Eukaryotic protein synthesis is a highly coordinated process involving the interaction of a large number of factors that participate in the assembly of ribosomes and aminoacylated tRNAs onto mRNA templates, followed by their correct and efficient translation into proteins. Regulation of protein synthesis is mainly achieved at the initiation step, and occurs through modulation of translation factor levels or activity.1 Small molecule ligands that inhibit the process of translation have provided exquisite insight into ribosome function and translation factor activity in both prokaryotes and eukaryotes.2-4 In addition, a growing number of studies indicate that deregulation of protein synthesis is a major contributor to carcinogenesis and subsequent metastasis.5 For instance, overexpression of certain initiation factors has been associated with malignant transformation, whereas suppression of these same factors can restrict the transformed carcinogenic phenotype. Additionally, components of the translational machinery not solely associated with initiation are often overexpressed or mutated in cancers. Thus, the identification of small molecules able to modulate translational processes represents an endeavor, important not only in understanding the mechanics of translation but also in the development of new therapeutic candidates that exploit the unique translational features characteristic of cancer cells.
We previously reported the use of a high-throughput bicistronic mRNA translation screen to identify inhibitors of eukaryotic protein synthesis.6,7 The application of this translation screen leads to the discovery of several new modulators of translation initation.8,9 We now report here the application ofthis translation inhibition screen to identify natural product inhibitors of translation produced by two new endophytic actinomycetes.
During the course of our efforts to identify novel natural products from microorganisms from diverse and unique ecological niches,10-13 we isolated a collection of actinomycete strains from endophytes and soil samples originating from the Yunnan province of China. Subjection of crude extract libraries of this collection to translation inhibition assay indicated the presence of potential translation inhibitors in two of the crude extracts prepared from the endophytic actinomycete Streptomyces sps. YIM56132 from Carex baccaus and YIM56141 from Fagopyrum cumosum; both plants were isolated from the Xishuang-banna Perfecture of Yunnan, where they are widely used to treat arthritis. Both extracts also possessed components possessing moderate cytotoxic activity against selected human cancer cells and having similar HPLC profiles. Subsequent large-scale fermentation (6l) and bioassay-guided fractionation with extracts from both YIM56132 and YIM56141 yielded two fractions whose components inhibited translation. Thorough evaluation of each extract revealed the presence of cycloheximide (1),14 iso-cycloheximide (2),14 A-75943 (3),15 two new cycloheximide congeners-secocycloheximides A (4) and B (5), as well as actiphenol (6),16 AH-135Y (7)16 and phenatic acid A (8)16 in the YIM56141 extract, whereas 1, 2 and 4-8 were also found in the YIM56132 extract (Figure 1a). In addition, four previously known diketopiperazines 11-14 were also found in the YIM56141 extract (Supplementary Information). Dereplication revealed 3 and 4 to be new congeners of 1, and concomitant efforts revealed that 1 and 2 were mainly responsible for translation inhibition and cytotoxicity. We also found that 3 showed selective cytotoxicity against breast cancer cell lines and that 6 was a weak translation inhibitor. All other compounds were devoid of activity (Table 2). Both 6 and 7 were subjected to selective reduction by NaBH4 to afford the corresponding 8-hydroxy analogs 9 and 10 (Figure 1a and Supplementary Information). Interestingly, 9 is a significantly more potent inhibitor of translation than 6 (Table 2). This finding hints at the importance of C-8 oxidation state in determining the extent to which these natural products inhibit eukaryotic translation.
Figure 1.
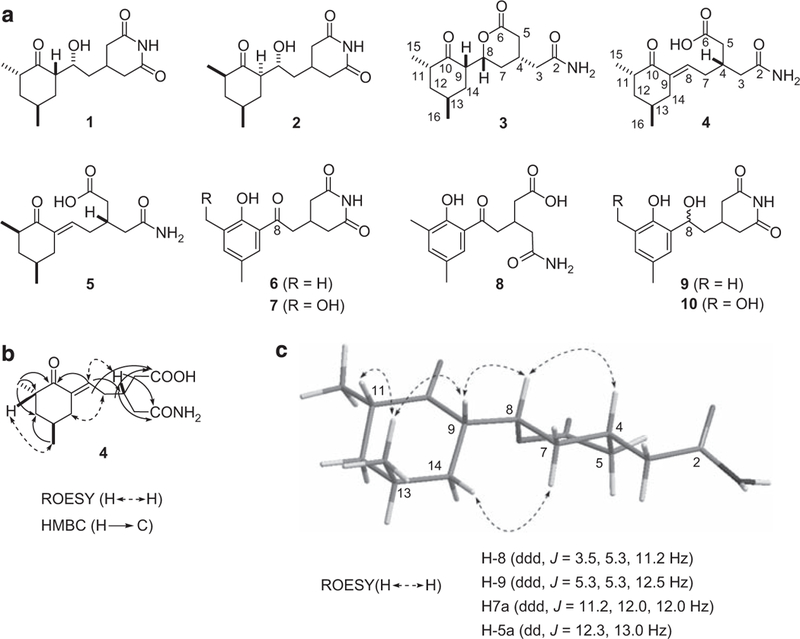
Cycloheximide (1) and new (4, 5, 9 and 10) and known (2, 3 and 6-8) congeners isolated from Streptomyces sps YIM56132 and YIM56141, as well as 8-hydroxy variants of 6 and 7 (9 and 10): (a) structures of 1-10; (b) key HMBC and ROESY correlations for 4; and (c) key ROESY correlations and coupling constants for 3 overlaid with MM2 force field energy minimized model.
Table 2.
Summary of biological activities for 1-10: (a) inhibition of Cap-dependent translation by 1, 2, 6 and 9 (μM) and cytotoxicity against MDA-MB-231, Du145, HCT-15 and MCF7 human cancer cell lines by 1-3 (μM)
| IC50 |
||||
|---|---|---|---|---|
| A Translation inhibitiona | 1 | 2 | 6 | 9 |
| FF luciferase | 0.95 ±0.05 | 0.33±0.10 | 30.40 ±2.56 | 1.31±0.03 |
| Ren luciferase | 3.54±0.62 | 0.39±0.30 | >50 | 7.05±0.80 |
| IC50 |
||||
| B Cytotoxicityb | 1 | 2 | 3 | |
| MDA-MB-231 | 0.3 ±0.02 | 0.33 ±0.01 | 2.28±0.07 | |
| Du145 | 1.86 ± 0.06 | 2.21 ±0.05 | >10 | |
| HCT-15 | 0.82 ±0.04 | 0.86 ±0.03 | >10 | |
| MCF7 | 0.28±0.01 | 0.31 ±0.01 | 2.47 ±0.07 | |
Abbreviations: FF, firefly; Ren, renilla.
Substrate for the translation inhibition assay was capped-(CAG)33FF/HCV/RenpA51 mRNA.
All other compounds showed no detectable inhibition at 20μm.
All other compounds showed no detectable toxicity at 10 μm.
Efforts to elucidate natural product structures from YIM56141 and YIM56132 relied heavily on HR-MS and NMR spectroscopy analyses and were facilitated by prior knowledge of 1. APCI-MS analysis of 1-3 afforded the same protonated [M+H]+ ion at m/z 282 and suggested a molecular formula of C15H23NO4. Combined with the 1H and 13C NMR and HR-MS data suggested that 1-3 are cycloheximide,14 iso-cycloheximide14 and A-75943 (see Morishita et al.15), respectively. The 1H and 13C NMR spectra of purified 1-3 were assigned on the basis of extensive 1D and 2D NMR (gCOSY, gHSQC and gHMBC) and HR-ESI-MS analyses, and comparison with the spectroscopic data previously reported (Table 1).
Table 1.
Summary of 1H and 13C NMR data for 4 and 5 in comparison with 3 in CD3OD
| Position |
3a |
4 |
5 |
|||
|---|---|---|---|---|---|---|
| 𝛿H (mult, J in Hz) | 𝛿C | 𝛿H (mult, J in Hz) | 𝛿C | Sh (mult, J in Hz) | 𝛿C | |
| 2 | 172.6 | 174.8 | 176.8 | |||
| 3a | 2.63-2.79 (2H, m) | 40.3 | 2.40 (m)b | 37.7 | 2.21 (m) | 39.9 |
| 3b | 2.28 (m)c | 2.34 (m)b | ||||
| 4 | 2.19 (m) | 27.2 | 2.50 (m) | 32.5 | 2.48 (m) | 33.7 |
| 5a | 2.21 (dd, 12.3, 13.0) | 35.4 | 2.28 (2H, m)c | 39.1 | 2.26 (2H, m)b | 41.5 |
| 5b | 2.74 (m) | |||||
| 6 | 175.2 | 176.1 | 179.2 | |||
| 7a | 1.37 (ddd, 11.2, 12.0, 12.0) | 31.9 | 2.21 (m)c | 31.3 | 2.24-2.31 (2H, m)b | 31.9 |
| 7b | 2.07 (m) | 2.34 (m) | ||||
| 8 | 4.70 (ddd, 3.5, 5.3 11.2) | 78.8 | 6.43 (brt, 6.5) | 134.4 | 6.57 (brt, 6.5) | 136.6 |
| 9 | 2.84 (ddd, 5.3, 5.3, 12.5) | 49.4 | 138.7 | 137.4 | ||
| 10 | 212.2 | 204.8 | 205.6 | |||
| 11 | 2.45 (m) | 28.9 | 2.41 (m)b | 43.9 | 2.60 (m) | 40.5 |
| 12a | 1.90 (m) | 42.5 | 1.27 (m) | 40.8 | 1.72 (m) | 38.2 |
| 12b | 1.60 (m) | 2.02 (m) | 1.77 (m) | |||
| 13 | 2.21 (m) | 41.3 | 1.91 (m)d | 30.3 | 2.05 (m) | 26.0 |
| 14a | 2.13 (m) | 34.9 | 1.91 (m)d | 35.9 | 2.17 (m) | 34.4 |
| 14b | 1.76 (m) | 2.86 (m) | 2.70 (m) | |||
| 15 | 1.26 (d, 7.0) | 17.2 | 1.11 (d, 6.4) | 14.9 | 1.13 (d, 6.5) | 16.3 |
| 16 | 0.94 (d, 6.5) | 13.6 | 1.08 (d, 6.5) | 21.2 | 1.09 (d, 6.5) | 20.3 |
1H and 13C assignments were based on 1H-1H COSY, HMBC and HSQC spectra, which corrected the misassignments reported previously in Morishita et al.15
Signals overlapped.
HR-ESI-MS analysis of 4 showed a [M+H]+ ion at m/z 282.17037, with the same molecular formula, C15H23NO4, as that of 1-3. Analysis of 1H and 13C NMR data for 4, along with HR-ESI-MS and 2D NMR (Supplementary Information) data indicated the presence of structural moieties characteristic of the cycloheximide, including cyclohexanone (δc 204.8), two Me carbons (δH 1.11 and 1.08) and two carbonyl carbons (δc 174.8 and δc 176.1) (Table 1). Apart from these moieties, immediately identifiable from the NMR spectroscopic data for 4 were resonances consistent with one trisubstituted double bond [δC 134.4 (d) and 138.7 (s); δH 6.43 (br t)]. Through careful analysis of the MS, HMBC and COSY spectra, the structure was deduced to be seco-anhydrocycloheximide14 as shown in Figure 1b on the basis of the key correlations of the vinylic proton H-8 (δH 6.43) with the ketone C-10 (δC 204.8) and methine C-4 (δC 32.5), of H-4 (δH 2.50) and H2-3 [(δH 2.40 (m), 2.28 (m)] with the carbonyl C-2 (δC 174.8) and of H-4 and H2-5 [δH 2.28 (m)] with the carbonyl C-6 (δC 176.1) (Figure 1b). The carbon NMR chemical shifts of the carbonyl moieties in the glutarimide ring are almost identical.14,17,18 The different chemical shifts of C-2 (δC 174.8) and C-6 (δC 176.1) in 4 further corroborated the presence of the seco-glutarimide ring. The double bond geometry was established by ROESY correlation of H-7b (δH 2.34) with H-14b (δH 2.86) (Figure 1b). The relative stereochemistry of C-11 and C-13 in 4 was established by analyzing information from the ROESY spectrum (Supplementary Information) and by comparison of its spectroscopic data with those of 1 and anhydrocy-cloheximide.14
The relative configuration at C-4 in 4 cannot be determined from spectroscopic data alone. Fortunately, we found that 4 was converted into 3 during silica gel chromatography using a chloroform-methanol gradient (from 100:1 to 4:1). This observation is easily rationalized by intramolecular Michael addition in a highly favored 6-exo-trig fashion (Supplementary Information). Given this rationale and the non-labile nature of C-4 in both 3 and 4, we reasoned that 3 and 4 share the same stereochemical configuration. Consequently, ROESY and coupling constant data were used to establish the relative stereochemistry of 3. The configurations for C-11 and C-13 are supported by strong ROESY correlations of H-11 (δH 2.45) and H-9 (δH 2.84), with H3-16 (δH 0.94) as shown in molecular models (Figure 1c), energy minimized using the MM2 force field in Chem3D overlaid with key correlations observed in the ROESY spectrum. Relayed ROESY correlations of H-4 (δH 2.19) with H-8 (δH 4.70), of H-8 with H-9 (δH 2.84) and of H-9 with H3-16, which were all supported by splitting patterns of H-8 and H-9 and small coupling constants (J=5.3 Hz) between H-8 and H-9, are consistent with the assigned relative configurations of C-4, C-8 and C-9 depicted in Figure 1c. These relative stereochemistry assignments are identical to those previously reported.15 Consequently, the complete structure of 3, hence the same stereochemistry at C-4 for 4, was deduced as shown in Figure 1a.
Secocycloheximide B (5) was assigned the same molecular formula, C15H23NO4, as that of 4, on the basis of its HR-ESI-MS and 1H and 13C NMR data. Careful 1D and 2D NMR analyses showed that 5 has the same cyclohexanone scaffold as 4. However, the dramatically different chemical shifts of C-11, C-12 and C-13 in the 13C NMR for 4 and 5 (Table 1) supports the notion that an epimerized C-11 differentiates 5 from other cyclohexanone-containing metabolites produced by cycloheximide producers.19 In addition, the stereochemistry of C-4 was tentatively assigned to be the same as that of 4 on the basis of shared biosynthetic machineries.
All compounds isolated were evaluated for their ability to inhibit eukaryotic translation in Krebs-2 extracts programmed with capped-(CAG)33FF/HCV/RenpA51 mRNA as described previously;7 from these efforts was derived a clear correlation of structure to activity (Table 2A). Both 1 and 9 inhibited translation from the firefly and renilla luciferase cistrons in a dose-dependent manner, whereas 6 showed only weak inhibitory activity. Interestingly, 9 is a significantly more potent translation inhibitor than its 8-oxo-variant 6 (Table 2B). This finding supports earlier work highlighting the importance of glutarimide side chain hydroxylation in endowing the glutarimide-containing polyketides, such as 1 and lactimidomycin, with biological activities surpassing those of non-hydroxylated congeners.20,21
All compounds isolated were also tested for cytotoxicity against human breast cancer MDA-MB-231, human prostate carcinoma Du145, human colon cancer HCT-15 and human breast adenocarcinoma MCF7 cell lines using previously described methods (Table 2B).22 Both 1 and 2 demonstrated comparable inhibitory activity against all the cell lines, suggesting the relative unimportance of C-11 stereochemistry, whereas 3 demonstrated activity only against breast cancer cell lines MDA-MB-231 and MCF7. The IC50s for 3 were ~ 10-fold higher than demonstrated for 1 and 2, suggesting that loss of the glutarimide, presence of the lactone moiety, or both, relative to 1 and 2, detrimentally impacts cytotoxic activity. All other compounds were found to be inactive against all the four cancer cell lines (no detectable cytotoxicity at 10 μM).
In summary, we have found two new endophytic cycloheximide producers Streptomyces sps. YIM56132 and YIM56141 from unique ecological niches using a novel bicistronic mRNA translation assay and have isolated two new cycloheximide congeners 4 and 5, which contain carboxylic acid and primary amide functionalities instead of an intact glutarimide. Most importantly, translation inhibition assays showed that the presence of a hydroxyl group at C-8 significantly improves inhibitory activity. This discovery could, therefore serve as a scaffold for the chemical synthesis and biosynthesis of more potent modified eukaryotic protein synthesis inhibitors.
The structural differences of 1 and 6 is that the latter replaces the cyclohexanone moiety of 1 with a substituted phenol and the C-8 hydroxyl moiety with a ketone at C-8. Thus, the dual production of both 1 and 6 in the same strain is biosynthetically intriguing, as both polyketides must be derived from the same biosynthetic machinery. It is, therefore not clear which compound precedes the other during biosynthesis, and which one may represent a shunt metabolite. Perhaps the most interesting insight into biosynthesis of glutarimide-containing antibiotics by Streptomyces sps. YIM56132 and YIM56141 is gleaned by the isolation of glutarimide ring opened-natural products 4, 5 and 8 from the wild-type strains. Isolation of these compounds raises the possibility that glutarimide installation might be catalyzed by a post-PKS enzyme encoded by the corresponding gene cluster. In light of the significant and novel bioactivities of glutarimide-containing polyketides such as 1, lactimidomycin, migras-tatin and iso-migrastatin, the questions raised here clearly warrant further attention, perhaps best addressed in continued studies of 1 and 6 biosynthesis in Streptomyces sps YIM56132 and YIM56141.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison for support in obtaining MS and NMR data. This work was supported in part by the NIH grant CA113297 (BS), NCIC grant #020066 (JP), Yunnan University funding KL080001 (LXZ), and the Chinese Ministry of Education 111 Project B08034 (YD).
Footnotes
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
References
- 1.Raught B, Gingras AC & Sonenberg N Regulation of ribosome recruitment in eukaryotes in Translational Control of Gene Expression (eds Sonenberg N, Hershey JWB, Mathews MB), 245–293 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2000). [Google Scholar]
- 2.Pestka S Inhibitors of protein synthesis, Academic Press, New York, 1977. [Google Scholar]
- 3.Vazquez D Inhibitors of protein biosynthesis. Mol. Biol. Biochem. Biophys. 30, 1–312 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Pelletier J & Peltz SW Therapeutic Opportunities in Translation in Translational Control in Biology and Medicine (eds Mathews M, Sonenberg N, & Hershey JWB) 855–895 (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY, 2007). [Google Scholar]
- 5.Watkins SJ & Norbury CJ Translation initiation and its deregulation during tumorigenesis. Br. J. Cancer 86, 1023–1027 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novac O, Guenier AS & Pelletier J Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 32, 902–915 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cencic R, Robert F & Pelletier J Identifying small molecule inhibitors of eukaryotic translation initiation. Methods Enzymol. 431, 269–302 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Bordeleau1 ME et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2, 213–220 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Bordeleau ME et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest 118, 2651–2660 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang SX et al. Erythronolides H and I, new erythromycin congeners from a new halophilic actinomycete Actinopolyspora sp. YIM90600. Org. Lett. 11, 1353–1356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang SX et al. Discovery and total synthesis of a new estrogen receptor heterodimerizing actinopolymorphol A from Actinopolymorpha rutilus. Org. Lett. 12, 3525–3527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell E et al. Identification and characterization of a novel estrogenic ligand actinopolymorphol A. Biochem. Pharmacol. 80, 1221–1229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Z et al. Bafilomycins produced by an endophytic actinomycete Streptomyces sp. YIM56209. J. Antibiot. 64, 159–162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffs P & McWilliams D Carbon-13 nuclear magnetic resonance study of the biosynthesis of cycloheximide, stereospecific incorporation of malonate into the glutarimide ring. J. Am. Chem. Soc. 103, 6185–6192 (1981). [Google Scholar]
- 15.Morishita T et al. A novel bone resorption inhibitor, A-75943 isolated from Streptomyces sp. SANK 61296. J. Antibiot. 51, 531–538 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Fukuda T, Matsumoto A, Takahashi Y, Tomoda H & Omura S Phenatic acids A and B, new potentiators of antifungal miconazole activity produced by Streptomyces sp. K03-0132. J. Antibiot. 58, 252–259 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Ju J, Seo JW, Her Y, Lim SK & Shen B New lactimidomycin congeners shed insight into lactimidomycin biosynthesis in Streptomyces amphibiosporus. Org. Lett. 9, 5183–5186 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Ju J, Lim SK, Jiang H, Seo JW & Shen B Iso-Migrastatin congeners from Streptomyces platensis and generation of a glutarimide polyketide library featuring the dorrigocin, lactimidomycin, migrastatin, and NK30424 scaffolds. J. Am. Chem. Soc. 127, 11930–11931 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Cheenpracha S et al. Yeast glycogen synthase kinase-3 pathway inhibitors from an organic extract of Streptomyces sp. J. Nat. Prod. 72, 1520–1523 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Ennis H L Structure-activity studies with cycloheximide and congeners. Biochem. Pharmacol. 17, 1197–1206 (1968). [DOI] [PubMed] [Google Scholar]
- 21.Schneider-Poetsch T et al. Inhibition ofeukaryotictranslation elongation bycyclohex- imideand lactimidomycin. Nat. Chem. Biol. 6, 209–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju J et al. Evaluation of new migrastatin and dorrigocin congeners unveils cell migration inhibitors with dramatically improved potency. Bioorg. Med. Chem. Lett. 18, 5951–5954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


