Abstract
Eliminating or silencing a gene’s level of activity is one of the classic approaches developmental biologists employ to determine a gene’s function. A recently developed method of gene perturbation called CRISPR-Cas, which was derived from a prokaryotic adaptive immune system, has been adapted for use in eukaryotic cells. This technology has been established in several model organisms as a powerful and efficient tool for knocking out or knocking down the function of a gene of interest. It has been recently shown that CRISPR-Cas functions with fidelity and efficiency in Ciona robusta. Here, we show that in C. robusta CRISPR-Cas mediated genomic knock-ins can be efficiently generated. Electroporating a tissue-specific transgene driving Cas9 and a U6-driven gRNA transgene together with a fluorescent protein-containing homology directed repair (FP-HDR) template results in gene-specific patterns of fluorescence consistent with a targeted genomic insertion. Using the Tyrosinase locus to optimize reagents, we first characterize a new Pol III promoter for expressing gRNAs from the C. savignyi H1 gene, and then adapt technology that flanks gRNAs by ribozymes allowing cell-specific expression from Pol II promoters. Next, we examine homology arm-length efficiencies of FP-HDR templates. Reagents were then developed for targeting Brachyury and Pou4 that resulted in expected patterns of fluorescence, and sequenced PCR amplicons derived from single embryos validated predicted genomic insertions. Finally, using two differentially colored FP-HDR templates, we show that biallelic FP-HDR template insertion can be detected in live embryos of the F0 generation.
Keywords: ascidian, Ciona, CRISPR-Cas9, homology-directed repair, genome editing
Introduction
In the few years since its development, CRISPR-Cas technology has proved to be an important biotechnological advancement used for a wide range of applications including genomic knock-outs (Jinek et al., 2012), transcriptional repression (Qi et al., 2013), and genomic knock-ins (Gokcezade et al., 2014; Kimura et al., 2014). Adapted from a prokaryotic adaptive immune system, CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated) relies on a Streptococcus pyogenes Cas9 nuclease that pairs with an artificial single guide RNA molecule containing a genomic target of 17–20 bp that provides sequence-specific nuclease activity to the Cas9-gRNA complex. CRISPR-Cas’s baseline effect, double-strand breaks that result in insertion-deletion mutations, has been demonstrated in nearly every experimental model where the components can be delivered (Ma and Liu, 2015). In the ascidian Ciona robusta (formerly C. intestinalis species A), CRISPR-Cas has been shown to efficiently cleave genes of interest via injection of Cas9 mRNA and gRNAs (Sasaki et al., 2014) and, through electroporation of plasmid transgene components (Gandhi et al., 2017; Stolfi et al., 2014).
When CRISPR-Cas reagents are combined with a homology directed repair (HDR) template, it is possible to generate precise genomic edits and sequence insertions by taking advantage of endogenous cellular HDR mechanisms (Gaj et al., 2013). Introduced sequences of DNA that include stretches of genomic homology flanking a designated mutation or insertion site are used by the cell as a template to repair the double-strand break created by the Cas9 nuclease. Single-strand oligodeoxynucleotides (ssODNs) between 100–200 bp with homology arms of ~50 bp have been shown to effectively introduce single bp changes, or to insert a FLAG-tag sequence (Singh et al., 2015). For larger modifications, e.g. the insertion of a fluorescent protein (FP) sequence (~750 nt), homology arms of the insert size or larger are more effective (Li et al., 2014).
Here, we take advantage of our ability to introduce DNA constructs into thousands of fertilized Ciona eggs via electroporation to develop a transgene-based approach to perform precise genomic modifications using CRISPR-Cas transgenes and transgenes containing FP-HDR templates. Using the melanocyte gene Tyrosinase (Tyr), in which introduced phenotypes are easily scorable, we describe the generation of FP-HDR reagents and discuss ways to optimize template-based HDR editing. Embryos and larvae that expressed fluorescent proteins in the predicted cells, a result of correct HDR-mediated insertions, were identified with fluorescent microscopy in vivo. Importantly, it was almost never the case that we detected cells that expressed fluorescent proteins in control experiments in which gRNA transgenes were omitted, suggesting that fluorescence was not due to cis-regulatory sequences contained within the FP-HDR templates and that the editing process requires all editing reagents to be present. We generated an alternative RNA polymerase (pol) III promoter from the Ciona savignyi H1 (CsH1) gene that effectively facilitated HDR in the melanocytes and show that a gRNAs transcribed from this promoter are expressed a cell cycle earlier than the U6 promoter. Although the H1 promoter is routinely used in vertebrates, its use in ascidians has not yet been reported. Furthermore, we adapted a technique (Gao and Zhao, 2014) that incorporates a pair ribozymes to enable the expression of gRNAs from tissue-specific RNA polymerase II promoters and show that this technique is efficient at eliciting FP-HDR genomic insertions in Ciona. After demonstrating that HDR-mediated editing was possible using various gRNA delivery methods, we analyzed how altering homology arm length affects HDR efficiency. To show that our CRISPR-Cas mediated FP-HDR genomic insertions are broadly applicable in Ciona and not specific to the Tyr locus, we demonstrate FP insertions at two additional loci: Brachyury (Bra), which is expressed exclusively in the notochord, and Pou4, which is specific to the peripheral nervous system (PNS). Confirmation of fluorescent protein insertions at the desired genomic locus was obtained by sequencing PCR amplicons, either from pools of a small number of embryos or from individual embryos. Finally, we used two differently colored FP-HDR templates to show that both alleles in the same cell can be targeted for HDR-mediated genomic insertions simultaneously.
Materials and Methods
Adult C. robusta were collected from marinas in Mission Bay, San Diego, and kept in a recirculating refrigerated tank under constant illumination to allow the accumulation of gametes. Varying amounts of transgene DNAs used for electroporations were pooled together, ethanol precipitated and the pellets dissolved in 0.77M Mannitol. Electroporations were carried out essentially as previously reported (Zeller, 2018; Zeller et al., 2006a) with no relevant modifications, and electroporated embryos were left to develop at 16–18°C in 0.45μm-filtered seawater supplemented with penicillin/streptomycin (10 U/mL and 10μg/mL, respectively) for the duration of the experiment. Embryos developing past mid tailbud stage were anesthetized with MS-222 (Sigma) at 12–15hpf. In this study, a biological replicate refers to an independent electroporation on a different day using gametes derived from different adult animals. All imaging was performed on a Zeiss AxioPlan 2E Imaging epifluorescence microscope. Fluorescent images were captured with a Hamamatsu ORCA-Flash 4.0 V2 monochromatic camera and brightfield images were captured with an AxioCam ICc1 color camera.
Immunohistochemistry was performed as described in Joyce Tang et al. (2013). Briefly, embryos were fixed in 2% paraformaldehyde, washed in PBST and briefly permeabilized in 100% methanol. Embryos were incubated at 4°C overnight with primary antibody (Rabbit-anti-GFP, Thermo Fisher Scientific, A11122; Mouse anti-acetylated tubulin antibody, Sigma, T7451) at 1:2000, washed thoroughly and incubated at 4°C overnight with secondary antibody (Alexa Fluor 546, A11003; Alexa Fluor 488, A-11001: Thermo Fisher Scientific,) at 1:500. Embryos were washed thoroughly, incubated for 10min in 50% glycerol +300nM DAPI, and then transferred to 70% glycerol for imaging.
Whole mount in situ hybridization was performed essentially as in (Satou et al., 2001). We designed and cloned a plasmid that would generate a probe complimentary to the Tyr gRNA sequence that was ~870 nucleotides in length; cloning details described below. Due to the short nature of the gRNA sequence being hybridized to, hybridization was performed at 50°C for 48hrs. After colorimetric development embryos were fixed briefly in 4% paraformaldehyde, washed 2X in PBST, and transferred to 70% glycerol for imaging.
Molecular cloning
All transgenes used in this study were cloned into our custom vector, pZapANX (pZANX), which is a pSP72 plasmid with a cryptic mesenchymal promoter sequence deletion (pSP72–1.27; Zeller et al 2006). Cas9 and the gRNA were obtained from pX330-U6-Chimeric_BB-CBh-hSpCas9 (Cong et al., 2013), Addgene (#42230). F+E gRNA modifications were performed to the gRNA backbone as described in Chen et al. (2013). All C. robusta and C. savignyi fragments were amplified using genomic DNA isolated from sperm. PCR products were generated with Q5 DNA polymerase from New England Biolabs using standard PCR conditions. Transgene names, descriptions and expression patterns are summarized in Table 1.
Table 1.
Descriptions of transgenes that are referenced in the figure legends.
| Transgene Name | Description | Expression Pattern |
|---|---|---|
| Tyr::Cas9 | Tyr cis-reg DNA drives expression of Cas9 | melanocytes |
| Fkh::Cas9 | FoxD cis-reg DNA drives expression of Cas9 | notochord precursors |
| EpiB::Cas9 | EpiB cis-reg DNA drives expression of Cas9 | epidermis |
| SoxB1::Cas9 | SoxB1 cis-reg DNA drives expression of Cas9 | epidermis |
| U6::gRNA(Tyr) | CrU6 cis-reg DNA drives expression of Tyr-targeting gRNAs | ubiquitous |
| H1::gRNA(Tyr) | CsH1 cis-reg DNA drives expression of Tyr-targeting gRNAs | ubiquitous |
| Tyr::RZ-gRNA(Tyr)-RZ | Tyr cis-reg DNA drives expression of the ribozyme-flanked gRNA(Tyr) sequence | melanocytes |
| H1::gRNA(Bra-Cod) | CsH1 cis-reg drives expression of Bra coding region-targeting gRNAs | ubiquitous |
| H1::gRNA(Bra-UTR) | CsH1 cis-reg drives expression of Bra UTR-targeting gRNAs | ubiquitous |
| U6::gRNA(Pou4-UTR) | CrU6 cis-reg drives expression of Pou4 UTR targeting gRNAs | ubiquitous |
| U6::gRNA(GFP) | CrU6 cis-reg drives expression of GFP targeting gRNAs | ubiquitous |
| Tyr FP-HDR(2.0 kB) | YFP flanked by 2.0 kB arms of Tyr DNA | N/A |
| Tyr FP-HDR(1.0 kB) | YFP flanked by 1.0 kB arms of Tyr DNA | N/A |
| Tyr FP-HDR(500bp) | YFP flanked by 500 bp arms of Tyr DNA | N/A |
| Tyr FP-HDR(CFP) | CFP flanked by 1.0 kB arms of Tyr DNA | N/A |
| Tyr FP-HDR(mCherry) | mCherry flanked by 1.0 kB arms of Tyr DNA | N/A |
| Bra FP-HDR | YFP flanked by ~1.3 kB arms of Bra DNA | N/A |
| Pou4 FP-HDR | YFP flanked by ~1.2 kB arms of Pou4 DNA | N/A |
Cloning Cas9 transgenes
Briefly, cis-regulatory DNA was amplified from genomic DNA, digested and purified, and ligated with the Cas9 coding region and pZANX to create the Cas9 transgenes. Tyr (KH.C12.469) cis-regulatory DNA was amplified with 5′-CGCTCTAGAACCATTTTTCATGCCTGACAAATGGCGG −3′ and 5′-CGCGGTACCATGGTTGGAAGTGCTAGACAAATGTGTG −3′. Fkh (KH.C8.396) cis-regulatory DNA was generated with 5′-GCGGCTAAGTCGGCAGTTTCCAAG-3′ and 5′-GACAACATCATTTTTGTACTTGCAAAAGTAATCC-3′. EpiB (KH.C7.154) cis-regulatory DNA was amplified with 5′-GTTTATACATTGCAGTCAAACGAAGCACC-3′ and 5′-CGTTTTCCGTAATTAAAATAGTACCATG-3′. SoxB1 (KH.C1.99) cis-regulatory DNA was amplified with 5′-AGCGTTGCGTGGCCTGCCTCCAGTCG-3′and 5′-CTGTAAAGTAGAAGTTTCAGTTAAAAATC-3′.
Cloning gRNA transgenes
The putative Ciona H1 RNA was first identified from a computational screen for non-coding RNAs (Piccinelli et al., 2005) and has also been annotated in the Ensembl Ciona genome browser as gene ENSCINT00000030147. U6::gRNA and H1::gRNA transgenes were constructed by first amplifying the CrU6 (Fwd: 5′-TGTACCACCACGTCGCAGACG-3′; Rev 5′-GATGGGTCTTCGAGAAGACCTGTTTAAGAGCTATGCTGGAAACAGCATAG-3′) and CsH1 (Fwd: 5′-CTAAAATAAATGTACAGACCTTGGTCATG-3′; Rev: 5′-GCCTGTACTAAATACAAAAGATAGTGTTTTTTAACCGGGTCTTCGAGAAGACCTG-3′) promoters from C. robusta and C. savignyi genomic DNA, respectively. The gRNA sequence was amplified from pX330 with forward primers (U6: 5′-CTGTTTAAGAGCTATGCTGGAAACAGCATAGCAAGTTAAAATAAGGCTAGTCCG-3′; H1: 5′-CCGGGTCTTCGAGAAGACCTGTTTAAGAGCTATGC-3′) and a common reverse primer (5′-GGCACCGAGTCGGTGCTTTTTTGTTTTAGAGGATTCGCG-3′). Overlap PCR was performed to create the empty U6::gRNA and H1::gRNA vectors into which desired targets could be cloned. The sequence of the H1 promoter is as follows: atgcttagctagctagctagctagctgatgcttagctagctagctagctagctgactgatcgatcgtagagcttacgactcagc.
gRNA design began by visually searching for suitable targets that were closest to the desired point of YFP insertion. We performed a BLAST search of each potential target sequence+NGG against the C. robusta genome; targets with potential off-target binding were not considered for use. Targets were added to the U6/H1::gRNA transgenes by BbsI digestion followed by insertion of a phosphotased, complementary pair of annealed oligos that included BbsI overhangs as in Gandhi et al. (2017). Using the CRISPOR website, http://crispor.tefor.net (Haeussler et al., 2016), we assessed specificity and efficacy scores which are reported in Table 2.
Table 2.
gRNA specificity scores for targeted genomic regions
| gene targeted | gRNA sequence used in this study | specificity score | Doench ‘16 efficiency | Mor.-Mateos efficiency |
|---|---|---|---|---|
| Tyr | TGTGGGAGAAAGTGAAATCC | 91 | 51 | 32 |
| Bra-Cod | CGTCACCGGGGGTCATGCAA | not found | 65 | 62 |
| Bra-UTR | CATGATAGATCGAGTGTTTA | 99 | 20 | 33 |
| Pou4 | GTTTCTTCAATTTCGGTGCG | 100 | 59 | 34 |
To generate Tyr::RZ-gRNA(Tyr)-RZ first we used a primer set (Fwd: 5′-GTGAGGACGAAACGAGTAAGCTCGTCGGGTCTTCGAGAAGACCTGTTTAAG-3′; Rev: 5′-GGCACCGAGTCGGTGCTTTTTTGGCCGGCATGGTCCCAGCCTCCTCGCTGG-3′) to amplify the gRNA sequence from our empty U6::gRNA adding ribozyme sequence to either end. We then performed a second PCR using a second primer set (Fwd: 5′-GTGAGGACGAAACGAGTAAGCTCGTCGGGTCTTCGAGAAGACCTGTTTAAG-3′; Rev: 5′- GGCATGGTCCCAGCCTCCTCGCTGGCGCCGGCTGGGCAACATGCTTCGG-3′) using the previous PCR product as template to add additional ribozyme sequence. This fragment was cloned into pZANX. This plasmid was then digested with BbsI and annealed Tyr target oligos were inserted as described above. This incomplete transgene was used as a template for the final PCR reaction, which amplified the final fragment using a third primer set (Fwd: 5′-CCCACACTGATGAGTCCGTGAGGACGAAACG-3′; Rev: 5′-GGCTGGGCAACATGCTTCGGCATGGCGAATGGGAC-3′). This final PCR added the remaining ribozyme sequence. We then combined this fragment with the Tyr cis-reg element and cloned them into pZANX. The complete sequence of the Tyr-targeting gRNA flanked by ribozymes is as follows: cggtacccccacactgatgagtccgtgaggacgaaacgagtaagctcgtctgtgggagaaagtgaaatccgtttaagagctatgctggaaacagcatagcaagttaaaataaggctagtccgttatcaacttgaaaaagtggcaccgagtcggtgcttttttggccggcatggtcccagcctcctcgctggcgccggctgggcaacatgcttcggcatggcgaatgggac (Tyr target in bold; gRNA sequence underlined)
To generate a longer-than-natural probe to detect the short gRNA transcripts for in situ hybridizations we performed an overlap PCR using a 720 bp GFP fragment and the 120 bp Tyr gRNA fragment as template. The GFP fragment, which lacked internal poly-T tracks which would cause Pol III termination, was amplified with the following primers: 5′- actctcggcatggacgagctgtacaagtaaggtaccgcgc-3′ and 5′-ccggtgaacagctcctcgcccttgctcaccattctaaaacaaaaaagc-3′; the Tyr gRNA fragment was amplified with the following primers: 5′- ggcaccgagtcggtgcTTTTTTgttttagaatggtgagc-3′ and 5′- cgcggtaccTGTGGGAGAAAGTGAAATCCGTTTAAGAGC −3′. These fragments were used as templates in an overlap PCR creating a GFP-gRNA fusion 840 bp in length that was then cloned into pBS2KS+. This plasmid was sequenced and then linearized with Acc65I. The antisense Digoxigenin-labeled probe was synthesized with T7 RNA polymerase.
Cloning FP-HDR transgenes
The Tyr FP-HDR(2.0 kB) 5′ arm was amplified with the following primers: 5′-CGTCAGCAGGTTATTCCCTGACGCAG-3′ and 5′-GCAGCCGTTGACGTCATTACGTTACC-3′, and the 3′ arm was amplified with the following primers: 5′- CCTGGTTCGTGAACTTGGCCCG −3′ and 5′- GTGTCATGACGTCACTTCGTTCTTCTCC −3′. These fragments were cloned into pZANX upstream and downstream of GFP.
To generate Tyr FP-HDR(1.0 kB) and Tyr FP-HDR(500 bp), we used Tyr FP-HDR(2.0 kB) as a template for PCR. A fragment that included 1.0 kB homology arms 5′ and 3′ of GFP was generated with 5′- GGTGTCGCAGGTGAGCACAATAAC −3′ and 5′- GCAATGCTACGTGTATCTAGCCG −3, and a fragment that included 500 bp homology arms 5′ and 3′ of GFP was generated with 5′- CCTATATCTGCACTGTAATGTATGCG −3′ and 5′- CAATGAAGTATAAATGTGTTCTAACATAC −3. The fragments were each cloned into pZANX.
Removing GFP from Tyr FP-HDR(1.0 kB) and inserting either the CFP or mCherry sequences generated Tyr FP-HDR(CFP) and Tyr FP-HDR(mCherry).
To generate Bra FP-HDR, the 5′ arm was amplified with a primer set (5′-CAGGAGACAAGGTTCATTGCTGTTACTGC-3′ and 5′-CGCCGCTTACGCCACCTTCTTTG-3) that amplified 1.3kB upstream of the last codon. PAM mutations to the last Bra coding exon were introduced to the fragment with an additional primer set (5′-GGGCGTCATGCAACGTCCACAAGAAG-3′ and 5′-GGGCGTCATGCAACGTCCACAAGAAG-3). The 3′ arm was amplified (Fwd: 5′-TGACGTCACAATGCGAATATAATTATCG-3′ and Rev: 5′- CCCAACACCCTCCATCATACATTCAA-3′) and UTR PAM mutations (27nt downstream of the stop codon) introduced with primers (Fwd: 5′-CAAAGGTTGATTGTCATGATAGATCGAGTGTTTAACCCATGTTCAAGG-3′ and Rev: 5′-CGTCAACTTACAAACGTGACCAAACCTTGAACATGGGTTAAACACTCG-3′). The 5′ fragment was cloned upstream of YFP and the 3′ arm was cloned downstream of YFP into pZANX.
To generate Pou4 FP-HDR, the 5′ arm was amplified with a primer set (5′- CACCCACCCCGCCCCAACC −3′ and 5′- ACATAATCACGTCCTGAAATATTAAATAAATAAAC −3) amplifying 1.3kB upstream of the last codon. The 3′ arm was amplified (5′- TAGAATATGCTGTGACCTCTTCAATGC −3′ and 5′- ACCGAGTCGTTAATATTTCATTTTTTAATG −3) and UTR mutations were introduced with primers (Fwd: 5′-GCACTTTTCACTTCTTCTTGTTTCTTCAATTTCGGTGCGACCTCG-3′ and Rev: 5′-GGAACTGAATAGAAACTTGAAGTCGAGGTCGCACCGAAATTGAAGAAAC-3). The 5′ fragment was cloned upstream of YFP and the 3′ arm was cloned downstream of YFP into pZANX.
Cloning and sequencing of genomic insertions
The Brachyury-YFP PCR fragment was amplified from pools of 10 YFP-positive embryos using a primer that annealed outside the Bra FP-HDR template (5’-CAGGAGACAAGGTTCATTGCTGTTACTGC-3’) and a GFP AS primer (5’- TGACGTCACAATGCGAATATAATTATCG −3’). This fragment was used as a template for a second PCR with the GFP AS primer and a primer that was nested inside of the first amplicon yet still annealed outside of the Bra FP-HDR 5′ arm of homology (diagrammed in Supplementary Fig. 2). The Pou4-YFP fragment was generated from single YFP-positive embryos with a primer that annealed outside of the Pou4 FP-HDR template (5’-CCGCCAACATTTAACTCGTCGGTGATGATG-3’) and the GFP AS primer. The Pou4-YFP and Bra-YFP fragments were cloned into pBS2KS+ and sequenced from both ends with T3 Fwd and T7 Rev primers.
Results
To detect HDR in ascidian larvae, we designed our FP-HDR templates to have the fluorescent protein coding region in-frame with the coding region of Tyr, Bra, or Pou4. We hypothesized that if HDR occurred we would be able to detect fluorescence produced from the resulting fusion protein. FP-HDR templates were designed to fuse a fluorescent protein either in-frame with a truncated protein, Tyr, in which we predicted the function of the Tyr protein would be impaired or eliminated; or in-frame with a full-length protein to maintain its function, as with Bra and Pou4. Because we used fluorescence as our detection scheme, it was important to design HDR templates that lacked any functional cis-regulatory elements to reduce the possibility of false-positives. For each gene targeted, all three transgene components were required; only rarely did we score embryos in control groups as exhibiting fluorescent protein expression..
The Tyrosinase locus can be efficiently targeted for FP-HDR
The Ciona robusta tyrosinase gene encodes the rate-limiting enzyme of the melanin synthesis pathway in the two larval melanocytes of the central nervous system (CNS), the ocellus and otolith. Disruption of the Tyr gene eliminates melanin synthesis and produces albino larvae (Jiang et al., 2005; Nishiyama and Fujiwara, 2008). We first generated a transgene that drove ubiquitous expression of gRNAs with the Ciona robusta U6 promoter (CrU6) (Nishiyama and Fujiwara, 2008) that targeted the coding region of the Tyr locus (U6::gRNA(Tyr)). We also generated a transgene that drove Cas9 expression exclusively in the melanocytes by using 1.6 kB of Tyr cis-regulatory DNA (Tyr::Cas9). Electroporation of these constructs resulted in albino embryos, indicating that the Tyr locus was targeted and suggesting that the CRISPR-Cas9 system worked as expected (Supplementary Fig. 1A). To test if we could precisely edit the genome by inserting a specific sequence into the Tyr locus, we constructed an HDR template that inserted a green fluorescent protein (GFP) coding sequence plus a stop codon amino-terminal to the Tyr transmembrane domain (Fig. 1A). This FP-HDR template (Tyr FP-HDR(2.0 kB)) contained 5′ and 3′ homology arms that were each 2.0 kB in length and upon successful recombination the resulting fusion was expected to produce a truncated, non-functioning Tyr protein that would generate GFP fluorescence in the melanocytes. We electroporated equal masses of Tyr::Cas9, U6::gRNA(Tyr), and Tyr FP-HDR(2.0 kB) into fertilized eggs and cultured embryos to larval stage. Because we occasionally detected autofluorescence in the channel used to detect GFP expression, we confirmed this observation by detecting GFP protein with an anti-GFP antibody and a secondary antibody imaged in a channel in which autofluoresnce was not detectable. The larval melanocytes provide a straightforward way to score efficiency of the resulting FP insertions because in any electroporation, larvae had 0/2, 1/2, or 2/2 melanocytes expressing GFP (Fig. 1B). Of 308 larvae examined (4 biological replicates) an average of 58.6% of the embryos exhibited one fluorescing melanocyte while 16.5% had fluorescence in both melanocytes (Fig. 1C). In control groups that lacked the gRNA transgene (n=192, 4 biological replicates), we scored one embryo having fluorescence in one pigment cell (Fig. 1C). These results demonstrated that the CRISPR-Cas system was effective and efficient at inserting a fluorescent protein sequence at the Tyr locus when including an FP-HDR template.
Figure 1. GFP is efficiently inserted into the C. robusta Tyrosinase locus using CRISPR-Cas mediated HDR.
(A) Schematics of HDR construct. The red X indicates the genomic location where gRNAs were targeted; this target site does not exist within the recombination template. Tyr FP-HDR templates have a stop codon at the 3′ end of the GFP sequence (red octagon) producing a truncated Tyr protein fused in-frame with GFP. The same gRNA target sequence was used in each experiment. (B) Examples of embryos with 0 (top), 1 (middle), or 2 (bottom) fluorescing melanocytes; a result of recombination with the HDR template. GFP fluorescence is detected by anti-GFP immunohistochemistry at larval stage. (C) Varying the gRNA delivery technique by electroporating 8 μg each of Tyr::Cas9, Tyr FP-HDR(2.0 kB) and either the CrU6::gRNA(Tyr), CsH1::gRNA(Tyr) or Tyr::RZ-gRNA(Tyr)-RZ resulted in varying percentages of melanocytes expressing GFP. Electroporating CrU6::gRNA(Tyr) resulted in an average of (n=308; 4 biological replicates) 16.5% of embryos displaying fluorescence in both pigment cells, 58.6% of the embryos with one pigment cell fluorescing, and 24.9% of embryos displayed no fluorescence. H1::gRNA(Tyr) resulted in an average of (n=200; 3 biological replicates) 16.4% of embryos displaying fluorescence in both pigment cells, 60.7% of the embryos with one pigment cell fluorescing, and 22.9% of embryos displayed no fluorescence. Electroporating 8 ug each of Tyr::Cas9, Tyr FP-HDR(2.0 kB), and Tyr::RZ-gRNA(Tyr)-RZ resulted in an average of (n=312; 4 biological replicates) 15.1% of embryos that had fluorescence in both pigment cells, 66.3% of the embryos in one pigment cell, and 18.6% of embryos displayed no fluorescence. In control electroporations that did not include a gRNA transgene, one embryo was scored as having GFP expression. In “C”, the different symbols represent different experiments; groups of either triangles, squares, X’s, circles or diamonds represent percentages of embryo phenotypes collected from the same electroporation. The different colors (orange, yellow, grey) indicate percentages of embryos with either 2, 1, or 0 fluorescent melanocytes, respectively. Significance was tested by MANOVA analysis on raw embryo counts followed by pairwise ANOVA with Tukey post-hoc analysis using R. Significant differences were observed for the case of embryos with 1 fluorescent melanocyte when comparing the ribozyme-driven sgRNA to either the U6-driven sgRNA (p adj = 0.007) or the H1-driven sgRNA (p adj = 0.028). There was no difference observed between the U6- or H1-driven sgRNAs (p adj = 0.742). There were no statistically significant differences in the abilities of these sgRNAs to produce embryos with 2 fluorescent melanocytes. (D) gRNAs are expressed from the H1 promoter at the 8-cell stage, a cell cycle earlier than the U6 promoter. 15 μg of either U6::gRNA(Tyr) or H1::gRNA(Tyr) was electroporated into fertilized eggs and embryos were collected and fixed for in situ hybridization every 20 minutes starting at the 4-cell stage until the 64-cell stage. gRNAs expressed from the U6 promoter (upper row) were detected first at the 16-cell stage, while we detected the earliest gRNA transcription from the H1 promoter at the 8-cell stage (bottom row). Expression depicted in images is mosaic due to the nature of electroporations.
The ascidian H1 promoter is effective at expressing gRNAs and facilitating FP-HDR recombination
In mammalian systems, the U6 RNA pol III and the H1 RNA pol III promoters are used for expressing short RNAs that do not require post-transcriptional processing, including gRNAs (Ranganathan et al., 2014). The H1 promoter generates the H1 RNA which is a component of the RNAse P complex (Bartkiewicz et al., 1989). We identified putative H1 promoters in Ciona (see materials and methods) and tested their expression in embryos. Like their mammalian counterparts, the H1 promoters in the two Ciona species are short, only ~100bp long, compared to the Ciona U6 promoter which is ~1.0 kB long. Because the H1 promoter is one-tenth the size of the U6 promoter, more copies of H1 transgenes are present compared to U6 transgenes for the same mass of DNA, which may result in higher levels of gRNA expression. The CsH1 promoter only required a single point mutation to introduce the appropriate restriction enzyme site for cloning gRNAs, compared to the C. robusta promoter; therefore we cloned the CsH1 promoter by amplifying a 97 bp fragment upstream of the H1 sequence. We designed the H1 transgene to express the same Tyr-targeting gRNA sequence (H1::gRNA(Tyr)) as the U6 transgene. We first compared the temporal and spatial activity of the CrU6 and CsH1 promoters by in situ hybridization and observed that while both promoters expressed gRNAs ubiquitously, transcripts from the CsH1 promoter were expressed a full cell cycle earlier than those from the CrU6 promoter (8-cells vs. 16-cells, respectively; Figure 1D). Like the U6 driver, electroporating H1::gRNA and Tyr::Cas9 resulted in albino embryos (data not shown). To examine efficacy of the CsH1 promoter in facilitating HDR, we electroporated equal masses of H1::gRNA(Tyr), Tyr::Cas9, and Tyr FP-HDR(2.0 kB) and detected GFP expression by immunofluorescence at larval stage. Expressing gRNAs with this promoter resulted in an average of 60.7% of embryos expressing YFP in a single melanocyte and 16.4% of embryos expressing YFP in both melanocytes (n=200, 3 biological replicates; Fig. 1C). In control electroporations that excluded H1::gRNA, we detected no fluorescent melanocytes.
Ribozyme-flanked gRNAs can be expressed tissue-specifically with RNA pol II promoters
In Ciona, RNA pol III promoters are not as well characterized as RNA pol II promoters, which often have detailed temporal and spatial expression data (Sierro et al., 2006; Wang and Christiaen, 2012). To avoid concerns that the pol III promoters might not express at high levels or at all in particular cell types of interest, and to prevent expressing guide RNAs in non-targeted cells, we adapted an approach that flanks the gRNAs by ribozymes (RZ) (Gao and Zhao, 2014) and allows them to be expressed by tissue-specific RNA pol II type promoters. By flanking the gRNA with RZs that self cleave at the exact 5′ and 3′ end of the gRNA sequence, the gRNA is consequently “cleaved-out” from the full RZ-gRNA-RZ transcript, thus freeing a precisely formed gRNA molecule to pair with Cas9. We generated a transgene in which the Tyr promoter was used to drive expression of a transcript that included a Hammerhead RZ (Prody et al., 1986) 5′ of the gRNA as well as an HDV RZ (Ferre-D’Amare et al., 1998) 3′ of the gRNA sequence (Tyr::RZ-gRNA(Tyr)-RZ). We examined the RZ construct function by electroporating an equal mass of the Tyr::Cas9 and Tyr::RZ-gRNA(Tyr)-RZ and then assaying for loss of melanization in the melanocytes. This resulted in albino embryos, demonstrating that like the U6 and H1 gRNA drivers, the RZ-flanked gRNA transgene also facilitated CRISPR-Cas targeting of the Tyr locus (data not shown). To examine if the activity of the RZ-flanked gRNA transgene translated to a change in FP-HDR insertion efficiency, we electroporated equal amounts of the Tyr::Cas9, Tyr::RZ-gRNA(Tyr)-RZ and the Tyr FP-HDR construct. Driving ribozyme-flanked gRNAs with the Tyr cis-regulatory element resulted in an average of 66.3% of embryos expressing YFP in a single melanocyte and 15.1% of embryos expressing YFP in both melanocytes (n=312, 4 biological replicates; Fig. 1C). Omitting the RZ construct produced no embryos with fluorescing melanocytes. These results demonstrate the effectiveness of using RNA pol II promoters to express gRNAs in the developing Ciona embryo in a cell-type specific manner.
Reducing the homology arm lengths affects FP-HDR efficiency
It has been reported in other systems that reducing homology arm length below a certain limit can reduce the efficiency of HDR (Li et al., 2014; Song and Stieger, 2017). To determine if homology arm length affected HDR efficiency in our system, we constructed two additional Tyr FP-HDR templates with homology arms of approximately 1.0 kB (Tyr FP-HDR(1.0 kB)), or 0.5 kB (Tyr FP-HDR(500 bp)) that targeted GFP to the same location in the Tyr locus (Fig. 2A). Electroporations were performed using an equal mass ratio of Tyr::Cas9, U6::gRNA(Tyr) and either Tyr FP-HDR(2.0 kB), Tyr FP-HDR(1.0 kB), or Tyr FP-HDR(500 bp). Compared to the 2.0 kB control group, Tyr FP-HDR(1.0 kB) produced approximately the same percentage (n=496; 4 biological replicates) of embryos expressing YFP in one or two pigment cells (59.5% and 21.4%, respectively; Fig. 2B). However, Tyr FP-HDR(500 bp) (n=477; 4 biological replicates), only 48.1% expressed YFP in one pigment cell and only 7.8% of embryos expressed YFP in both pigment cells. Although not statistically significant (p adj = 0.09, Figure 2) the drop-off in efficiency of the 0.5kB construct to produce embryos expressing YFP in both pigments cells suggests that homology arms below this length may be significantly less able to produce knockins. These results were consistent with studies from other model systems that showed a dependence of HDR efficiency on homology arm length (Li et al., 2014) and led us to construct all future FP-HDR templates with homology arm lengths of at least 1.0 kB.
Figure 2. FP-HDR homology arm length and the efficiency of YFP insertion.
(A) Schematic of constructs designed to test homology arm length efficiencies. The only difference between each template was the length of homology arm from their 5′ and 3′ ends, insertion location and gRNA used were identical. (B) Shortening the arms of homology to 500 bp reduced the percentage of cells expressing GFP compared to arm lengths of 1.0 kB and 2.0 kB. Electroporations were performed (4 biological replicates) with 8 μg of each construct: Tyr::Cas9 and U6::gRNA(Tyr) and either Tyr FP-HDR(2.0 kB), Tyr FP-HDR(1.0 kB), or Tyr FP-HDR(500bp). Compared to the 2.0 kB control group (16.6% with two YFP+ melanocytes, 61.7% with one YFP+ melanocyte, 21.7% with no YFP+ melanocytes; n=575), the 1.0 kB transgene resulted in approximately the same percentage of embryos expressing YFP in two, one, or zero pigment cells (19.1%, 59.5%, and 21.4% respectively, n=496). However, in the 500 bp HDR template group (n=477), the percentage of embryos with two YFP+ melanocytes was reduced to 7.8%, and percentage of embryos with one YFP+ melanocyte was reduced to 48.1% while the percentage of embryos with no YFP+ melanocytes increased to 44.1%. In “B”, the different symbols represent different experiments; groups of either triangles, squares, circles or diamonds represent percentages of embryo phenotypes collected from the same electroporation. The different colors (orange, yellow, grey) indicate percentages of embryos with either 2, 1, or 0 fluorescent melanocytes, respectively. Significance was tested by MANOVA analysis on raw embryo counts followed by pairwise ANOVA with Tukey post-hoc analysis using R. There were no significant differences between the tested homology arm lengths, however there was indication that the 500 bp arm was less efficient at producing embryos with both melanocytes expressing YFP compared to the 2 kb arm (p adj = 0.09). This suggests that arms below 500 bp are likely to be less efficient at mediating knockins than longer arms.
The Brachyury and Pou4 loci can be efficiently targeted for FP-HDR
To demonstrate the broad applicability of our optimized reagents, we chose two genes expressed in divergent tissues: Bra, which is expressed exclusively in the developing notochord; and Pou4, which is expressed exclusively in the PNS. We first developed reagents to target and disrupt Bra and Pou4 to examine functionality of our CRISPR-Cas reagents and then we designed FP-HDR templates to Bra and Pou4 to determine if our CRISPR-Cas FP-HDR technology broadly effective in Ciona.
Brachyury encodes a T-box transcription factor required for the differentiation and patterning of the embryonic notochord (Corbo et al., 1997; Di Gregorio, 2017; Yasuo and Satoh, 1993), and disruption of this gene causes major notochordal defects (Chiba et al., 2009). In initial experiments, we drove gRNAs with the CsH1 promoter targeted against two positions within the Bra coding region and expressed Cas9 with a 1.6 kB Forkhead (Fkh) promoter (Fkh::Cas9) that drives expression in the notochord precursor cells one cell cycle prior to the separation of the notochord from the endoderm (Yagi et al., 2004). The resulting embryos displayed a stubby-tail phenotype, consistent with the phenotype of a null mutant, suggesting that our CRISPR-Cas reagents targeted Bra (Supplementary Fig. 1B). In those experiments we titrated the amount of the Fkh::Cas9 driver and determined that 4 μg was the optimal amount to electroporate, because electroporating greater amounts of this driver (6 μg or 8 μg), when combined with a negative control gRNA targeting a GFP sequence, resulted in embryos displaying a stubby tail phenotype (Supplementary Fig. 1C). These results highlight the importance of optimizing the delivery of CRISPR reagents to minimize undesirable phenotypes.
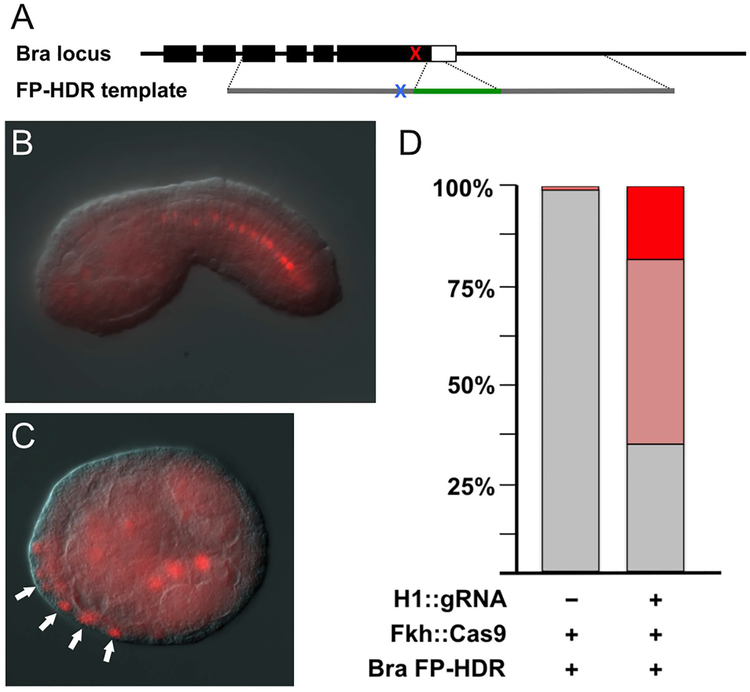
The Bra FP-HDR template was constructed with a yellow fluorescent protein (YFP) sequence inserted in-frame at the 3′ end of the Bra coding sequence designed to produce a Brachyury-YFP fusion protein (Fig. 3A) and the gRNA targeted the last coding exon of Bra. The protospacer-adjacent motif (PAM) was mutated in the FP-HDR template without altering the Bra coding sequence to reduce or eliminate Cas9-gRNA ribonucleoprotein binding. We electroporated 4 μg of Fkh::Cas9 and 10 μg each of H1::gRNA(Bra-Exon) and Bra FP-HDR and let the embryos develop to tailbud stage. To score HDR efficiency, we binned anti-GFP immunostained embryos into three groups: no YFP expression, 1–30% expression, and >30% expression. Combined averages (3 biological replicates of 75–100 embryos scored for each replicate) revealed 48.6% of embryos had fluorescent nuclei in up to 30% of their notochord cells, while 19.3% of embryos had fluorescent nuclei in >30% of their notochord cells when the exon-targeting gRNA constructs were included (Fig. 3B, 3D). We scored one embryo in control electroporations without the gRNA construct that had fluorescence in the notochord. When we expressed gRNAs that targeted the 3′UTR of Bra (H1::gRNA(Bra-UTR)), HDR efficiency was reduced and fluorescence was seen in non-notochordal cells (Fig. 3C). We also generated a transgene driving the same exon-targeting gRNAs as H1::gRNA(Bra-exon) but using the CrU6 promoter. Electroporating this construct with Fkh::Cas9 and Bra FP-HDR failed to produce cells expressing YFP. These observations suggest that combinations of gRNA drivers and FP-HDR templates should be tested and optimized to minimize off-target effects and maximize FP-HDR of targeted loci.
Figure 3. YFP is efficiently inserted into the C. robusta Brachyury locus using CRISPR-Cas mediated HDR resulting in YFP expression in the notochord.
(A) Schematic of Brachyury HDR template. The red X indicates genomic site in the Bra coding region where gRNAs were targeted; the blue X indicates conservatively mutated PAM in the HDR template upstream of the YFP sequence (green). (B) An example of a tailbud embryo with fluorescing notochord nuclei. Fertilized eggs were electroporated with 4 ug of Fkh::Cas9, 10 ug of CsH1::gRNA(Bra-Cod) targeting the 3′ end of the Bra coding region, and 10 ug of the Bra FP-HDR template. YFP was detected by immunohistochemistry with an anti-GFP antibody. (C) Correct combinations of Cas9 drivers and gRNA drivers are required for proper genome editing. Electroporating 4 μg of Fkh::Cas9 and 8 μg each of Bra FP-HDR and H1::gRNA(Bra-UTR) resulted in YFP being expressed in a non-notochordal lineage. Image shows an early gastrula embryo with YFP expression in several Fkh-expressing cells (arrows) that are not of the notochord lineage. (D) Quantitation of embryos with YFP fluorescing notochord cells. Percentage of embryos (3 biological replicates, n=260) with no fluorescing notochord cells (32.1%, gray); percentage of embryos with 1–30% of notochord cells fluorescing (48.6%, pink); percentage of embryos with >30% of notochord cells fluorescing (19.3%, red). Excluding H1::gRNA(Bra-Cod) resulted in scoring one embryo expressing YFP.
Subsequently, we designed CRISPR-Cas transgenes for FP-HDR to the Pou4 gene. Pou4 is a transcription factor that in Ciona is involved in the differentiation of ciliated epidermal sensory neurons (ESN) of the larval epidermis, including caudal, trunk and sensory neurons of the larval palps (Candiani et al., 2005; Joyce Tang et al., 2013). In initial experiments, we expressed Cas9 in the epidermis with an EpiB promoter (EpiB::Cas9) (Zeller et al., 2006b) together with U6-driven gRNAs targeting the Pou4 coding region. We hypothesized that disrupting Pou4 would prevent cilia formation because of the role it plays in the regulatory network governing the formation of ESNs (Joyce Tang et al., 2013). Embryos immunostained for acetylated tubulin had reduced numbers of ESNs, demonstrating that our Pou4-targeting constructs were functional and suggesting that Pou4 is indispensible for ESN differentiation (Supplementary Fig. 1D).
We then designed a Pou4 FP-HDR template with approximately 1.3 kB arms of homology and inserted YFP in-frame at the 3′ end of the Pou4 coding sequence. We could not identify an optimal targeting sequence near the end of the Pou4 coding region so we selected a target in the Pou4 3′UTR ~200 bp from the stop codon and mutated the target’s PAM sequence in the Pou4 FP-HDR template (Fig. 4A). We electroporated equal masses of EpiB::Cas9, U6::gRNA(Pou4-UTR) and Pou4 FP-HDR, let the embryos develop to larval stage, and detected YFP expressing cells using immunohistochemistry. For simplification of scoring efficiencies, we examined only caudal ESNs and did not assess abundance of YFP expressing cells in the palps or trunk. We binned the embryos into three groups: no caudal ESNs displaying YFP expression, 1–5 pairs of caudal ESNs displaying YFP expression (1–30% expressing), and 6–15 pairs of caudal ESNs displaying YFP expression (>30% expressing). In a typical electroporation, most (~85%) embryos had at least one ESN expressing YFP; 55% of the embryos expressed YFP in 1–5 pairs of CESNs and 30% had 6–15 pairs of CESNs expressing YFP (Fig. 4B, 4E). To examine whether fusing YFP to the C-terminal end of Pou4 would disrupt its function and thus disrupt cilia projections, we immunostained cilia with an acetylated tubulin antibody. Localization of fluorescent nuclei coincided with the presence of cilia at each ESN pair, indicating that recombination likely generated a functional Pou4-YFP fusion protein (Fig. 4D) as disrupting Pou4 expression disrupts cilia formation (Supplementary Fig. 1D). It is worth highlighting the observation that although Cas9 was expressed throughout the epidermis with the EpiB promoter, YFP fluorescence occurred only in ESNs. It is probable that the Pou4 locus was targeted for HDR in numerous epidermal cells, in addition to ESN cells. However, fluorescence was only observed in ESNs, consistent with the correct insertion of YFP into the Pou4 locus and subsequently expressed as a Pou4-YFP fusion protein and not simply doe to a random insertion of the construct into other genomics locations within epidermal cells..
Figure 4. YFP is efficiently inserted into the C. robusta Pou4 locus using CRISPR-Cas mediated HDR resulting in YFP expression in the ESNs.
(A) Schematic of Pou4 HDR template. A red X indicates genomic site in the Pou4 3′ UTR where gRNAs were targeted; a blue X indicates mutated target sequence in template upstream of the YFP sequence (green). Pou4 was targeted for FP-HDR by electroporating 8 μg each of EpiB::Cas9, U6::gRNA(Pou4-UTR), and Pou4 FP-HDR. (B) Example of larval stage embryo with 8/18 caudal ESN pairs expressing YFP (red, arrowheads). Palp ESNs also express YFP. Embryos were fixed and antibody stained for GFP (red) and anti-acetylated tubulin to label ESN cilia (green). (C) Expressing Cas9 with the SoxB1 promoter resulted in YFP expression in ESNs and ectopic expression of YFP. 8 μg each of SoxB1::Cas9, U6::gRNA(Pou4-UTR), and Pou4 FP-HDR was electroporated and embryos were fixed and stained for GFP and acetylated tubulin. Arrows point to non-ESN epidermal cells expressing YFP; arrowheads point to ESN nuclei expressing YFP. (D) ESN cilia (green, arrow) projecting from fluorescing ESN nuclei pair (red, arrow heads) indicate functional Pou4-YFP fusion protein. (E) Quantitation of embryos with YFP fluorescing ESNs. Combined percentages (n= 373; 3 biological replicates) of embryos with fluorescing ESNs. Percentage of embryos with no ESNs expressing YFP (15%, gray); percentage of embryos with 1–30% ESNs fluorescing (55%, pink); percentage of embryos with >30% of ESNs fluorescing (30%, red). In no-gRNA controls, we scored 1 embryos as having 1–30% fluorescent ESNs.
In developing and optimizing the reagents for Pou4 FP-HDR, we also generated a transgene driving Cas9 with a different epidermal promoter from the SoxB1 gene (SoxB1::Cas9) (Stolfi et al., 2014). We found that electroporating SoxB1::Cas9 and U6::gRNA(Pou4-UTR) with the Pou4 FP-HDR template produced YFP expression in non-ESN epidermal cells (Fig. 4C). In addition, we constructed a transgene that drove Pou4-targeting gRNAs from the H1 promoter (H1::gRNA(Pou4-UTR)). We found that electroporating H1::gRNA(Pou4-UTR) with EpiB::Cas9 and the Pou4 FP-HDR template was completely ineffective at generating FP-HDR (data not shown). These observations again indicate that care must be taken to optimize not only the choice of targeting sequence but also the drivers used to express Cas9 and the gRNAs.
Sequencing PCR amplicons from single embryos demonstrates genomic FP-HDR integration
In each of the experiments examining FP-HDR mediated genome insertions, control groups never expressed FP, demonstrating our FP-HDR templates are transcriptionally silent and that targeting-gRNAs are necessary to produce predicted patterns of fluorescence. To more conclusively demonstrate that FP-HDR had occurred, we generated PCR amplicons from the region surrounding the site of genomic editing and sequenced the resulting fusion sequences. We generated primer sets designed to amplify fragments that should exist only if FP-HDR template recombination occurred: one primer was designed to anneal to YFP and the other primer was designed to anneal to an area of the genome outside of the sequence of homology in the FP-HDR template (Supplementary Fig. 2). To generate amplicons revealing YFP integration at the Bra locus, we performed the same electroporation as above with Fkh::Cas9, H1::gRNA(Bra-Cod), and Bra FP-HDR. We live-sorted tailbud-stage embryos, pooled ten embryos that had 30% or more of their notochord cells fluorescing, and subjected those pools to PCR. Sequencing analysis of cloned PCR amplicons demonstrated a correct YFP insertion into the genome from the Bra HDR template at the correct genomic location (Supplementary Fig. 3).
To generate amplicons revealing YFP integration at the Pou4 locus, we electroporated equal masses of EpiB::Cas9, U6::gRNA(Pou4-UTR), and Pou4 FP-HDR and collected larval stage embryos that had ~50% of their ESNs fluorescing. Only ESN nuclei displayed fluorescence, but because Cas9 was expressed throughout the epidermis, it was likely that Cas9-directed FP-HDR occurred throughout the entire epidermis. Because the epidermis is a tissue that constitutes approximately 30% of the larval cell count, we reasoned there might be enough copies of modified genomes in a single embryo to detect the FP-HDR insertion and as such subjected individual embryos to PCR. Sequence analysis of cloned PCR amplicons from single YFP-positive Pou4 FP-HDR embryos demonstrated correct FP-HDR insertion into the genome (Supplementary Fig. 3). We were unable to produce this PCR product when embryos from control electroporations were used as template. Additionally, we performed control PCRs that attempted to amplify the same fragment using the Pou4 FP-HDR transgene and purified genomic DNA as template but were unsuccessful at generating amplicons. That we were successful at generating these amplicons only from embryos with predicted patterns of fluorescence suggests that the sequenced PCR product was produced from a genomic locus that had undergone recombination with the FP-HDR template.
Transgene-mediated biallelic knock-in
In other model systems, CRISPR-Cas9 has been shown to mediate HDR of both alleles (Chang et al., 2013; Gratz et al., 2014). To examine if we could detect biallelic FP-HDR in Ciona, we constructed two additional FP-HDR templates for Tyr that contained either cyan fluorescent protein (CFP) (Tyr FP-HDR(CFP)) or mCherry (Tyr FP-HDR(mCherry)) in place of GFP. We reasoned construction of two differently colored FP-HDR templates would allow us to detect instances of biallelic incorporation: mCherry expression alone (one or both alleles targeted), CFP expression (one or both alleles targeted), or, both mCherry and CFP expressed together in a single cell (one allele incorporating the CFP template and one allele incorporating the mCherry template). We electroporated Tyr::Cas9 and Tyr::RZ-gRNA(Tyr)-RZ along with equal amounts of Tyr FP-HDR(CFP) and Tyr FP-HDR(mCherry). For the purpose of determining events of biallelic FP-HDR, we scored embryos as having no fluorophore expression, mCherry expression, CFP expression, or both mCherry and CFP expression. This time point is several hours earlier than what we reported in Figure 1 and thus the CRISPR reagents had significantly less time to mediate HDR. Because of this we rarely observed embryos with both pigment cells expressing a fluorescent protein. Electroporating these DNAs (2 biological replicates, 179 total embryos scored) resulted in an average of 27.5% of embryos expressed only mCherry in at least one of the two melanocytes, 32.5% of embryos expressed only CFP in at least one melanocyte, and 4.5% of embryos expressed both fluorescent proteins in a single cell (Fig. 5A, 5B). We did not observe instances of CFP and mCherry expressed together in both melanocytes of a single embryo (which would imply that the four Tyr alleles of both melanocytes had experienced HDR). These results suggest that our CRISPR-Cas FP-HDR templates mediated biallelic fluorophore insertion at the Tyr locus in at least 4.5% of the cells targeted. Because this assay was unable to detect instances where both alleles in a single cell had incorporated either only CFP or only mCherry, we can speculate that the overall rate of biallelic targeting may be closer to 12%, i.e. ~3X the measured rate.
Figure 5. A FP can be inserted into both alleles of the Tyr locus simultaneously using CRISPR-Cas 9 mediated HDR.
Fertilized eggs were electroporated with 8ug each of Tyr::Cas9 and Tyr::RZ-gRNA(Tyr)-RZ and with 4 ug each of Tyr FP-HDR(CFP) and Tyr FP-HDR(mCherry). (A) Representative live embryos (13 hpf) showing CFP and/or mCherry expression in melanocytes. Top row: embryo with melanocyte displaying both CFP and mCherry in a single melanocyte (both alleles targeted); middle row: embryo with melanocyte displaying mCherry only (one allele targeted); bottom row: embryo with melanocyte displaying CFP only (one allele targeted). CFP channel (right column), mCherry channel (middle column), composite channels (left column). (B) Percentages of embryos expressing both fluorophores (purple, 4.5%), only mCherry expressing cells (red, 27.5%), only CFP expressing cells (blue, 32.5%), and percentage of embryos that expressed no fluorescence (gray, 35.5%). Two biological replicates, n=179.
Discussion
The ability to precisely edit the genomes of a variety of organisms has greatly expanded the usefulness of these systems as models for human development and disease (Cardi et al., 2017; Chen et al., 2014; Singh et al., 2017). Ascidians are gaining prominence as excellent models for examining a wide variety of biological problems including regeneration (Spina et al., 2017), Alzheimer’s studies (Virata and Zeller, 2010), and climate change research (Lopez et al., 2017). The ascidian embryo also provides a rapid and useful system for the analysis of complex regulatory elements (Farley et al., 2015; Kusakabe, 2005; Satou and Imai, 2015). In this study, we demonstrated CRISPR-Cas-mediated HDR is efficient at inserting a desired sequence into a specific genomic location in the ascidian C. robusta. To demonstrate FP-HDR is widely applicable in Ciona, we targeted three genes expressed in divergent tissues: Tyrosinase, a membrane-tethered enzyme that is the rate limiting step in the melanization of the two larval CNS melanocytes; Brachyury, a transcription factor expressed in the mesoderm; and Pou4, a transcription factor expressed in the PNS. We determined three aspects of the system need to be optimized: the type of promoter for expressing gRNAs, the promoter used for expressing Cas9, and the gRNA sequence selected to target the gene of interest and facilitate HDR. Our results demonstrate that each of these components must be combinatorically tested to produce correct FP-HDR insertion and FP expression. We confirmed genomic recombination of the FP-HDR transgenes by sequencing PCR amplicons obtained from pools of embryos, and from single embryos. Most notably, we demonstrated biallelic genome editing mediated by FP-HDR templates occurs and can be detected in living F0 larvae.
We successfully modified three genes expressed in different cell types and at different developmental stages. Tyrosinase is expressed in only two melanocytes, the otolith and ocellus, which are likely involved in photo- and geotactic responses during larval swimming (Nicol and Meinertzhagen, 1991). Disrupting Tyrosinase function caused either loss of pigmentation and/or pigment replacement by fluorescence in one or both CNS melanocytes of the larva. This easily scorable phenotype made it straightforward to optimize the CRISPR-Cas FP-HDR components. Brachyury (mesoderm) and Pou4 (ectoderm) encode well-characterized transcription factors, allowing easily identifiable FP-HDR phenotypes against established gene expression data. For each of these genes, we were able to demonstrate successful FP-HDR suggesting that CRISPR-Cas-mediated recombination is generally applicable throughout the genome.
While optimizing the delivery of the editing reagents, we evaluated three different means of expressing gRNAs for FP-HDR: expression from the CrU6 RNA pol III promoter, expression from a newly characterized CsH1 RNA pol III promoter, and expression from an RNA pol II promoter that drove ribozyme-flanked gRNAs. We found that all three gRNA delivery systems efficiently mediated HDR at the Tyr locus. However, HDR efficiency was much more varied at the other two loci. At the Bra locus, only H1 driven gRNAs, and not U6 driven gRNAs, were effective at mediating HDR. Conversely, the opposite pattern was observed at the Pou4 locus: U6 was effective, while H1 was not. In human and mouse studies, the U6 and H1 promoters are both used to drive gRNAs as well as other small RNA molecules such as shRNAs. In the case of expressing shRNAs, the U6 promoter is thought to be more effective than the H1 promoter (Makinen et al., 2006). However, to express gRNAs for eliciting CRISPR-Cas activity in mammalian cell culture, the U6 and H1 resulted in similar efficiencies (Ranganathan et al., 2014). It is not clear why an apparent difference in activity exists in Ciona. When generating transgenic ascidians via electroporation, the amount of DNA that can be electroporated while still maintaining a high percentage of normally developing embryos is limited to less than about 125 ng/μL (Zeller et al., 2006a). The H1 promoter is 10% the length of the U6 promoter, so more copies of an H1 transgene can be delivered per mass of DNA compared to U6 transgenes as the H1-driven transgene is about 75% the mass of the U6-driven transgene. It other applications where it would be desirable to multimerize several Pol III promoters into a single plasmid vector to drive multiple gRNAs simultaneously, many more copies of the H1 transgene could be incorporated simply considering the length of the constructs, compared to a single copy of the U6 promoter.; about four complete H1-gRNA transgenes occupy the same sequence space as just the U6 promoter.
There is also likely to be cell-type specific differences in the abilities of these promoters to drive gRNAs For example, in the melanocytes, because H1/U6 efficiency differences were statistically insignificant, it may be that the H1 promoter is less transcriptionally active than the U6 promoter in that cell type and thus the increased number of H1 plasmids resulted in the same activity as the lower amount of U6 plasmids. In the notochord, the U6 promoter’s inability to produce FP-HDR activity may result from insufficient transcriptional output in that tissue, while H1 has a sufficient capacity for promoting required levels of transcription. The inverse is true for the epidermis, where H1 was unable to facilitate FP-HDR and U6 worked well. Directly testing the transcriptional output of these promoters on a tissue by tissue basis would be challenging, and this variability we report further supports the claim that several combinations of reagents may need to be tested for a gene of interest. Lastly, although a number of studies have developed scoring algorithms to design active gRNAs for genome cleavage (Gandhi et al., 2017; Haeussler et al., 2016), it is likely that additional design rules will need to be derived to mediate efficient HDR. Based on the optimizations reported here, we suggest driving ribozyme-flanked gRNAs from tissue-specific RNA pol II promoters. Although these constructs are more difficult to generate, the ability to control where gRNAs are expressed, or when they are expressed if using an inducible promoter, makes them a more attractive choice, in our opinion, for gRNA delivery. In addition, the ability to control which cells express gRNAs can help to minimize any adverse effects resulting from expressing reagents in cells that are not desired targets.
Titrating the amount of Cas9 protein and identifying appropriate gRNAs for each specific targeted locus is also essential. If null-phenotypes for a given gene are known, this is easily accomplished by performing a series of electroporations in which the Cas9 delivery transgene amount is varied and the resulting embryos/larvae carefully examined for non-specific phenotypic changes. To interpret these effects, it is important to have previously characterized the expression pattern of the gene being targeted. For example, in all three genes targeted in this report, we and others had previously characterized the spatiotemporal expression of each gene and thus could expect a known pattern of fluorescence that would indicate a successful editing event after FP insertion, In our opinion, it is important to determine the maximal amounts of deliverable Cas9 protein, both alone or co-expressed with a control gRNA, that will not introduce altered phenotypes in embryos/larvae. We recommend when possible to design gRNAs that target the coding regions of genomic loci. Ascidian genomes are highly polymorphic (Dehal et al., 2002) but there should be a reduced frequency of polymorphisms in coding regions compared to non-coding regions. Because we are using the expression of fluorescent protein fusions to indicate successful genome editing, it is important to ensure that the FP-HDR template alone has no inherent transcriptional activity, as this would confound the experimental outcome. Therefore, it is important to run controls in which only the FP-HDR template is electroplated into fertilized eggs to ensure that is it transcriptionally silent. Because of the requirement for the HDR template to be transcriptionally silent (to allow detection after recombination and expression), this approach will not likely work to insert sequences near the beginning of a gene or in regions where cis-regulatory elements are likely to part of the HDR template. For these types of insertions, some sort of selectable marker system will need to be established and optimized for use in ascidians.
One of the more noteworthy findings from this study is the ability to use CRISPR-Cas mediated FP-HDR to simultaneously target both alleles of a specific genetic locus in the F0 generation. By using two different FP-HDR templates containing either CFP or mCherry, we could show that both alleles of the Tyrosinase gene could be targeted together within the same cell of a living Ciona embryo. As we reported, about 4% of embryos in which we targeted both alleles had melanocytes that fluoresced both cyan and red, suggesting that each allele had been edited. Given that we are unable to detect biallelic knock-in events in melanocytes that express CFP/CFP or mCherry/mCherry, it is likely that 4% is an underestimate and that 10–12% of the resulting embryos/larvae have had both alleles edited in one of their melanocytes. In some cases, it may be more advantageous to generate knock-in lines of animals that carry genomic edits at specific loci. Here it should be possible to combine our knock-in approach with recently reported methods to induce germ cell mutations via forced regeneration following larval tail ablation (Yoshida et al., 2017) to generate a line of animals containing the desired genomic edit/insertion. It would still be possible to easily determine single versus double allele edits by generating lines that have either a CFP or mCherry knock-ins and then crossing these lines together; single allelic knock-ins would glow with a single color, while those cells in which a double knock-in was present would glow with both inserted FP colors. By performing the appropriate crosses in this manner, it would be straightforward to separate out the resulting allelic modifications and easily determine whether one or both alleles were targeted in the experiment.
We see several implications based on the results presented in this study. The FP-HDR technology, as others have reported (Gaj et al., 2013), can be used to insert a fluorophore sequence in-frame with a gene of interest, effectively tagging the protein allowing researchers to follow not only the advent but also the cellular location of that particular protein. The tagged proteins provided by the FP-HDR approach should be expressed at endogenous levels and reflect a more accurate of expression and cellular location than transgenically expressed FP-tagged genes. This is often difficult to achieve when transgene drivers are used, primarily because all cis-regulatory elements should be present; transgenes likely lack some of these elements, particularly if the transgenes used contain minimal elements required for expression.
CRISPR-Cas-mediated knock-ins to transcription factors would also benefit ChIP-Seq studies in ascidians, as the use of tagged proteins obviates the current lack of antibodies that recognize ascidian transcription factors and would allow IP to be performed against the motif of a fusion protein. Previous ChIP-Seq studies in ascidians have been performed on embryos in which transgenes express tagged TF proteins (Kubo et al., 2010; Oda-Ishii et al., 2016). However, when these TF fusion proteins are expressed transgenically, the level of TF-fusion expression is unknown and in some cases outside of the native cell lineage (Kubo et al., 2010). Driving high expression levels of a TF can cause it to bind at low affinity sites that are normally not bound, while the binding of a TF expressed outside of its endogenous expression pattern may also confound results (Mahen et al., 2014). The use of endogenously expressed GFP-tagged TFs has been reported in other systems (Niu et al., 2011; Samuel et al., 2014), and the use of the FP-HDR approach described here should facilitate the widespread adoption of ChIP-Seq in ascidians.
Ascidians are excellent models for studying gene regulation during development, and as simple chordates offer insight into more complicated vertebrate development. A significant potential application of this technology is to edit the germline. A recent report that demonstrated mutations introduced into embryonic somatic tissues can be inherited by regenerated germ cells (Yoshida et al., 2017) introduced the mutations using a TALEN-based approach (Sasakura et al., 2017; Wright et al., 2014); these components are far more difficult to generate and test than CRISPR-based systems. Our CRISPR-Cas9 mediated approach for precisely modifying the ascidian genome, coupled with forced germ cell regeneration, is likely to accelerate the ability to generate stable knock-in lines in ascidians and will be of great benefit to the ascidian community.
Supplementary Material
Supplemental Figure 1. CRISPR-Cas reagents targeting Tyr, Bra and Pou4 are effective at eliciting null-phenotypes. (A) The CsH1 promoter is effective at expressing gRNAs and disrupting the Tyr gene resulting in unpigmented melanocytes. 8 μg of Tyr::Cas9 and H1::gRNA(Tyr) were electroporated and embryos developed to larval stage. Examples of embryos with 1 (left) or both (right) unmelanized melanocytes. (B) Targeting the Bra locus results in embryos displaying a phenotype consistent with a described null phenotype (Chiba et al., 2009). Electroporating 4 μg Fkh::Cas9 and 8 μg H1::gRNA(Bra) results in embryos with stubby tails. (C) Negative control electroporations indicate 8 μg of Fkh::Cas9 is an excessive amount. Electroporating 8 μg of Fkh::Cas9 and 8 μg of a negative control H1::gRNA(GFP) results in embryos displaying a phenotype consistent with a described null phenotype, whereas electroporating 4 μg Fkh::Cas9 and 8 μg of H1::gRNA(GFP) resulted in normal development (image not shown). (D) Targeting the Pou4 locus results in a reduction of ESN cilia. Embryos electroporated with 8 μg EpiB::Cas9 and 4 μg each of four different U6-driven gRNA transgenes targeting the Pou4 coding region had reduced numbers of ESN cilia (2 total ESNs, top, green). The average number of cilia (dorsal/ventral) was reduced to 3.9/2.3, vs. 8.5/7.2 in control group (n>100 per group). Embryos electroporated with only the EpiB::Cas9 transgene had normal numbers of ESNs (bottom). White arrowheads indicate ESNs.
Supplemental Figure 2. Schematic of Pou4 HDR template and amplification strategy as an example of how genomic insertions were verified. (A) Red X indicates genomic site where gRNAs were targeted, dark blue X indicates mutated PAM site in the Pou4 FP-HDR plasmid preventing CRISPR-Cas binding. Light blue represents pZapANX vector. (B) Diagram of hypothesized recombined genome with YFP insert (green). Orange arrows indicate locations of FWD and REV primers used to generate a PCR amplicon that should be produced only if the genome has incorporated the FP-HDR template. Amplicons were cloned into pBS2KS+ and sequenced with T7 and T3 primers to examine both ends of the fragment.
Supplemental Figure 3. Sequencing Brachyury FP-HDR and Pou4 FP-HDR PCR amplicons demonstrates genomic FP-HDR integration. A total of 10 amplicons from 10 different Bra-YFP+ batches of embryos and 8 amplicons from 8 different Pou4-YFP+ embryos were cloned into pBSK; 4 Bra clones were sequenced and 4 Pou4 clones were sequenced. Each of the examined sequences was nearly identical to the sequences reported above. The primers used (orange) generated sequences that span from a genomic region outside of the homology arm (black) through the template arm (grey; primers used to generate homology arm in blue) and into the YFP sequence (green). The polylinker fusing Bra (top) or Pou4 (bottom) to YFP is shown in lowercase. The sequences shown above are one of four sequences derived from eight cloned Bra-YFP or Pou4-YFP amplicons. SNPs found within the sequenced clones are indicated as bold grey.
Acknowledgements
We thank Christina Niemeyer for useful comments on the manuscript and thank Karl Garcia and Tiffany Hoang for assistance with molecular cloning. This work was supported by NSF grants IOS-0951347, IOS-1557448 and NIH grant 1R21DC01318001A1 (NIDCD) to RWZ.
This work was supported by NSF grants IOS-0951347, IOS-1557448 and NIH grant 1R21DC01318001A1 (NIDCD) to RWZ.
References
- Bartkiewicz M, Gold H, Altman S, 1989. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev 3, 488–499. [DOI] [PubMed] [Google Scholar]
- Candiani S, Pennati R, Oliveri D, Locascio A, Branno M, Castagnola P, Pestarino M, De Bernardi F, 2005. Ci-POU-IV expression identifies PNS neurons in embryos and larvae of the ascidian Ciona intestinalis. Dev Genes Evol 215, 41–45. [DOI] [PubMed] [Google Scholar]
- Cardi T, D’Agostino N, Tripodi P, 2017. Genetic Transformation and Genomic Resources for Next-Generation Precise Genome Engineering in Vegetable Crops. Front Plant Sci 8, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ, 2013. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 23, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B, 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tang L, Xiang H, Jin L, Li Q, Dong Y, Wang W, Zhang G, 2014. Advances in genome editing technology and its promising application in evolutionary and ecological studies. Gigascience 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Jiang D, Satoh N, Smith WC, 2009. Brachyury null mutant-induced defects in juvenile ascidian endodermal organs. Development 136, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW, 1997. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589–602. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS, 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157–2167. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, 2017. T-Box Genes and Developmental Gene Regulatory Networks in Ascidians. Curr Top Dev Biol 122, 55–91. [DOI] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS, 2015. Suboptimization of developmental enhancers. Science 350, 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Zhou K, Doudna JA, 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395, 567–574. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF 3rd, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S, Haeussler M, Razy-Krajka F, Christiaen L, Stolfi A, 2017. Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona. Dev Biol 425, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y, 2014. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol 56, 343–349. [DOI] [PubMed] [Google Scholar]
- Gokcezade J, Sienski G, Duchek P, 2014. Efficient CRISPR/Cas9 plasmids for rapid and versatile genome editing in Drosophila. G3 (Bethesda) 4, 2279–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM, 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, Joly JS, Concordet JP, 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Tresser JW, Horie T, Tsuda M, Smith WC, 2005. Pigmentation in the sensory organs of the ascidian larva is essential for normal behavior. J Exp Biol 208, 433–438. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce Tang W, Chen JS, Zeller RW, 2013. Transcriptional regulation of the peripheral nervous system in Ciona intestinalis. Dev Biol 378, 183–193. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S, 2014. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 4, 6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Suzuki N, Yuan X, Nakai K, Satoh N, Imai KS, Satou Y, 2010. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development 137, 1613–1623. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, 2005. Decoding cis-regulatory systems in ascidians. Zoolog Sci 22, 129–146. [DOI] [PubMed] [Google Scholar]
- Li K, Wang G, Andersen T, Zhou P, Pu WT, 2014. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS One 9, e105779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CE, Sheehan HC, Vierra DA, Azzinaro PA, Meedel TH, Howlett NG, Irvine SQ, 2017. Proteomic responses to elevated ocean temperature in ovaries of the ascidian Ciona intestinalis. Biol Open 6, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Liu F, 2015. Genome Editing and Its Applications in Model Organisms. Genomics Proteomics Bioinformatics 13, 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahen R, Koch B, Wachsmuth M, Politi AZ, Perez-Gonzalez A, Mergenthaler J, Cai Y, Ellenberg J, 2014. Comparative assessment of fluorescent transgene methods for quantitative imaging in human cells. Mol Biol Cell 25, 3610–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen PI, Koponen JK, Karkkainen AM, Malm TM, Pulkkinen KH, Koistinaho J, Turunen MP, Yla-Herttuala S, 2006. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J Gene Med 8, 433–441. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA, 1991. Cell counts and maps in the larval central nervous system of the ascidian Ciona intestinalis (L.). J Comp Neurol 309, 415–429. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Fujiwara S, 2008. RNA interference by expressing short hairpin RNA in the Ciona intestinalis embryo. Dev Growth Differ 50, 521–529. [DOI] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, Slightham C, Jiang L, Hyman AA, Kim SK, Waterston RH, Gerstein M, Snyder M, Reinke V, 2011. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 21, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Ishii I, Kubo A, Kari W, Suzuki N, Rothbacher U, Satou Y, 2016. A Maternal System Initiating the Zygotic Developmental Program through Combinatorial Repression in the Ascidian Embryo. PLoS Genet 12, e1006045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinelli P, Rosenblad MA, Samuelsson T, 2005. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res 33, 4485–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G, 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231, 1577–1580. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA, 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan V, Wahlin K, Maruotti J, Zack DJ, 2014. Expansion of the CRISPR-Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat Commun 5, 4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel A, Housset M, Fant B, Lamonerie T, 2014. Otx2 ChIP-seq reveals unique and redundant functions in the mature mouse retina. PLoS One 9, e89110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Yoshida K, Hozumi A, Sasakura Y, 2014. CRISPR/Cas9-mediated gene knockout in the ascidian Ciona intestinalis. Dev Growth Differ 56, 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y, Yoshida K, Treen N, 2017. Genome Editing of the Ascidian Ciona intestinalis with TALE Nuclease. Methods Mol Biol 1630, 235–245. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, 2015. Gene regulatory systems that control gene expression in the Ciona embryo. Proc Jpn Acad Ser B Phys Biol Sci 91, 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, Nakayama A, Nishino A, Sasakura Y, Satoh G, Shimotori T, Shin IT, Shoguchi E, Suzuki MM, Takada N, Utsumi N, Yoshida N, Saiga H, Kohara Y, Satoh N, 2001. Gene expression profiles in Ciona intestinalis tailbud embryos. Development 128, 2893–2904. [DOI] [PubMed] [Google Scholar]
- Sierro N, Kusakabe T, Park KJ, Yamashita R, Kinoshita K, Nakai K, 2006. DBTGR: a database of tunicate promoters and their regulatory elements. Nucleic Acids Res 34, D552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E, 2015. A mouse geneticist’s practical guide to CRISPR applications. Genetics 199, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Braddick D, Dhar PK, 2017. Exploring the potential of genome editing CRISPR-Cas9 technology. Gene 599, 1–18. [DOI] [PubMed] [Google Scholar]
- Song F, Stieger K, 2017. Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks. Mol Ther Nucleic Acids 7, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina EJ, Guzman E, Zhou H, Kosik KS, Smith WC, 2017. A microRNA-mRNA expression network during oral siphon regeneration in Ciona. Development 144, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Gandhi S, Salek F, Christiaen L, 2014. Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development 141, 4115–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virata MJ, Zeller RW, 2010. Ascidians: an invertebrate chordate model to study Alzheimer’s disease pathogenesis. Dis Model Mech 3, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Christiaen L, 2012. Transcriptional enhancers in ascidian development. Curr Top Dev Biol 98, 147–172. [DOI] [PubMed] [Google Scholar]
- Wright DA, Li T, Yang B, Spalding MH, 2014. TALEN-mediated genome editing: prospects and perspectives. Biochem J 462, 15–24. [DOI] [PubMed] [Google Scholar]
- Yagi K, Satou Y, Satoh N, 2004. A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development 131, 1279–1288. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Satoh N, 1993. Function of vertebrate T gene. Nature 364, 582–583. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Hozumi A, Treen N, Sakuma T, Yamamoto T, Shirae-Kurabayashi M, Sasakura Y, 2017. Germ cell regeneration-mediated, enhanced mutagenesis in the ascidian Ciona intestinalis reveals flexible germ cell formation from different somatic cells. Dev Biol 423, 111–125. [DOI] [PubMed] [Google Scholar]
- Zeller RW, 2018. Electroporation in Ascidians: History, Theory and Protocols. Advances in experimental medicine and biology 1029, 37–48. [DOI] [PubMed] [Google Scholar]
- Zeller RW, Virata MJ, Cone AC, 2006a. Predictable mosaic transgene expression in ascidian embryos produced with a simple electroporation device. Dev Dyn 235, 1921–1932. [DOI] [PubMed] [Google Scholar]
- Zeller RW, Weldon DS, Pellatiro MA, Cone AC, 2006b. Optimized green fluorescent protein variants provide improved single cell resolution of transgene expression in ascidian embryos. Dev Dyn 235, 456–467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. CRISPR-Cas reagents targeting Tyr, Bra and Pou4 are effective at eliciting null-phenotypes. (A) The CsH1 promoter is effective at expressing gRNAs and disrupting the Tyr gene resulting in unpigmented melanocytes. 8 μg of Tyr::Cas9 and H1::gRNA(Tyr) were electroporated and embryos developed to larval stage. Examples of embryos with 1 (left) or both (right) unmelanized melanocytes. (B) Targeting the Bra locus results in embryos displaying a phenotype consistent with a described null phenotype (Chiba et al., 2009). Electroporating 4 μg Fkh::Cas9 and 8 μg H1::gRNA(Bra) results in embryos with stubby tails. (C) Negative control electroporations indicate 8 μg of Fkh::Cas9 is an excessive amount. Electroporating 8 μg of Fkh::Cas9 and 8 μg of a negative control H1::gRNA(GFP) results in embryos displaying a phenotype consistent with a described null phenotype, whereas electroporating 4 μg Fkh::Cas9 and 8 μg of H1::gRNA(GFP) resulted in normal development (image not shown). (D) Targeting the Pou4 locus results in a reduction of ESN cilia. Embryos electroporated with 8 μg EpiB::Cas9 and 4 μg each of four different U6-driven gRNA transgenes targeting the Pou4 coding region had reduced numbers of ESN cilia (2 total ESNs, top, green). The average number of cilia (dorsal/ventral) was reduced to 3.9/2.3, vs. 8.5/7.2 in control group (n>100 per group). Embryos electroporated with only the EpiB::Cas9 transgene had normal numbers of ESNs (bottom). White arrowheads indicate ESNs.
Supplemental Figure 2. Schematic of Pou4 HDR template and amplification strategy as an example of how genomic insertions were verified. (A) Red X indicates genomic site where gRNAs were targeted, dark blue X indicates mutated PAM site in the Pou4 FP-HDR plasmid preventing CRISPR-Cas binding. Light blue represents pZapANX vector. (B) Diagram of hypothesized recombined genome with YFP insert (green). Orange arrows indicate locations of FWD and REV primers used to generate a PCR amplicon that should be produced only if the genome has incorporated the FP-HDR template. Amplicons were cloned into pBS2KS+ and sequenced with T7 and T3 primers to examine both ends of the fragment.
Supplemental Figure 3. Sequencing Brachyury FP-HDR and Pou4 FP-HDR PCR amplicons demonstrates genomic FP-HDR integration. A total of 10 amplicons from 10 different Bra-YFP+ batches of embryos and 8 amplicons from 8 different Pou4-YFP+ embryos were cloned into pBSK; 4 Bra clones were sequenced and 4 Pou4 clones were sequenced. Each of the examined sequences was nearly identical to the sequences reported above. The primers used (orange) generated sequences that span from a genomic region outside of the homology arm (black) through the template arm (grey; primers used to generate homology arm in blue) and into the YFP sequence (green). The polylinker fusing Bra (top) or Pou4 (bottom) to YFP is shown in lowercase. The sequences shown above are one of four sequences derived from eight cloned Bra-YFP or Pou4-YFP amplicons. SNPs found within the sequenced clones are indicated as bold grey.