20.●●.Kas SM, de Ruiter JR, Schipper K, Annunziato S, Schut E, Klarenbeek S, Drenth AP, van der Burg E, Klijn C, Ten Hoeve JJ, Adams DJ et al. : Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat Genet 2017, 49: 1219–1230. [DOI] [PubMed] [Google Scholar]; This screen utilized the Rosa26Lox66SBLox71 and T2/Onc mouse lines on a Cdh1 deficient background to identify genes that promote the development of invasive lobular breast carcinoma. This study is notable because it revealed driver mutations that were not readily identified in human datasets, namely heterozygous inactivating insertions in Myh9, and because it prioritized driver genes that are present in large windows of amplifications in human breast cancer.
- 21.Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z, Shroff AS, Dickins RA, Vakoc CR, Bradner JE, Stock W et al. : Mll3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer cell 2014, 25: 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Will B, Steidl U: Combinatorial haplo-deficient tumor suppression in 7q-deficient myelodysplastic syndrome and acute myeloid leukemia. Cancer Cell 2014, 25: 555–557. [DOI] [PubMed] [Google Scholar]
- 23.Rangel R, Lee SC, Hon-Kim Ban K, Guzman-Rojas L, Mann MB, Newberg JY, Kodama T, McNoe LA, Selvanesan L, Ward JM, Rust AG et al. : Transposon mutagenesis identifies genes that cooperate with mutant pten in breast cancer progression. Proc Natl Acad Sci U S A 2016, 113: E7749–E7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Jenjaroenpun P, Pillai AM, Ivshina AV, Ow GS, Efthimios M, Zhiqun T, Tan TZ, Lee SC, Rogers K, Ward JM et al. : Transposon insertional mutagenesis in mice identifies human breast cancer susceptibility genes and signatures for stratification. Proc Natl Acad Sci U S A 2017, 114: E2215–E2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suarez-Cabrera C, Quintana RM, Bravo A, Casanova ML, Page A, Alameda JP, Paramio JM, Maroto A, Salamanca J, Dupuy AJ, Ramirez A et al. : A transposon-based analysis reveals rasa1 is involved in triple-negative breast cancer. Cancer Res 2017, 77: 1357–1368. [DOI] [PubMed] [Google Scholar]
- 26.Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P, Largaespada DA, Scheetz TE, Jenkins NA, Copeland NG: A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 2009, 69: 8150–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riordan JD, Keng VW, Tschida BR, Scheetz TE, Bell JB, Podetz-Pedersen KM, Moser CD, Copeland NG, Jenkins NA, Roberts LR, Largaespada DA, et al. : Identification of rtl1, a retrotransposon-derived imprinted gene, as a novel driver of hepatocarcinogenesis. PLoS Genet 2013, 9: e1003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell KA, Keng VW, York B, Reineke EL, Seo D, Fan D, Silverstein KA, Schrum CT, Xie WR, Mularoni L, Wheelan SJ et al. : A sleeping beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad SciUSA 2012, 109: E1377–E1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta S, Putluri N, Long W, Zhang B, Wang J, Kaushik AK, Arnold JM, Bhowmik SK, Stashi E, Brennan CA, Rajapakshe K et al. : Coactivator SRC-2-dependent metabolic reprogramming mediates prostate cancer survival and metastasis. J Clin Investig 2015, 125: 1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suresh S, Durakoglugil D, Zhou X, Zhu B, Comerford SA, Xing C, Xie XJ, York B, O’Donnell KA: SRC-2-mediated coactivation of anti-tumorigenic target genes suppresses myc-induced liver cancer. PLoS Genet 2017, 13: e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bard-Chapeau EA, Nguyen AT, Rust AG, Sayadi A, Lee P, Chua BQ, New LS, de Jong J, Ward JM, Chin CK, Chew V et al. : Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet 2014, 46: 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riordan JD, Feddersen CR, Tschida BR, Jackson P, Keng VW, Linden MA, Amin K, Stipp CS, Largaespada DA, Dupuy AJ: Chronic liver injury alters driver mutation profiles in hepatocellular carcinoma. Hepatology 2018, 67: 924–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tschida BR, Temiz NA, Kuka TP, Lee LA, Riordan JD, Tierrablanca CA, Hullsiek R, Wagner S, Hudson WA, Linden MA, Amin K et al. : Sleeping beauty insertional mutagenesis in mice identifies drivers of steatosis-associated hepatic tumors. Cancer Res 2017, 77: 6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA: High-resolution genome-wide mapping of transposon integration in mammals. Mot Cett Biot 2005, 25:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li MA, Pettitt SJ, Eckert S, Ning Z, Rice S, Cadinanos J, Yusa K, Conte N, Bradley A: The piggybac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration sites. Mol Cell Biol 2013, 33: 1317–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang Q, Kong J, Stalker J, Bradley A: Chromosomal mobilization and reintegration of sleeping beauty and piggybac transposons. Genesis 2009, 47: 404–408. [DOI] [PubMed] [Google Scholar]
- 37.Wilson MH, Coates CJ, George AL Jr: Piggybac transposon-mediated gene transfer in human cells. Mol Ther 2007,15: 139–145. [DOI] [PubMed] [Google Scholar]
- 38.●●.Friedrich MJ, Rad L, Bronner IF, Strong A, Wang W, Weber J, Mayho M, Ponstingl H, Engleitner T, Grove C, Pfaus A et al. : Genome-wide transposon screening and quantitative insertion site sequencing for cancer gene discovery in mice. NatProtoc 2017, 12: 289–309. [DOI] [PubMed] [Google Scholar]; This reference implemented a semi-quantitative transposon insertion site sequencing method using acoustic DNA shearing (QiSeq). Multiple SB and PB mouse lines were used including Rosa26PB, Rosa26LSL-PB, and ATP1–3 on a KrasLSL-G12D background.
- 39.●●.Rad R, Rad L, Wang W, Strong A, Ponstingl H, Bronner IF, Mayho M, Steiger K, Weber J, Hieber M, Veltkamp C et al. : A conditional piggybac transposition system for genetic screening in mice identifies oncogenic networks in pancreatic cancer. Nat Genet 2015, 47: 47–56. [DOI] [PubMed] [Google Scholar]; This reference utilized a conditional PB mouse model to identify novel oncogenic networks, including Foxp1 as an oncogenic transcription factor in pancreatic cancer. It also helped elucidate the genetic basis of different histologic subtypes of pancreatic cancer. These results implicate altered regulation of Fign, which encodes a member of the AAA-ATPase superfamily, in the development of hepatoid tumors, a rare pancreatic cancer subtype. This study used the Rosa26LSL-PB and ATP1-S2 mouse lines on a KrasLSL-G12D background.
- 40.Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, Australian Pancreatic Cancer Genome Initiative, Chang DK et al. : Sleeping beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA 2012, 109: 5934–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grutzmann R, Aust D, Rummele P, Knosel T et al. : The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 2012, 486: 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Donnell KA, An W, Schrum CT, Wheelan SJ, Boeke JD: Controlled insertional mutagenesis using a LINE-1 (ORFeus) gene-trap mouse model. Proc Natl Acad Sci USA 2013, 110: E2706–E2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCreery MQ, Halliwill KD, Chin D, Delrosario R, Hirst G, Vuong P, Jen KY, Hewinson J, Adams DJ, Balmain A: Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat Med 2015, 21: 1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riordan JD, Drury LJ, Smith RP, Brett BT, Rogers LM, Scheetz TE, Dupuy AJ: Sequencing methods and datasets to improve functional interpretation of sleeping beauty mutagenesis screens. BMC Genomics 2014, 15: 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.●.Mann KM, Newberg JY, Black MA, Jones DJ, Amaya-Manzanares F, Guzman-Rojas L, Kodama T, Ward JM, Rust AG, van der Weyden L, Yew CC et al. : Analyzing tumor heterogeneity and driver genes in single myeloid leukemia cells with SBCapSeq. Nat Biotechnol 2016, 34: 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference described a powerful method for detecting clonal insertion events in a myeloid leukemia mouse model and for identifying cooperating events in individual tumor cells. This method, called SBCapSeq, represents a refinement of previous transposon sequencing methods.
- 46.Temiz NA, Moriarity BS, Wolf NK, Riordan JD, Dupuy AJ, Largaespada DA, Sarver AL: RNA sequencing of sleeping beauty transposon-induced tumors detects transposon-RNA fusions in forward genetic cancer screens. Genome Res 2016, 26: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.●●.de la Rosa J, Weber J, Friedrich MJ, Li Y, Rad L, Ponstingl H, Liang Q, deQuiros SB, Noorani I, Metzakopian E, Strong A et al. : A single-copy sleeping beauty transposon mutagenesis screen identifies new pten-cooperating tumor suppressor genes. Nat Genet 2017, 49: 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference combined targeted gene inactivation to single-copy transposon mobilization to identify novel genes that cooperate with Pten in suppressing prostate, breast, and skin tumorigenesis. A single-copy SB transposon limited the number of insertions to one per cell, which reduced the number of passenger insertions. Transposition occurred simultaneously in the same cells that underwent Pten inactivation, enhancing the sensitivity of the screen. Finally, the gene trap promoted only inactivating mutations but not activating mutations, facilitating the identification of putative tumor suppressors but not oncogenes.
- 48.Ahmad I, Mui E, Galbraith L, Patel R, Tan EH, Salji M, Rust AG, Repiscak P, Hedley A, Markert E, Loveridge C et al. : Sleeping beauty screen reveals Pparg activation in metastatic prostate cancer. Proc Natl Acad Sci USA 2016, 113: 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriarity BS, Otto GM, Rahrmann EP, Rathe SK, Wolf NK, Weg MT, Manlove LA, LaRue RS, Temiz NA, Molyneux SD, Choi K et al. : A sleeping beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet 2015, 47: 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandzic T, Larsson J, He L, Kundu S, Ban K, Akhtar-Ali M, Hellstrom AR, Schuh A, Clifford R, Blakemore SJ, Stretford JC et al. : Transposon mutagenesis revealsfludarabine resistance mechanisms in chronic lymphocytic leukemia. Clin Cancer Res 2016, 22: 6217–6227. [DOI] [PubMed] [Google Scholar]
- 51.Perna D, Karreth FA, Rust AG, Perez-Mancera PA, Rashid M, lorio F, Alifrangis C, Arends MJ, Bosenberg MW, Bollag G, Tuveson DA et al. : BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci U S A 2015, 112: E536–E545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudalska R, Dauch D, Longerich T, McJunkin K, Wuestefeld T, Kang TW, Hohmeyer A, Pesic M, Leibold J, von Thun A, Schirmacher P et al. : In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med 2014, 20: 1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.●.Morrissy AS, Garzia L, Shih DJ, Zuyderduyn S, Huang X, Skowron P, Remke M, Cavalli FM, Ramaswamy V, Lindsay PE, Jelveh S et al. : Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016, 529: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference developed a SB mouse model of metastatic medulloblastoma (on a Ptch+/− background) treated with the standard therapy of tumor resection followed by radiotherapy in children with this disease. Sequencing of SB-induced tumors revealed a poor degree of overlap between the primary mouse tumors and the tumors that recurred after treatment, similar to findings from human tumor data. Additional analysis showed that the dominant clone at recurrence arose in part through clonal selection of a minor clone that was present at the time of diagnosis. The authors advocated that sequencing of biopsies at recurrence should guide clinical trials and treatment decisions.
- 54.Molyneux SD, Waterhouse PD, Shelton D, Shao YW, Watling CM, Tang QL, Harris IS, Dickson BC, Tharmapalan P, Sandve GK, Zhang X et al. : Human somatic cell mutagenesis creates genetically tractable sarcomas. Nat Genet 2014, 46: 964–972. [DOI] [PubMed] [Google Scholar]
- 55.Koso H, Takeda H, Yew CC, Ward JM, Nariai N, Ueno K, Nagasaki M, Watanabe S, Rust AG, Adams DJ, Copeland NG et al. : Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. Proc Natl Acad Sci USA 2012, 109: E2998–E3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kodama T, Newberg JY, Kodama M, Rangel R, Yoshihara K, Tien JC, Parsons PH, Wu H, Finegold MJ, Copeland NG, Jenkins NA: Transposon mutagenesis identifies genes and cellular processes driving epithelial–mesenchymal transition in hepatocellular carcinoma. Proc Natl Acad Sci U S A 2016,113: E3384–E3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.●.Guo Y, Updegraff BL, Park S, Durakoglugil D, Cruz VH, Maddux S, Hwang TH, O’Donnell KA: Comprehensive ex vivo transposon mutagenesis identifies genes that promote growth factor independence and leukemogenesis. Cancer Res 2016, 76: 773–786. [DOI] [PubMed] [Google Scholar]; This reference, which represents one of the largest ex vivo screens performed to date, identified genes that promote growth factor independence and transformation of Ba/F3 cells, an IL-3-dependent murine pro-B cell line. Recurrent insertions were identified in the JAK/STAT and MAPK pathway genes in addition to a large number of genes that are mutated or associate with survival of leukemia patients but had not previously been linked to these pathways.
- 58.●.Chen HJ, Wei Z, Sun J, Bhattacharya A, Savage DJ, Serda R, Mackeyev Y, Curley SA, Bu P, Wang L, Chen S et al. : A recellularized human colon model identifies cancer driver genes. Nat Biotechnol 2016, 34: 845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]; This ex vivo screen utilized a recellularized human colon model to identify genes that drive colorectal cancer progression. This advance provided a unique tissue-engineering method that enabled the implementation of forward genetic screening in human tissues under physiologic conditions.
- 59.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F: Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F: Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber J, Ollinger R, Friedrich M, Ehmer U, Barenboim M, Steiger K, Heid I, Mueller S, Maresch R, Engleitner T, Gross N et al. : CRlSPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc Natl Acad Sci US A 2015, 112: 13982–13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu C, Qi X, Du X, Zou H, Gao F, Feng T, Lu H, Li S, An X, Zhang L, Wu Y et al. : Piggybac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc Natl Acad Sci USA 2017, 114: 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, Krauthammer M et al. : In vivo identification of tumor-suppressive PTEN ceRNAS in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011, 147: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbott KL, Nyre ET, Abrahante J, Ho YY, Isaksson Vogel R, Starr TK: The candidate cancer gene database: a database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res 2015, 43(Database issue):D844–D848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Newberg JY, Mann KM, Mann MB, Jenkins NA, Copeland NG: SBCDDB: sleeping beauty cancer driver database for gene discovery in mouse models of human cancers. Nucleic Acids Res 2018, 46(D1):D1011–D1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dorr C, Janik C, Weg M, Been RA, Bader J, Kang R, Ng B, Foran L, Landman SR, O’Sullivan MG, Steinbach M et al. : Transposon mutagenesis screen identifies potential lung cancer drivers and cul3 as a tumor suppressor. Mol Cancer Res 2015,13: 1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heltemes-Harris LM, Larson JD, Starr TK, Hubbard GK, Sarver AL, Largaespada DA, Farrar MA: Sleeping beauty transposon screen identifies signaling modules that cooperate with STAT5 activation to induce B-cell acute lymphoblastic leukemia. Oncogene 2016, 35: 3454–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mann MB, Black MA, Jones DJ, Ward JM, Yew CC, Newberg JY, Dupuy AJ, Rust AG, Bosenberg MW, McMahon M, Print CG et al. : Transposon mutagenesis identifies genetic drivers of BRAF (v600e) melanoma. Nat Genet 2015, 47: 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montero-Conde C, Leandro-Garcia LJ, Chen X, Oler G, Ruiz-Llorente S, Ryder M, Landa I, Sanchez-Vega F, La K, Ghossein RA, Bajorin DF et al. : Transposon mutagenesis identifies chromatin modifiers cooperating with Ras in thyroid tumorigenesis and detects ATXN7 as a cancer gene. Proc Natl Acad Sci U S A 2017, 114: E4951–E4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morris SM, Davison J, Carter KT, O’Leary RM, Trobridge P, Knoblaugh SE, Myeroff LL, Markowitz SD, Brett BT, Scheetz TE, Dupuy AJ et al. : Transposon mutagenesis identifies candidate genes that cooperate with loss of transforming growth factor-beta signaling in mouse intestinal neoplasms. Int J Cancer 2017, 140: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takeda H, Rust AG, Ward JM, Yew CC, Jenkins NA, Copeland NG: Sleeping beauty transposon mutagenesis identifies genes that cooperate with mutant Smad4 in gastric cancer development. Proc Natl Acad Sci USA 2016, 113: E2057–E2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeda H, Wei Z, Koso H, Rust AG, Yew CC, Mann MB, Ward JM, Adams DJ, Copeland NG, Jenkins NA: Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet 2015, 47: 142–150. [DOI] [PubMed] [Google Scholar]
- 73.Wu J, Keng VW, Patmore DM, Kendall JJ, Patel AV, Jousma E, Jessen WJ, Choi K, Tschida BR, Silverstein KA, Fan D et al. : Insertional mutagenesis identifies a STAT3/Arid1b/beta-catenin pathway driving neurofibroma initiation. Cell Rep 2016, 14: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moriarity BS, Largaespada DA: Sleeping beauty transposon insertional mutagenesis based mouse models for cancer gene discovery. Curr Opin Genet Dev 2015, 30: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]