The Pharmacogene Variation Consortium (PharmVar) continues the mission of the Human Cytochrome P450 (CYP) Nomenclature database to provide the clinical and research communities a repository and standardized nomenclature of pharmacogene variation. To improve the utility of this resource, PharmVar has established a database and gene expert panels and expanded beyond CYPs to include other clinically important pharmacogenes. This report describes the evolution from static CYP variation tables to a resource with growing functionality describing pharmacogene variation.
Where PharmVar Started
The Human Cytochrome P450 (CYP) Nomenclature database provided static Web‐based tables listing CYP genetic variation (archived at https://www.pharmvar.org/htdocs/archive/index_original.htm). Although universally accepted and widely used, these tables are now outdated. The Pharmacogene Variation Consortium (PharmVar) was launched in September 2017 with the mission to continue providing haplotype information and transforming the original tables into a modern and interactive resource.1
To that end, PharmVar established a state‐of‐the‐art database and gene expert panels from its membership base to facilitate the review of submissions, develop standard operating procedures, standardize the submission/review process, and designate haplotypes. Table 1 itemizes the genes that have been transitioned to the database and details PharmVar member demographics and site use. Close partnership with the Pharmacogenomics Knowledge Base (PharmGKB) ensures consistency of data display, use, and dissemination across other databases and stakeholders, such as the Clinical Pharmacogenetics Implementation Consortium (CPIC).
Table 1.
PharmVar gene and membership demographics
| Genes | Star alleles | Haplotypes per genea | PharmVar members | PharmVar usersb | PharmVar site visitsb |
|---|---|---|---|---|---|
| CYP2A13 | *1‐*10 | 10 | Total | ||
| CYP2C8 | *1‐*14 | 14 | 95c | 2,204 | 3,779 |
| CYP2C9 | *1‐61 d | 61d | United States and Canada | ||
| CYP2C19 | *1‐*35 | 33e | 59 | 847 | 1,486 |
| CYP2D6 | *1‐*122 c | 106d,e | Europe | ||
| CYP2F1 | *1‐*6 | 6 | 18 | 449 | 812 |
| CYP2J2 | *1‐*10 | 10 | Asia | ||
| CYP2R1 | *1, *2 | 2 | 9 | 717 | 1,172 |
| CYP2S1 | *1‐*5 | 5 | Africa | ||
| CYP2W1 | *1‐*6 | 6 | 3 | 21 | 34 |
| CYP3A7 | *1‐*3 | 3 | South America | ||
| CYP3A43 | *1‐*3 | 3 | 4 | 94 | 154 |
| CYP4F2 | *1‐*3 | 3 | Australia and New Zealand | ||
| 2 | 50 | 81 | |||
| NUDT15 | *1‐*19 | 19 | |||
Gene and PharmVar member data are as of October 8, 2018. Only those genes transferred into the PharmVar database are listed. The following genes (number of alleles in parentheses) will be transferred: CYP1A1 (13), CYP1A2 (21), CYP1B1 (26), CYP2A6 (412), CYP2B6 (38), CYP2E1 (8), CYP3A4 (322), CYP3A5 (92), while CYP4A11, CYP4A22, CYP4B1, TBXAS1 (CYP5A1), PTGIS (CYP8A1), CYP19A1, CYP21A2 and CYP26A1 will receive legacy status. PharmVar membership demographics are provided by major geographical region. Website statistics are collected with Google Analytics and represent use over the period of 1 month (September 8 through October 8, 2018). PharmVar, Pharmacogene Variation Consortium.
aNot counting suballeles. bExcluding hits generated by Children's Mercy (PharmVar) and Stanford (PharmGKB). cIncluding 18 Steering Committee members and pending applications. dAdditional alleles have been defined and are awaiting release. eOne or more star alleles may have been removed or consolidated; the count of alleles does not correspond to the latest star allele number.
The PharmVar Database
PharmVar database features are built with the goal of facilitating access to important and often‐used information in a meaningful and consistent way. To that end, PharmVar displays allelic data consistently across genes (see STANDARDS document under GENES menu) and maps each gene to current versions of genomic and transcript reference sequences (RefSeq) and genome builds GRCh37 and GRCh38. The variant view shows coordinates across all references for two count modes, from the sequence start or the ATG start codon. The variant page also lists all haplotypes on which the variant can be found and provides the rs identification (ID) and link to the dbSNP database.
Each gene has its own page with important information (Figure 1 ); this section is followed by the listing of allelic variants defined to date. The impact of sequence variations (SVs; amino acid changing and splicing) is provided along with a selection of references. Functional information is cross‐referenced from functionality tables available through the PharmGKB (https://www.pharmgkb.org/page/pgxGeneRef). However, function can be substrate specific, and generalizations to other substrates should be made with caution.
Figure 1.
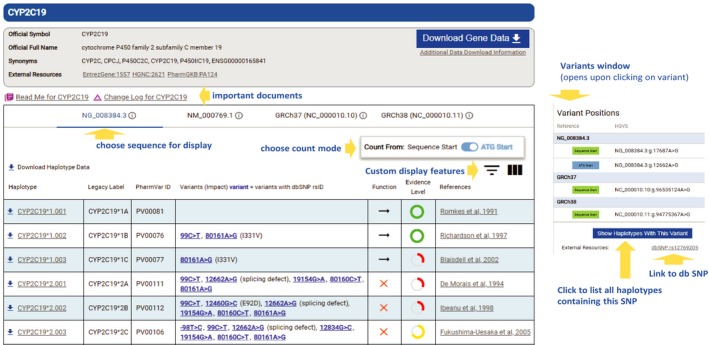
PharmVar gene page view for CYP2C19. The top portion of each gene page provides general information and links to important documents, including the READ ME and CHANGE LOG documents (some genes also have documentation for structural variation). In the middle section, the user can choose to visualize positions of sequence variations on the genomic or cDNA reference sequences (NG_008384.3 and NM_000769.1) or on the GRCh37 or GRCh38 genome build (information buttons link each to NCBI). The user also has the option between two count modes (start of the sequence or the ATG start codon). In the lower section, all defined alleles and suballeles are listed with their respective haplotype designation, legacy label, PharmVar ID, and sequence variations (those with rs IDs are highlighted in blue), followed by their impact (e.g., splicing defect or amino acid change) in parentheses, function (if available through the PharmGKB), evidence level, and selected reference(s). Download options include to download all gene data or individual haplotypes. On clicking a sequence variant of interest, the variant window (shown to the right) slides in “at a glance” information, including its rs ID and link to dbSNP.
As a gene transitions from its original table format into the PharmVar database, revisions may be required to standardize display (e.g., some coordinates for insertions/deletions have shifted because these were right aligned or because mapping now uses the current RefSeq). Original comments and notes are removed, which may trigger revisions for allele definitions (e.g., a note stating that “12662A>G is likely part of all CYP2C19*2 alleles” was removed, and 12662A>G was included in all *2 definitions). Inadvertent errors are being corrected (e.g., c.1425A>T was added to all CYP2C9*3 haplotypes because it was omitted when this allele was first defined ≈20 years ago). In addition, intronic single‐nucleotide polymorphisms have been removed from several CYP2C9 and CYP2C19 alleles; none of these are known to affect function and only represented some of many intronic single‐nucleotide polymorphisms. Finally, some haplotypes may be redesignated during the curation process. For instance, the nonfunctional CYP2D6*14A allele was revised to CYP2D6*114, whereas the decreased function CYP2D6*14B allele was retained as CYP2D6*14. Changes are detailed in each gene's CHANGE LOG document.
The PharmVar database also offers several download options, which greatly simplifies integration of allelic data into informatics workflows. The user can download the entire PharmVar database content or selected variants of interest in sequence (fasta or vcf file) or tab‐separated value formats. Furthermore, the database is versioned and dated. PharmVar members have the option to receive email notifications when a new version is released, and automated change log reports will be available in the future.
PharmVar works with the National Center for Biotechnology Information (NCBI) to develop locus reference genomic (LRG) sequences (https://www.lrg-sequence.org/) for pharmacogenes. LRGs are stable references that are developed for reporting sequence variants with clinical implications. The benefit of using LRGs is that these will not change over time (as opposed to RefSeqs), thereby providing the field with “gold standard” references for test reporting. PharmVar also seeks to update RefSeqs in certain instances (e.g., the NCBI created separate RefSeqs for CYP1A1 and CYP11A2, which were originally located in opposite directions on a single RefSeq).
New PharmVar Haplotype Versioning System
The Human CYP Nomenclature webpages initially defined suballeles using letters (A, B, C, etc.), but eventually stopped accepting these submissions. This triggered authors to publish variants using a suffix (e.g., *2var1 and *2var2), making it difficult, if not impossible, to keep track of allelic variation. Suballeles denote haplotypes that carry one or more SVs in addition to those defining a haplotype. Although these SVs are known or predicted to be inconsequential, they may interfere with genotyping assays and may lead to inaccurate result interpretations.2 Furthermore, a comprehensive catalog of allelic variation that includes suballeles will facilitate next‐generation sequencing (NGS)–based data interpretation using bioinformatic algorithms, such as Astrolabe3 or Stargazer,4 that assign diplotypes using star nomenclature. To provide the field with the most comprehensible data repository, PharmVar has resumed cataloging suballeles. To accommodate potentially large numbers of suballeles of major haplotypes (e.g., CYP2D6*2 or CYP2D6*4), a suballele numbering system has been introduced that replaces the use of letters. For example, CYP2D6*2A and CYP2D6*2B are now displayed as CYP2D6*2.001 and CYP2D6*2.002, respectively. Details can be found in the ALLELE DESIGNATION AND EVIDENCE LEVEL CRITERIA document under the SUBMISSION menu.
PharmVar Introduces Evidence Levels
For smaller genes (10–15 kb) including CYP2D6, haplotypes can be verified experimentally using allele‐specific long‐range polymerase chain reaction or NGS‐based single‐molecule sequencing techniques. In contrast, definitions for haplotypes for larger genes, such as CYP2C9 or CYP2C19, are often based on computationally inferred predictions. Although Sanger sequencing is still considered the gold standard, advances in NGS technology are yielding increasingly robust data. PharmVar accepts NGS‐based submissions, given that certain quality control criteria are fulfilled. Regardless of the sequencing method used, haplotype definitions can be based on “definitive” or “moderate” evidence (e.g., computational inference). Many haplotypes defined before PharmVar may lack crucial pieces of information now required for haplotype definition; these are considered of “limited” evidence. The evidence level of a haplotype may be important information for investigators and in clinical implementation. Detailed information regarding the methods used for haplotype determination is collected in the submission process and detailed in the ALLELE DESIGNATION AND EVIDENCE LEVEL CRITERIA document. PharmVar also accepts supporting submissions of the same haplotype to build a stronger body of evidence in support of a haplotype. This type of information, not systematically captured before PharmVar, is used to assign definitive, moderate, and limited evidence levels to each haplotype, reflecting the strength of evidence on which a definition is based.
Introduction of the PharmVar ID
The challenge of allele naming arises from the conflict between the desire to have consistent, stable identifiers for alleles and having meaningful names, allowing allele grouping by major allele, function, or relevant variant. Users expect an assigned allele name to have meaning on its own (i.e., all suballeles have the same or similar function). Given that it is impossible to know all of the eventual functional implications of each allele at the time a name is assigned, those two goals are not easily and simultaneously achieved. Thus, we either accept that allele names do not always represent the best familial grouping for an allele, or we accept that in the face of new knowledge, an allele name will sometimes have to change. PharmVar offers a solution to this dilemma by splitting those duties (stable identification vs. functional/familial grouping). Identification will be handled through the introduction of the PharmVar ID, a numeric identifier similar to the dbSNP's rs ID, while continuing to use star allele names, which might not be permanent, to show the functional grouping. This approach allows the identification of an allele to be fixed while its functional interpretation can change over time as more information becomes available. We recognize that it is unrealistic to expect the entire community to change how alleles are referred to, especially in the short‐term. However, because PharmVar needs to be able to revise allele designations in some cases to reflect a more appropriate classification, we suggest citing PharmVar IDs alongside star allele names moving forward (e.g., CYP2D6*46.002 (PV00182)).
Expanding the Repertoire of Genes
Building nomenclature for new genes faces numerous challenges, including the existence of star‐designated alleles in the absence of a formal nomenclature system, designations that do not conform to PharmVar criteria, the use of designations without a publication record, and insufficient published information, among others. Some of these challenges were encountered for NUDT15, which was chosen as the first PharmVar non‐CYP gene because of its contribution to thiopurine toxicity.5 NUDT15 genetic testing is increasingly being performed in concert with TPMT and, thus, nomenclature is a prerequisite for standardized reporting. Other CPIC genes for which no formal nomenclature exists (e.g., DPYD and SLCO1B1) will follow. Each gene will likely present unique challenges, which will be addressed by expert gene panelists and Steering Committee input.
PharmVar Keeps Evolving
Finally, PharmVar continues to evolve by improving display, developing new features (e.g., a comparative allele viewer tool and custom data displays), and adding complementary information (e.g., linking to Coriell samples known to contain a particular haplotype). PharmVar welcomes feedback and suggestions to further the evolution of this resource.
Funding
This work was funded by the National Institutes of Health for the Pharmacogene Variation Consortium (R24GM123930) and PharmGKB (R24GM61374).
Conflict of Interest
The authors declared no competing interests for this work.
Acknowledgments
We thank all Pharmacogene Variation Consortium Steering Committee members for their critical review of this article and all experts serving on gene panels. We also thank Scott Casey for website design and development.
References
- 1. Gaedigk, A. et al The Pharmacogene Variation (PharmVar) Consortium: incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature database. Clin. Pharm. Ther. 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nofziger, C. & Paulmichl, M. Accurately genotyping CYP2D6: not for the faint of heart. Pharmacogenomics 19, 999–1002 (2018). [DOI] [PubMed] [Google Scholar]
- 3. Twist, G. et al Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole‐genome sequence. Genomic Med. (2016). http://www.nature.com/articles/npjgenmed20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee, S.B. et al Stargazer: a software tool for calling star alleles from next‐generation sequencing data using CYP2D6 as a model. Genet. Med. 10.1038/s41436-018-0054-0. [e‐pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang, J.J. et al Pharmacogene variation consortium gene introduction: NUDT15 . Clin. Pharm. Ther. 10.1002/cpt.1268. [e‐pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]


