Abstract
The content of the beta subunit of the ATP synthase (β-F1-ATPase), which forms the catalytic site of the enzyme ATP synthase, is reduced in muscle of obese humans, along with reduced capacity for ATP synthesis. We studied 18 young (37 ± 8 years old) subjects of which nine were lean (BMI = 23 ± 2 kg/m2) and nine were obese (BMI = 34 ± 3 kg/m2) to determine the fractional synthesis rate (FSR) and gene expression of β-F1-ATPase, as well as the specific activity of the ATP synthase. FSR of β-F1-ATPase was determined using a combination of isotope tracer infusion and muscle biopsies. Gene expression of β-F1-ATPase and specific activity of the ATP synthase were determined in the muscle biopsies. When compared to lean, obese subjects had lower muscle β-F1-ATPase FSR (0.10 ± 0.05 vs 0.06 ± 0.03 %/hr; P < 0.05) and protein expression (P < 0.05), but not mRNA expression (P > 0.05). Across subjects, abundance of β-F1-ATPase correlated with the FSR of β-F1-ATPase (P < 0.05). The specific activity of muscle ATP synthase was lower in the obese when compared to lean subjects (0.035 ± 0.004 vs 0.042 ± 0.007 arbitrary units; P < 0.05), but this difference was not significant after the activity of the ATP synthase was adjusted to the β-F1-ATPase content (P > 0.05). Obesity impairs the synthesis of β-F1-ATPase in muscle at the translational level, reducing the content of β-F1-ATPase in parallel with reduced capacity for ATP generation via the ATP synthase complex.
Keywords: protein synthesis, mitochondria, adiposity, insulin resistance
Introduction
Obesity is linked to insulin resistance to glucose metabolism in skeletal muscle, but the effects of obesity on the regulation of energy metabolism and production rate of associated proteins in skeletal muscle are less clear. Magnetic resonance spectroscopy studies have shown that obese (Newcomer et al., 2001), and insulin-resistant (Petersen et al., 2004) individuals have lower capacity for muscle adenosine triphosphate (ATP) turnover in vivo. Lower capacity for muscle ATP production in humans with obesity has also been reported in studies using in vitro mitochondrial respiration assays (Abdul-Ghani et al., 2009; Chanseaume et al., 2010). Furthermore, lower concentration of ATP has been reported in the skeletal muscle of obese individuals (Lennmarken et al., 1986). In muscle, the mitochondrial enzyme ATP synthase is responsible for more than 90% of the ATP produced in that tissue, and recent evidence indicates that it plays a key role in metabolic disorders by regulating substrate metabolism (Formentini et al., 2017). From a functional standpoint, the beta subunit of the ATP synthase (β-F1-ATPase) forms the catalytic site of the ATP synthase, and therefore has direct role among the rest of the proteins of the enzyme in determining the capacity for ATP production. Characteristically, lower β-F1-ATPase expression is associated with lower ATP content in liver of animal models of diabetes (Wang et al., 2014).
Previous research has documented that obese, insulin-resistant, individuals have reduced content of β-F1-ATPase in skeletal muscle (Tran et al., 2016; Hojlund et al., 2010; Hwang et al., 2010). Reduced β-F1-ATPase content in the muscle of individuals with excess adiposity or insulin resistant could, therefore, contribute to the reduced capacity for ATP synthesis previously observed in the muscle of such individuals (Newcomer et al., 2001; Petersen et al., 2004; Abdul-Ghani et al., 2009; Chanseaume et al., 2010). In addition, antibodies directed against the β-F1-ATPase inhibit ATP generation in vitro (Moser et al., 2001), further underlying the critical role of β-F1-ATPase in the synthesis of ATP. The mechanisms that contribute to the reduced content of β-F1-ATPase in the skeletal muscle of obese individuals are not known. Furthermore, whether the production rate of β-F1-ATPase in skeletal muscle can directly be linked to impaired activity of the ATP synthase enzyme remains speculative.
Measurements of the content of β-F1-ATPase in skeletal muscle provide only a static snapshot of the metabolism of β-F1-ATPase in muscle. On the other hand, measurement of the rate of production of β-F1-ATPase in skeletal muscle provides direct information on the turnover rate of the β-F1-ATPase in muscle. Therefore, the underlying mechanism(s) behind the reduced content of muscle β-F1-ATPase in obesity can be directly investigated only by performing dynamic, real time, measurements, such as by determining the rate of synthesis of muscle β-F1-ATPase, in vivo. Recent studies have reported fractional synthesis rate (FSR) of human muscle β-F1-ATPase under a variety of circumstances (Murphy et al., 2018; Shankaran et al., 2016), but none of them have investigated the effects of obesity. We have established a procedure that allows quantifying the FSR of muscle β-F1-ATPase based on the incorporation of a stable isotope amino acid tracer in the protein and in combination with the use of high-performance liquid chromatography tandem mass spectrometry (LC/MS/MS) (Everman et al., 2011).
We hypothesized that reduced content of β-F1-ATPase in muscle of obese individuals is associated with reduced rate of synthesis of β-F1-ATPase. In addition to its importance in maintaining the quantity of muscle β-F1-ATPase, a given rate of synthesis of β-F1-ATPase is also important in maintaining the quality of the protein by renewing the β-F1-ATPase pool within skeletal muscle. Because older molecules of a protein are more prone to metabolic damage than newer molecules of the protein (Jaleel et al., 2010), decrease in the rate of synthesis of muscle β-F1-ATPase can lead to accumulation of older β-F1-ATPase molecules that are in a modified/less functional state. Therefore, we also hypothesized that reduced synthesis rate of muscle β-F1-ATPase is observed together with reduced specific activity of the ATP synthase enzyme.
Methods
Ethical approval
All study procedures conformed to the standards set by the Declaration of Helsinki and were approved by the Institutional Review Board at Mayo Clinic (IRB #: 12–004000). Risks stemming from participation in the study were described to each study participant before obtaining written consent. The study was registered at ClinicalTrials.gov (NCT01824173).
Subjects
Subjects were asked to participate in the study if they were apparently healthy as determined by an initial screening over the phone and had body mass index (BMI) of > 30 kg/m (i.e., obese) or < 25 kg/m2 (i.e., lean). Exclusion criteria for study participation included diabetes, heart disease, peripheral vascular disease, history of liver or kidney disease, smoking, participation in a weight-loss regimen, and use of medications or nutritional supplements. Only subjects that took part in various physical activities ≤ 2 per week were invited in the study (i.e., sedentary individuals). A comprehensive screening was later performed at the Clinical Studies Infusion Unit (CSIU) at Mayo Clinic in Scottsdale, Arizona, to determine further eligibility for participation in the study. It included a medical history/physical examination, resting electrocardiogram, standard blood and urine tests, as well as an oral glucose tolerance test (OGTT). Plasma glucose and insulin concentration values during the OGTT were used to estimate the subjects’ insulin sensitivity by calculating the Matsuda insulin-sensitivity index as described previously (Matsuda and DeFronzo, 1999).
Body composition was determined using bioelectrical impedance analysis (BIA 310e, Biodynamics Corp., Shoreline, WA), and the waist-to-hip ratio was calculated by measuring the circumferences at the level of the umbilicus and the buttocks. Peak oxygen uptake (VO2peak) was measured using a cycle ergometer, and by incrementally increasing the workload (20 Watts/min) to volitional exhaustion while continuously monitoring the expired gases (MedGraphics Metabolic Cart, Saint Paul, MN).
Experimental protocol
Subjects that qualified for the study took part in a single infusion trial performed at the CSIU. Prior to their participation in the infusion trial, subjects were instructed to avoid any exercise, maintain a regular diet, and avoid alcohol consumption during the three days leading to the infusion trial. Subjects arrived at the CSIU at z7 AM after an overnight fast. Compliance with the diet and exercise instructions indicated above was confirmed with each study participant before the initiation of the experimental procedures.
Experimental procedures were initiated by first inserting two catheters: one into a hand vein for obtaining blood samples and another one into an antecubital vein of the opposite arm for infusion of a stable isotope of an amino acid. L-[2,3,3,4,5,5,5,6,6,6-2H10]leucine (Cambridge Isotope Laboratories, Inc., MA) was infused (constant rate, 0.15 μmol/kg FFM/min; prime, 6.4 μmol/kg FFM) throughout the experimental protocol to enrich the muscle free amino acid pool with labeled leucine and measure the rate of synthesis of muscle proteins. Synthesis rate of muscle proteins was measured by determining the stable isotopic enrichment of the proteins with d9-leucine, and as we have previously described (Tran et al., 2015). Biopsy samples were collected from the vastus lateralis muscle at 120 and 300 mins after the start of the labeled leucine infusion. Following removal of visible fat and connective tissue, samples were blotted dry and stored in liquid nitrogen until analyses. Blood samples were collected for the determination of labeled leucine and various blood chemistry variables prior to the initiation of the infusion and again at 110, 115, 140, 260, 280, and 300 mins following the start of the labeled leucine infusion.
Analyses of samples
Preparation of samples for stable isotope enrichment determination
The isotopic enrichment of blood and mixed-muscle protein with d9-leucine, expressed as molar percent excess (MPE), was measured using HPLC-MS/MS, by first isolating the amino acids and following procedures similar to those we have used in the past (Tran et al., 2015). Blood samples collected during the infusion study was transferred into glass tubes containing 15% sulfosalicylic acid (SSA) and the tubes were vortexed several times. These tubes were then centrifuged at 2,500 x g for 15 min at 4°C, and the supernatant containing the amino acids was collected. These amino acids were isolated using cation-exchange columns (AG 50W-8× 100–200-mesh; Bio-Rad Laboratories, Inc., CA), which were conditioned with 3 ml of 2N NH4OH and 3 ml of 1N HCl prior to the addition of the samples (i.e., 500 ul of the blood/SSA mixture supernatant). The amino acids were eluted from the columns using 8 ml of 2N NH4OH. Proteins in the muscle samples (i.e., z15 mg of tissue) were precipitated using 500 uL of 5% SSA. The samples were homogenized, centrifuged at 2,500 x g for 45 min at 4°C, and the resulting pellet was collected. The samples were washed with 500 ul of 5% SSA two more times, and prior to collection of the final pellet that included the muscle proteins. This pellet was first washed with 1 ml of ethanol and then with 1 ml of ethyl ether, and it was then let to dry overnight at 50°C. Proteins in these samples were hydrolyzed with 6 N HCl at 110°C over 24 hours. The protein hydrolysate was added to cation-exchange columns (AG 50W-8× 200–400-mesh; Bio-Rad Laboratories, Inc., CA), following conditioning of the columns as described for blood (above), and in order to isolate/purify the amino acids from the proteins in the muscle samples. The amino acids were eluted from the columns with 8 ml of 2N NH4OH.
Tryptic peptides for the determination of d9-leucine enrichment of β-F1-ATPase were produced after purification of the muscle β-F1-ATPase (Everman et al., 2011). Specifically, following homogenization of the muscle (see “Protein Immunoblot Assays” section below), muscle β-F1-ATPase was purified from the homogenate using immunoprecipitation. First, a mouse monoclonal β-F1-ATPase-specific antibody (Cat# ab5432, RRID:AB_304883, Abcam) was coupled to protein G agarose beads, and by rotation of the antibody/bead mixture for 1 hr at room temperature. After antibody-bead conjugation, 6 mg of whole-muscle lysate protein and homogenization buffer were added to the mixture, which was incubated overnight at 4°C. The beads were subsequently washed four times with PBS and the proteins were denatured and eluted from the beads by incubation for 30 min at 37°C in 17 µL of SDS-containing loading buffer. The immunoprecipitated protein was resolved using 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and a band corresponding to β-F1-ATPase was excised from the gel and cut into small pieces. The gel pieces were placed into microcentrifuge tubes and washed with 400 uL of deionized water. Coomassie stain was removed by washing two times with 300 uL of 50% acetonitrile (ACN) in 40 mM NH4HCO3, and the gel pieces were dehydrated with 100% ACN. The ACN was removed and the gel pieces were dried further in a vacuum centrifuge at 62°C for 30 min. Peptides in the samples were generated by using trypsin (250 ng) in 40 mM NH4HCO3 and the digestion left to continue overnight at 37°. The digestion was stopped using 5% formic acid (FA). Following incubation of the samples at 37 °C for 30 min and brief centrifugation for 1 min the collected supernatant was transferred to a clean polypropylene tube. The peptides were purified using solid-phase extraction (C18 ZipTip; Millipore) after sample loading in 0.05% heptafluorobutyric acid:5% FA and elution with 4 uL 50% ACN:1% FA and 4 uL 80% ACN:1% FA. The eluates were combined, and after the volume of the sample was reduced (i.e., z2 µL) by vacuum centrifugation, 20 µl of 0.05% heptafluorobutyric acid:1% FA:2%ACN was added to the sample.
Stable isotope enrichment determination by mass spectrometry
The isotopic enrichment of amino acids was measured by LC-MS/MS, using the isobutyl ester derivative of the amino acids (Tran et al., 2015). In the blood samples, selected reaction monitoring was applied for transitions of m/z 188 → 86 and 197 → 95 for the quantification of m + 0 and m + 9 leucine isotopes, respectively. For the determination of leucine isotopic enrichment in the muscle samples, transitions of m/z 190 → 88 (m + 2) and 197 → 95 (m + 9) were quantified and m + 9 / m + 0 enrichment was calculated using a corresponding calibration curve. (i.e, MPE vs m + 9 / m + 2).
The isotopic enrichment of the β-F1-ATPase was determined based the enrichment of the β-F1-ATPase134–143 peptide with d9-leucine, which was measured following mass spectrometry procedures we have previously developed (Everman et al., 2011). For the HPLC-ESI-MS/MS analysis we used a Thermo Electron Orbitrap Elite Velos Pro fitted with an EASY source (Thermo Electron, San Jose, CA). HPLC separations were accomplished with a linear gradient. Tandem mass spectra were extracted from Xcalibur “RAW” files and charge states were assigned using the Extract_MSN script (Thermo Fisher, CA). MS/MS spectra were analyzed using Mascot (Matrix Science, London, UK; version 2.4.1). The search parameters for the protein peptides were as follows: 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine, and tyrosine, and addition of d9-labeled leucine. Probability assessment of peptide assignments and protein identifications were made through use of Scaffold (version Scaffold_4.3.4, Proteome Software Inc., Portland, OR). The MS/MS peak areas of the following three fragment ions associated with the β-F1-ATPase134–143 peptide were quantified (+1 charge state): y6 (672.4), y7 (729.4) and y8 (828.5). The corresponding fragment ion MS/MS peak areas with +9 Da mass, containing d9-leucine from the β-F1-ATPase134–143 peptide were also quantified. Extracted ion chromatographic peak areas corresponding to d9-leucine labeled and unlabeled fragment ions from the β-F1-ATPase134–143 peptide were generated and they were used for the quantification of the enrichment of the β-F1-ATPase134–143 peptide with d9-leucine. The enrichment was calculated as the ratio of the sum of the areas of the three d9-leucine labeled fragment ions to the sum of the areas of the three corresponding unlabeled fragments ions of the β-F1-ATPase134–143 peptide. The β-F1-ATPase peptide d9-leucine enrichment was converted to amino acid d9-leucine enrichment (i.e., MPE) using a four-point d9-leucine enrichment standard curve. This standard curve was generated by measuring d9-leucine enrichment both at the peptide and amino acid levels (the latter after hydrolysis of the peptides as described for muscle proteins) in mixtures containing known concentrations of commercially-synthesized d9-leucine labeled and unlabeled peptides (AnaSpec, Fremont, CA) corresponding to the β-F1-ATPase134–143 sequence (i.e., same amino acid sequence as the one targeted in the muscle β-F1-ATPase samples).
Protein immunoblot assays
Muscle (z 70 mg) was homogenized in ice-cold freshly prepared buffer (1 ml/100 mg tissue) containing Tris-HCl (20 mM) pH 7.5, NaCl (150 mM), Na2EDTA (1 mM), EGTA (1 mM), NP-40 (1%), sodium deoxycholate (1%), sodium pyrophosphate (2.5 mM), β-glycerophosphate (1mM), Na3VO4 (1mM), and leupeptin (1ug/ml). Following centrifugation at 14,000 x g for 15 min at 4°C, aliquots of the supernatant were stored in −80°C for further sample analyses. Protein concentration in the muscle homogenates was determined by the method of Lowry (Lowry et al., 1951), and β-F1-ATPase abundance was quantified using immunoblotting (Tran et al., 2016). Specifically, supernatant from the muscle homogenate (40 ug of total protein) was diluted (1:1) in a 2X sample buffer and boiled for 5 min at 100ºC. Proteins were separated by SDS-PAGE using Any kD™ gradient polyacrylamide gels (Mini-PROTEAN, Bio-Rad Laboratories, Inc.). After electrophoresis, proteins were transferred to polyvinylidene difluoride membrane following the manufacturer’s protocol. The membrane was then incubated with primary antibody (Cat. # A-21351, RRID:AB_221512, Thermo Fisher Scientific Inc) overnight at 4°C to quantify total abundance of β-F1-ATPase. Prior to incubating the membrane with the primary antibody, the membrane was washed with TBST and blocked with TBST + 5% BSA at room temperature for one hour. The next day, the membrane was washed with TBST and incubated for 1 hour at room temperature with IgG HRP-linked secondary antibody (Cat# sc-516102, RRID:AB_2687626, Santa Cruz Biotechnology). Following visualization of the bands, the membrane was stripped of primary and secondary antibodies using Restore™ Western Blot Stripping Buffer (Cat. # 21059, ThermoFisher Scientific, Inc.) and was subsequently re-probed for GAPDH, which was employed as a loading control, using a GAPDH antibody (Cat. # 600–401-A33, RRID:AB_2107593, Rockland Immunochemicals). Following an overnight incubation with the GAPDH antibody, the membrane was washed and incubated with a secondary antibody (Cat# 7074, RRID:AB_2099233, Cell Signaling Technology). All resulting bands were visualized using the Clarity™ Western ECL Blotting Substrate kit (Bio-Rad, Hershey, PA), and imaged using the ImageQuant LAS 4000 (GE Healthcare, Wauwatosa, WI). Densitometry analysis was performed using the ImageJ software (National Institutes of Health, Bethesda, MD).
mRNA quantification assays
Determination of β-F1-ATPase mRNA expression was performed in muscle (z30 mg) homogenized in TRIzol® reagent, and after extraction of total RNA. The extracted RNA was purified using the RNeasy MinElute Cleanup Kit (Cat. # 74204, Qiagen Inc, Germantown, MD). The mRNA expressions of β-F1-ATPase and GAPDH were quantified by qRT-PCR. Synthesis of cDNA was performed from the isolated RNA using the ABI High Capacity cDNA Reverse Transcription kit (Cat. # 4368814, Thermo Fisher Scientific Inc.). Real time PCR was performed on an Applied Biosystems 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific Inc.) by utilizing TaqMan® gene expression assays for β-F1-ATPase and GAPDH (Thermo Fisher Scientific Inc.), and by following the manufacturer’s protocols.
ATP synthase specific activity
ATP synthase specific activity was measured in muscle homogenates using a commercially available enzyme activity assay kit (Cat. # ab109716, Abcam, Cambridge, MA). After addition of 50 ug of protein from muscle homogenate into microplate wells, the enzyme was immunocaptured within the wells using pre-coated antibodies. Its activity was measured as the rate of ATP hydrolysis to ADP, and where ultimately production of ADP is coupled to the oxidation of NADH to NAD+, and monitored as the decrease in absorbance at 340 nm (i.e., nm/minute/amount of sample loaded into the well). The quantity of the enzyme captured in each well was measured by adding an ATP synthase specific antibody conjugated with alkaline phosphatase. The quantity of the ATP synthase was measured as the change in absorbance at 405 (i.e., nm/minute/amount of sample loaded into the well). Therefore, ATP synthase specific activity was calculated as the ratio of the absorbance at 340 nm to that at 405 nm, and it is reported as arbitrary units.
Plasma biochemical assays
Plasma glucose concentrations were measured using an automated glucose analyzer (YSI 2300, Yellow Springs, OH), whereas those of plasma insulin were determined using an ELISA kit (ALPCO Diagnostics, Windham, NH). Concentrations of individual plasma non-esterified fatty acid (NEFA) concentrations were measured using HPLC-mass spectrometry (Persson et al., 2010).
Calculations
Protein FSR (%/hour) was determined by the precursor-product approach using the measured d9-leucine isotopic enrichment in blood and the muscle protein(s) of interest (i.e., β-F1-ATPase, mixed-muscle protein) (Tran et al., 2015). The specific activity of ATP synthase was calculated as arbitrary units describing the change in the optical density associated with the activity of the enzyme over the change in the optical density associated with the quantity of the enzyme (i.e., change in alkaline phosphatase activity). Measured β-F1-ATPase mRNA levels were normalized to GAPDH and responses between groups were compared using the comparative CT method (2-ΔΔCT).
Statistical analyses
Data were normally distributed based on the D’Agostino & Pearson and Shapiro-Wilk normality tests. Measurements between lean and obese groups were compared using the Student’s unpaired t-test. Pearson product-moment correlation coefficient (r) was calculated to test the strength of association between variables of interest. Significant difference was set at P ≤ 0.05, and data are presented as mean ± SD. Statistical analyses were performed using the GraphPad Prism software (version 7.0, GraphPad Software, La Jolla, CA).
Results
Subject characteristics
Based on the anthropometric characteristics and blood chemistry profiles of the lean and obese subjects showed in Table 1, the two groups represented typical populations of lean individuals and individuals with obesity. As expected, obese subjects were insulin resistant to glucose metabolism based on OGTT-estimated insulin sensitivity. However, the two groups did not differ in aerobic fitness based on their VO2peak responses (Table 1).
Table 1.
Subject characteristics
| Lean (3M/6F) |
Obese (5M/4F) |
|
|---|---|---|
| Age, y | 37 ± 8 | 37 ± 9 |
| Weight, kg | 66 ± 12 | 100 ± 13* |
| BMI, kg/m2 | 23 ± 2 | 34 ± 3* |
| Fat free mass, kg | 50 ± 11 | 66 ± 9* |
| Body fat, % | 25 ± 8 | 33 ± 8* |
| Waist-to-Hip ratio | 0.80 ± 0.05 | 0.91 ± 0.09* |
| VO2peak, ml/min | 1860 ± 719 | 2331 ± 616 |
| VO2peak, ml/kg/min | 27.7 ± 8.3 | 23.6 ± 6.3 |
| VO2peak, ml/kgFFM/min | 36.4 ± 7.8 | 35.0 ± 7.2 |
| Plasma glucose, mg/dl | 88 ± 8 | 98 ± 15 |
| Plasma insulin, uIU/ml | 4 ± 2 | 11 ± 6* |
| Matsuda-ISI | 9 ± 3 | 5 ± 4* |
| TSH, mU/l | 2.3 ± 1.8 | 1.9 ± 0.9 |
| Plasma lipids, mg/dl | ||
| Triglycerides | 78 ± 31 | 183 ± 138* |
| Total cholesterol | 188 ± 41 | 175 ± 30 |
| HDL-C | 71 ± 17 | 42 ± 9* |
| LDL-C | 102 ± 39 | 94 ± 19 |
| NEFA, umol/l | ||
| PUFA | 109 ± 41 | 108 ± 42 |
| SA | 190 ± 57 | 186 ± 55 |
| MUFA | 204 ± 91 | 184 ± 99 |
Values are means ± SD; BMI, body mass index; VO2peak, peak oxygen uptake; Matsuda-ISI, Matsuda insulin-sensitivity index determined during an oral glucose tolerance test; TSH, thyroid-stimulating hormone; HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol. PUFA, sum of concentrations of polyunsaturated NEFAs (i.e., EPA, linolenic acid, DHA, arachidonic acid, linoleic acid); SA, sum of concentrations of saturated NEFAs (i.e., myristic acid, palmitic acid, stearic acid); MUFA, sum of concentrations of monounsaturated NEFA (i.e., palmitoleic acid, oleic acid)
P < 0.05 vs lean.
β-F1-ATPase synthesis rate
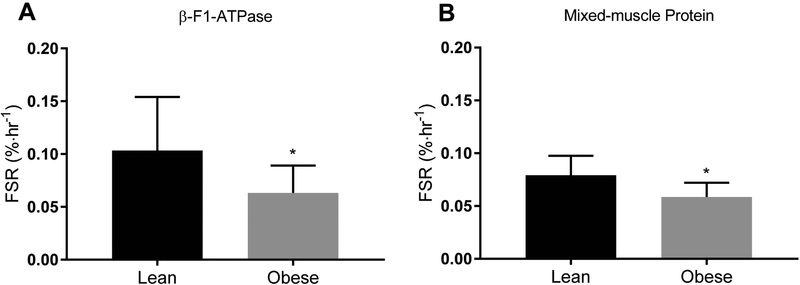
Average blood leucine enrichment (MPE) was not different between the lean (4.1 ± 0.8) and obese (4.3 ± 0.4) groups during the experimental period (P > 0.05). Synthesis rate of β-F1-ATPase was lower in the muscle of the obese subjects (Figure 1A; P < 0.05), along with lower synthesis rate of mixed-muscle protein (Figure 1B; P < 0.05). Mixed-muscle protein synthesis is the most commonly reported measure of protein synthesis in skeletal muscle reflecting the average rate of the synthesis of all proteins within the muscle homogenate. Data on mixed-muscle protein synthesis from some of the subjects studied in the present study have been recently published (Tran et al., 2018). Although not the focus of the present studies, the synthesis rate of mixed-muscle protein is provided herein for comparison purposes with that of β-F1-ATPase. The synthesis rate (%/hour) of β-F1-ATPase (0.104 ± 0.05) tended to be higher than that of the mixed-muscle protein (0.079 ± 0.02) in the lean (P = 0.08) but not in the obese (0.063 ± 0.03 vs 0.059 ± 0.01; P = 0.32) subjects. The ratio of β-F1-ATPase synthesis rate to mixed-muscle protein synthesis rate, however, did not differ between lean (1.3 ± 0.6) and obese (1.1 ± 0.5) subjects (P > 0.05). Across subjects, synthesis rate of β-F1-ATPase correlated with that of mixed-muscle protein (r = 0.46; P = 0.05). Furthermore, synthesis rate of β-F1-ATPase correlated inversely with the plasma insulin concentration (r = −0.49; P < 0.05), but not with the insulin sensitivity to glucose metabolism (i.e., Matsuda index; P > 0.05).
Figure 1.
Fractional synthesis rate (FSR) of the mitochondrial protein β-F1-ATPase (A) and mixed-muscle protein (B) in skeletal muscle of lean and obese subjects. Values are mean ± SD. *P < 0.05 versus lean (unpaired two-sided t test).
β-F1-ATPase gene expression
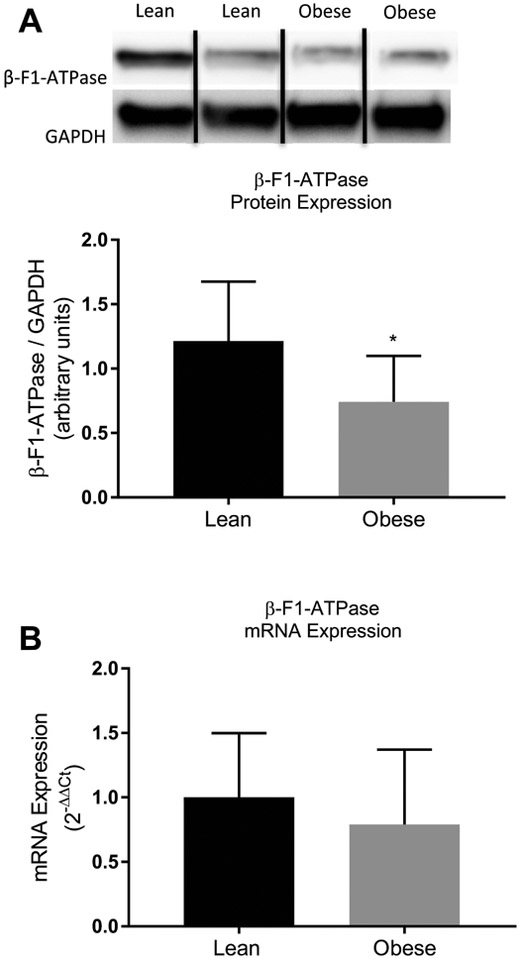
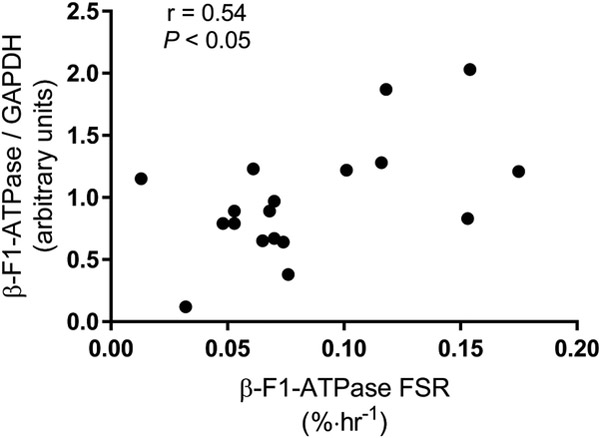
As expected, protein expression of β-F1-ATPase was lower in the obese subjects (Figure 2A; P < 0.05). However, mRNA expression of β-F1-ATPase was not different between lean and obese subjects (Figure 2B; P = 0.32). Across subjects, protein expression of β-F1-ATPase correlated significantly with the synthesis rate of β-F1-ATPase (Figure 3; P < 0.05), and in a way that when the content of the β-F1-ATPase was normalized to the synthesis rate of β-F1-ATPase, the content of β-F1-ATPase was not different between lean (13.4 ± 4.8) and obese (18.6 ± 27.4) subjects (P > 0.05).
Figure 2.
Protein (A) and mRNA (B) expression of β-F1-ATPase in skeletal muscle from lean and obese subjects. Representative Western blots are shown for protein expression. Dividing lines between blots indicate blots from different gels or different parts of the same gel. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. mRNA expression was determined using the comparative CT method (i.e., 2-ΔΔCT). Values are mean ± SD. *P < 0.05 versus lean (unpaired two-sided t test).
Figure 3.
Pearson product-moment correlation (r) between fractional synthesis rate (FSR) of mitochondrial protein β-F1-ATPase and abundance of β-F1-ATPase in skeletal muscle across lean and obese subjects. GAPDH, glyceraldehyde-3-phosphate dehydrogenase was used as a loading control to normalize β-F1-ATPase abundance.
ATP synthase specific activity
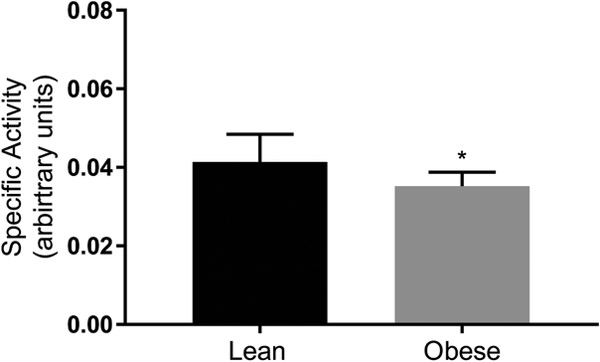
The specific activity of the ATP synthase complex was lower in the obese subjects (Figure 4; P < 0.05). However, the specific activity of the enzyme was not different between the lean and obese subjects when adjusted to the β-F1-ATPase expression (P > 0.05). There was no correlation between the synthesis rate of β-F1-ATPase and the specific activity of the ATP synthase (P > 0.05).
Figure 4.
ATP synthase specific activity in skeletal muscle of lean and obese subjects. ATP synthase activity was measured as the rate of ATP hydrolysis to ADP, ultimately coupled to the oxidation of NADH to NAD+ and monitored as the change in absorbance at 340 nm (i.e., nm/minute/amount of sample loaded into the well). The quantity of the enzyme was measured using an ATP synthase specific antibody conjugated with alkaline phosphatase, and by monitoring the change in absorbance at 405 (i.e., nm/minute/amount of sample loaded into the well). ATP synthase specific activity was calculated as the ratio of the absorbance at 340 nm to that at 405 nm and it is reported as arbitrary units. Values are mean ± SD. *P < 0.05 versus lean (unpaired two-sided t test).
Discussion
The rate of synthesis determined in vivo provides the most direct measure of the rate of biogenesis and renewal of any given protein, in addition to determining the content of that protein. The main findings of the present study are that obese, insulin-resistant individuals have reduced rate of synthesis of β-F1-ATPase in skeletal muscle together with reduced abundance of muscle β-F1-ATPase. However, reduced synthesis rate of β-F1-ATPase could not be attributed to lower muscle β-F1-ATPase mRNA expression. In parallel with lower synthesis rate and content of β-F1-ATPase, we also show that the specific activity of the ATP synthase is lower in the muscle of obese, insulin-resistant humans.
Mixed-protein synthesis traditionally measured in skeletal muscle reflects an average rate of synthesis across all muscle proteins, whose individual rates, however, differ considerably within the skeletal muscle (Jaleel et al., 2008; Shankaran et al., 2016). Previous research has shown almost two-fold higher synthesis rate of mixed-mitochondrial protein when compared to the synthesis rate of mixed-muscle protein in skeletal muscle of young, healthy humans (Rooyackers et al., 1996). The synthesis rate of the mitochondrial β-F1-ATPase in the present study was, however, only 35% higher than that of the mixed-muscle protein. This observation is in line with findings in rodents, and where the synthesis rate of β-F1-ATPase is closer to that of mixed-muscle protein when compared to other mitochondrial proteins (Jaleel et al., 2008). The FSR of β-F1-ATPase in our healthy sedentary lean humans (0.10 ± 0.05 %/hour) was higher than that previously reported in either healthy sedentary (z 0.05 %/hour) (Shankaran et al., 2016) or older overweight/obese (z 0.03 %/hour) (Murphy et al., 2018) humans. In the latter studies, FSR of β-F1-ATPase was measured using oral consumption of deuterated water. Although measurement of protein FSR using deuterated water compares well under certain circumstances with that using the traditional approach that is based on the incorporation of labeled amino acids into protein (Wilkinson et al., 2015b), the deuterated water approach appeared previously to either overestimate (Murphy et al., 2018) or underestimate (Wilkinson et al., 2015b) muscle protein FSR. There is still considerable debate with respect to the details describing the application of the deuterated water approach to accurately measure muscle protein FSR (Fluckey et al., 2015; Wilkinson et al., 2015a). To our knowledge, our study is the first to report the FSR of human muscle β-F1-ATPase using the rather well-established amino acid tracer technique to measure the synthesis rate of a protein in skeletal muscle.
By measuring the synthesis rate of β-F1-ATPase we show that the production rate of β-F1-ATPase is a defect contributing to the reduced content of β-F1-ATPase in the skeletal muscle of obese, insulin-resistant humans. The synthesis rate of β-F1-ATPase showed significant positive correlation with that of mixed-muscle protein across all subjects, and the ratio of β-F1-ATPase synthesis rate to mixed-muscle protein synthesis rate did not differ between lean and obese subjects. These findings suggest that underlying mechanism(s) responsible for the decrease in the synthesis rate of β-F1-ATPase in muscle of humans with obesity might relate directly to those that decrease the synthesis rate of overall protein in the muscle of these individuals. Although the biological mechanism(s) that suppress the synthesis rate at the overall protein level in obese humans are currently unknown, our findings reveal a clear discordance between message expression and synthesis rate specifically for the β-F1-ATPase. Therefore, measurements of the mRNA content of any individual type of protein in muscle of obese humans does not necessarily give accurate description on protein biogenesis occurring in vivo. Our findings show that in obese, insulin-resistant humans expression of β-F1-ATPase is impaired specifically at the level of mRNA translation.
Primary role of mitochondrial proteins in muscle is to defend against challenges in energy homeostasis by generating ATP. We found that the specific activity of the ATP synthase is reduced in muscle of obese individuals, which is in line with recent findings on the activity of this enzyme determined in primary culture of skeletal muscle cells from obese subjects (Formentini et al., 2017). Moreover, previous research has documented reduced rate of ATP synthesis determined in vivo in muscle of individuals with increased body fat (Newcomer et al., 2001) and insulin resistance (Petersen et al., 2004). Our findings show that obesity, and the associated insulin resistance, impair specifically the function of the mitochondrial ATP synthase, and that this effect is seen together with reduced abundance of its catalytic β-F1-ATPase subunit. Although the specific activity of the ATP synthase was reduced in muscle samples from the obese subjects, it was not different between the lean and the obese subjects when adjusted to the immunoblot-determined β-F1-ATPase expression. This latter observation would be consistent with a notion that the capacity of the ATP synthase enzyme is not altered in obesity/insulin resistance, unless the content of its individual subunits is reduced in the metabolic setting of human obesity. Moreover, lack of correlation between the synthesis rate of β-F1-ATPase and the specific activity of the ATP synthase suggests that the synthesis, and thus renewal, rate of β-F1-ATPase is not a direct determinant of the capacity of the ATP synthase to generate ATP. In line with this observation, discordance between mixed-mitochondrial protein synthesis rate (i.e., increased) and mitochondrial oxidative capacity (i.e., decreased) has been previously shown in muscle of rodents (Chanseaume et al., 2007). Therefore, our overall findings show that lower capacity for ATP synthesis in muscle of obese, insulin-resistant humans is the result of a decrease in the abundance of muscle β-F1-ATPase, rather than impaired turnover rate of β-F1-ATPase, per se, in the muscle of these individuals.
It is likely that the rates of synthesis of other subunits of the muscle ATP synthase complex previously shown also to be lower in obesity, such as the α-F1-ATPase (Formentini et al., 2017; Hojlund et al., 2010), are reduced in obese, insulin-resistant humans. Given the amount of tissue biopsy required to determine the stable isotope enrichment of individual proteins using the experimental approach of the present study, it was not feasible to measure the synthesis rate across all the subunits of the ATP synthase complex. Nevertheless, recent evidence shows that not all of the currently characterized 17 subunits forming the ATP synthase complex (Fujikawa et al., 2015) are decreased in obesity (Formentini et al., 2017). Such evidence points to distinct metabolic regulation of the content of the individual proteins forming the ATP synthase complex in skeletal muscle. Furthermore, and although a coordinated regulation might occur at the transcriptional level, our data show that responses can ultimately be modified, and as we show with the β-F1-ATPase, at the translational level in determining the content of the individual ATP synthase subunits in muscle. Our study is, therefore, novel in its ability to provide a comprehensive understanding of the regulation of the catalytic β-F1-ATPase subunit of the ATP synthase complex by linking its synthesis rate to the function of the overall ATP synthase complex via reduced abundance of β-F1-ATPase. It is reasonable to conclude that reduced abundance of muscle β-F1-ATPase is due to lower synthesis rate of muscle β-F1-ATPase, given the significant correlation we observed between muscle β-F1-ATPase content and muscle β-F1-ATPase synthesis rate across the study subjects.
In the present study, the synthesis rate of β-F1-ATPase did not correlate with the insulin sensitivity, as estimated from the OGTT-determined Matsuda insulin sensitivity index (Matsuda and DeFronzo, 1999). This implies that insulin sensitivity to glucose metabolism is not a direct factor on regulating the synthesis rate of mitochondrial β-F1-ATPase under basal/non-insulin-stimulated conditions. On the other hand, we have previously shown that exposure of primary muscle cells to fatty acids reflecting a plasma free fatty acid profile representative of human obese phenotype decreases the protein expression of β-F1-ATPase (Tran et al., 2016). Under those conditions, the decrease in β-F1-ATPase expression was observed together with increased expression of a specific microRNA (i.e., miR-127–5p) (Tran et al., 2016). Also, experiments in β-cell lines show that lipotoxicity, which is a common manifestation in skeletal muscle of individuals with obesity (Coen et al., 2010), decreases β-F1-ATPase expression in parallel with decrease in intracellular ATP concentrations (Kohnke et al., 2007). Such evidence suggests that the metabolic environment associated specifically with the lipid metabolism in obesity/insulin resistance may be a primary factor contributing to the reduced muscle β-F1-ATPase by interfering with the β-F1-ATPase mRNA translation, and ultimately suppressing the capacity for ATP synthesis in humans with obesity.
Impaired ATP synthase activity may not only decrease muscle ATP levels as previously reported in obesity (Lennmarken et al., 1986), but it may also play role in determining insulin sensitivity. Although whether impairments in mitochondrial function are cause or consequence of insulin resistance is still controversial, impaired ATP synthase activity linked to lower β-F1-ATPase content reduces fatty acid oxidation in skeletal muscle cells resulting in lipid accumulation in the cytosol (Formentini et al., 2017). Reduced oxidation of fatty acids is indeed a manifestation of impaired substrate metabolism in muscle of obese individuals (Kim et al., 2000), resulting in accumulation of lipid mediators such as diacylglycerols in the cytosol that interfere with the insulin signaling (Montgomery and Turner, 2015). Impaired ATP synthase function may also increase reactive oxygen species (ROS) production, and similar to the increase in ROS production when the ATP synthase is experimentally inhibited in vitro by oligomycin (Roy et al., 2008). In turn, ROS production can interfere with insulin signaling in muscle (Henriksen et al., 2011). Furthermore, decreased capacity for ATP synthesis and associated lipid oxidation in the muscle of obese individuals can lead to decreased capacity for physical activity (Rogge, 2009), which is evident in people with obesity (Cooper et al., 2000), decreasing total daily energy expenditure and exasperating weight gain in individuals with obesity (Ravussin et al., 1988).
Studies employing animal models show that physical exercise can upregulate the content of β-F1-ATPase in muscle (Gonzalez et al., 2000). Also, exercise training in lean, healthy humans upregulates muscle ATP synthase flux (Kacerovsky-Bielesz et al., 2009). Given this evidence, and despite studying sedentary individuals with aerobic fitness (i.e., VO2peak) that was comparable between groups, we cannot exclude the possibility that subjects with obesity had lower overall fitness and/or physical activity levels. Therefore, and given that it was not experimentally feasible to characterize the everyday physical activity patterns of our subjects in a definitive manner, our study is possibly limited in its ability to attribute the observed findings on β-F1-ATPase metabolism and ATP synthase activity on obesity, per se.
Conclusions
We show lower rate of synthesis of β-F1-ATPase, which forms the catalytic subunit of the enzyme responsible for ATP generation, in skeletal muscle of humans with obesity/insulin resistance. Lower rate of synthesis of β-F1-ATPase is associated with lower content of β-F1-ATPase, which in turn appears to contribute to reduced capacity for ATP generation via the ATP synthase in the muscle of humans with obesity/insulin resistance.
New Findings.
-
What is the central question of this study?
Humans with obesity have lower ATP synthesis in muscle along with lower content of the beta subunit of the ATP synthase (β-F1-ATPase), the catalytic component of the ATP synthase. Does lower synthesis rate of β-F1-ATPase in muscle contribute to these responses in humans with obesity?
-
What is the main finding and its importance?
Humans with obesity have lower synthesis rate of β-F1-ATPase and ATP synthase specific activity in muscle. These findings indicate that reduced production of subunits forming the ATP synthase in muscle may contribute to impaired generation of ATP in obesity.
Acknowledgments
The authors thank the nurses and recruitment staff at the Clinical Studies Infusion Unit at Mayo Clinic in Scottsdale, Arizona, for help with subject recruitment and the conduct of the experiments. We also thank Matthew R. Buras from the Mayo Clinic in Arizona Biostatistics Core for providing expertise related to the statistical aspects of the study.
Funding
The study was supported by National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant R01DK094062 (CSK) and the Mayo Clinic Metabolomics Resource Core through grant U24DK100469 from the NIH/NIDDK.
Footnotes
Competing interests
None declared.
References
- Abdul-Ghani MA, Jani R, Chavez A, Molina-Carrion M, Tripathy D & Defronzo RA (2009). Mitochondrial reactive oxygen species generation in obese non-diabetic and type 2 diabetic participants. Diabetologia 52(4), 574–582. doi: 10.1007/s00125-009-1264-4 [DOI] [PubMed] [Google Scholar]
- Chanseaume E, Barquissau V, Salles J, Aucouturier J, Patrac V, Giraudet C, Gryson C, Duche P, Boirie Y, Chardigny JM & Morio B (2010). Muscle mitochondrial oxidative phosphorylation activity, but not content, is altered with abdominal obesity in sedentary men: synergism with changes in insulin sensitivity. J Clin Endocrinol Metab 95(6), 2948–2956. doi: jc.2009–1938 [DOI] [PubMed] [Google Scholar]
- Chanseaume E, Giraudet C, Gryson C, Walrand S, Rousset P, Boirie Y & Morio B (2007). Enhanced muscle mixed and mitochondrial protein synthesis rates after a high-fat or high-sucrose diet. Obesity (Silver Spring) 15(4), 853–859. doi: 10.1038/oby.2007.582 [DOI] [PubMed] [Google Scholar]
- Coen PM, Dube JJ, Amati F, Stefanovic-Racic M, Ferrell RE, Toledo FG & Goodpaster BH (2010). Insulin resistance is associated with higher intramyocellular triglycerides in type I but not type II myocytes concomitant with higher ceramide content. Diabetes 59(1), 80–88. doi: 10.2337/db09-0988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AR, Page A, Fox KR & Misson J (2000). Physical activity patterns in normal, overweight and obese individuals using minute-by-minute accelerometry. Eur J Clin Nutr 54(12), 887–894. [DOI] [PubMed] [Google Scholar]
- Everman S, Yi Z, Langlais P, Mandarino LJ, Luo M, Roberts C & Katsanos CS (2011). Reproducibility of an HPLC-ESI-MS/MS method for the measurement of stable-isotope enrichment of in vivo-labeled muscle ATP synthase beta subunit. PLoS One 6(10), e26171. doi: 10.1371/journal.pone.0026171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckey JD, Lambert BS, Greene NP, Shimkus KL, Cardin JM, Riechman SE & Crouse SF (2015). Reply to letter to the editor: to D2O or not to D2O? What are the reasons we D2O it at all? Am J Physiol Endocrinol Metab 308(10), E928–931. doi: 10.1152/ajpendo.00136.2015 [DOI] [PubMed] [Google Scholar]
- Formentini L, Ryan AJ, Galvez-Santisteban M, Carter L, Taub P, Lapek JD Jr., Gonzalez DJ, Villarreal F, Ciaraldi TP, Cuezva JM & Henry RR (2017). Mitochondrial H(+)-ATP synthase in human skeletal muscle: contribution to dyslipidaemia and insulin resistance. Diabetologia 60(10), 2052–2065. doi: 10.1007/s00125-017-4379-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa M, Sugawara K, Tanabe T & Yoshida M (2015). Assembly of human mitochondrial ATP synthase through two separate intermediates, F1-c-ring and b-e-g complex. FEBS Lett 589(19 Pt B), 2707–2712. doi: 10.1016/j.febslet.2015.08.006 [DOI] [PubMed] [Google Scholar]
- Gonzalez B, Hernando R & Manso R (2000). Stress proteins of 70 kDa in chronically exercised skeletal muscle. Pflugers Arch 440(1), 42–49. [DOI] [PubMed] [Google Scholar]
- Henriksen EJ, Diamond-Stanic MK & Marchionne EM (2011). Oxidative stress and the etiology of insulin resistance and type 2 diabetes. Free Radic Biol Med 51(5), 993–999. doi: 10.1016/j.freeradbiomed.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojlund K, Yi Z, Lefort N, Langlais P, Bowen B, Levin K, Beck-Nielsen H & Mandarino LJ (2010). Human ATP synthase beta is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle. Diabetologia 53(3), 541–551. doi: 10.1007/s00125-009-1624-0 [DOI] [PubMed] [Google Scholar]
- Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Hojlund K, Yi Z & Mandarino LJ (2010). Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes 59(1), 33–42. doi: db09–0214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleel A, Henderson GC, Madden BJ, Klaus KA, Morse DM, Gopala S & Nair KS (2010). Identification of de novo synthesized and relatively older proteins: accelerated oxidative damage to de novo synthesized apolipoprotein A-1 in type 1 diabetes. Diabetes 59(10), 2366–2374. doi: 10.2337/db10-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleel A, Short KR, Asmann YW, Klaus KA, Morse DM, Ford GC & Nair KS (2008). In vivo measurement of synthesis rate of individual skeletal muscle mitochondrial proteins. Am J Physiol Endocrinol Metab 295(5), E1255–1268. doi: 10.1152/ajpendo.90586.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacerovsky-Bielesz G, Chmelik M, Ling C, Pokan R, Szendroedi J, Farukuoye M, Kacerovsky M, Schmid AI, Gruber S, Wolzt M, Moser E, Pacini G, Smekal G, Groop L & Roden M (2009). Short-term exercise training does not stimulate skeletal muscle ATP synthesis in relatives of humans with type 2 diabetes. Diabetes 58(6), 1333–1341. doi: 10.2337/db08-1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Hickner RC, Cortright RL, Dohm GL & Houmard JA (2000). Lipid oxidation is reduced in obese human skeletal muscle. Am J Physiol Endocrinol Metab 279(5), E1039–1044. [DOI] [PubMed] [Google Scholar]
- Kohnke R, Mei J, Park M, York DA & Erlanson-Albertsson C (2007). Fatty acids and glucose in high concentration down-regulates ATP synthase beta-subunit protein expression in INS-1 cells. Nutr Neurosci 10(5–6), 273–278. [DOI] [PubMed] [Google Scholar]
- Lennmarken C, Sandstedt S, von Schenck H & Larsson J (1986). Skeletal muscle function and metabolism in obese women. JPEN J Parenter Enteral Nutr 10(6), 583–587. doi: 10.1177/0148607186010006583 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL & Randall RJ (1951). Protein measurement with the Folin phenol reagent. J Biol Chem 193(1), 265–275. [PubMed] [Google Scholar]
- Matsuda M & DeFronzo RA (1999). Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22(9), 1462–1470. [DOI] [PubMed] [Google Scholar]
- Montgomery MK & Turner N (2015). Mitochondrial dysfunction and insulin resistance: an update. Endocr Connect 4(1), R1–R15. doi: 10.1530/EC-14-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser TL, Kenan DJ, Ashley TA, Roy JA, Goodman MD, Misra UK, Cheek DJ & Pizzo SV (2001). Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by angiostatin. Proc Natl Acad Sci U S A 98(12), 6656–6661. doi: 10.1073/pnas.131067798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CH, Shankaran M, Churchward-Venne TA, Mitchell CJ, Kolar NM, Burke LM, Hawley JA, Kassis A, Karagounis LG, Li K, King C, Hellerstein M & Phillips SM (2018). Effect of resistance training and protein intake pattern on myofibrillar protein synthesis and proteome kinetics in older men in energy restriction. J Physiol 596(11), 2091–2120. doi: 10.1113/JP275246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer BR, Larson-Meyer DE, Hunter GR & Weinsier RL (2001). Skeletal muscle metabolism in overweight and post-overweight women: an isometric exercise study using (31)P magnetic resonance spectroscopy. Int J Obes Relat Metab Disord 25(9), 1309–1315. doi: 10.1038/sj.ijo.0801673 [DOI] [PubMed] [Google Scholar]
- Persson XM, Blachnio-Zabielska AU & Jensen MD (2010). Rapid measurement of plasma free fatty acid concentration and isotopic enrichment using LC/MS. J Lipid Res 51(9), 2761–2765. doi: 10.1194/jlr.M008011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R & Shulman GI (2004). Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350(7), 664–671. doi: 10.1056/NEJMoa031314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravussin E, Lillioja S, Knowler WC, Christin L, Freymond D, Abbott WG, Boyce V, Howard BV & Bogardus C (1988). Reduced rate of energy expenditure as a risk factor for body-weight gain. N Engl J Med 318(8), 467–472. doi: 10.1056/NEJM198802253180802 [DOI] [PubMed] [Google Scholar]
- Rogge MM (2009). The role of impaired mitochondrial lipid oxidation in obesity. Biol Res Nurs 10(4), 356–373. doi: 10.1177/1099800408329408 [DOI] [PubMed] [Google Scholar]
- Rooyackers OE, Adey DB, Ades PA & Nair KS (1996). Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proc Natl Acad Sci U S A 93(26), 15364–15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Ganguly A, BoseDasgupta S, Das BB, Pal C, Jaisankar P & Majumder HK (2008). Mitochondria-dependent reactive oxygen species-mediated programmed cell death induced by 3,3’-diindolylmethane through inhibition of F0F1-ATP synthase in unicellular protozoan parasite Leishmania donovani. Mol Pharmacol 74(5), 1292–1307. doi: 10.1124/mol.108.050161 [DOI] [PubMed] [Google Scholar]
- Shankaran M, King CL, Angel TE, Holmes WE, Li KW, Colangelo M, Price JC, Turner SM, Bell C, Hamilton KL, Miller BF & Hellerstein MK (2016). Circulating protein synthesis rates reveal skeletal muscle proteome dynamics. J Clin Invest 126(1), 288–302. doi: 10.1172/JCI79639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L, Hanavan PD, Campbell LE, De Filippis E, Lake DF, Coletta DK, Roust LR, Mandarino LJ, Carroll CC & Katsanos CS (2016). Prolonged Exposure of Primary Human Muscle Cells to Plasma Fatty Acids Associated with Obese Phenotype Induces Persistent Suppression of Muscle Mitochondrial ATP Synthase beta Subunit. PLoS One 11(8), e0160057. doi: 10.1371/journal.pone.0160057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L, Kras KA, Hoffman N, Ravichandran J, Dickinson JM, D’Lugos A, Carroll CC, Patel SH, Mandarino LJ, Roust L & Katsanos CS (2018). Lower Fasted-State but Greater Increase in Muscle Protein Synthesis in Response to Elevated Plasma Amino Acids in Obesity. Obesity (Silver Spring) 26(7), 1179–1187. doi: 10.1002/oby.22213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L, Masters H, Roust LR & Katsanos CS (2015). A new method to measure muscle protein synthesis in humans by endogenously introduced d9-leucine and using blood for precursor enrichment determination. Physiol Rep 3(8), e12479. doi: 10.14814/phy2.12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen Z, Li S, Zhang Y, Jia S, Li J, Chi Y, Miao Y, Guan Y & Yang J (2014). Hepatic overexpression of ATP synthase beta subunit activates PI3K/Akt pathway to ameliorate hyperglycemia of diabetic mice. Diabetes 63(3), 947–959. doi: 10.2337/db13-1096 [DOI] [PubMed] [Google Scholar]
- Wilkinson DJ, Atherton PJ, Phillips BE, Greenhaff PL & Smith K (2015a). Application of deuterium oxide (D2O) to metabolic research: just D2O it? Depends just how you D2O it! Am J Physiol Endocrinol Metab 308(9), E847. doi: 10.1152/ajpendo.00581.2014 [DOI] [PubMed] [Google Scholar]
- Wilkinson DJ, Cegielski J, Phillips BE, Boereboom C, Lund JN, Atherton PJ & Smith K (2015b). Internal comparison between deuterium oxide (D2O) and L-[ring-13C6] phenylalanine for acute measurement of muscle protein synthesis in humans. Physiol Rep 3(7). doi: 10.14814/phy2.12433 [DOI] [PMC free article] [PubMed] [Google Scholar]