Abstract
Over the past half-century, we have gained significant insights into the molecular biology of long-term memory storage at the level of the synapse. In recent years, our understanding of the cellular architecture supporting long-term memory traces has also substantially improved. However, the molecular biology of consolidation at the level of neuronal systems has been relatively neglected. In this opinion article, we first examine our current understanding of the cellular mechanisms of synaptic consolidation. We then outline areas requiring further investigation on how cellular changes contribute to systems consolidation. Finally, we highlight recent findings on the cellular architecture of memory traces in rodents and how the application of new technologies will expand our understanding of systems consolidation at the neural circuit level. In the coming years, this research focus will be critical for understanding the evolution of long-term memories and for enabling the development of novel therapeutics which embrace the dynamic nature of memories.
Keywords: memory trace, consolidation, neural systems, engram
Memory: From Synapses to Systems
Memory is the canvas upon which we paint the portrait of our lives, and research in the last half-century has provided tremendous insight into this canvas. We now know that the consolidation of long-term memories requires synaptic plasticity; that this plasticity depends on key molecular signaling cascades; and that these cascades serve to strengthen particular synaptic connections to consolidate memories in discrete brain networks (for review see [1]). There has been particularly strong progress in identifying the electrophysiological [2], genetic [3], proteomic [4], and epigenetic [5, 6] underpinnings of long-term memory (LTM; see Glossary) consolidation [1, 7]. Recently, with the advent of techniques for labeling active neuronal ensembles, we have also acquired the ability to resolve the cellular architecture of the memory trace/engram in mammals at the level of neural circuits [8].
Three key events in the mid-20th century catalyzed the search for molecular mechanisms of LTM traces. First, the formalization of a theory by Donald Hebb in the late 1940s that LTMs require concomitant activation and strengthening of pre- and post-synaptic neurons [9]. Second, the identification by Milner and colleagues in the 1950s that the hippocampus is critical for forming explicit (i.e., episodic) LTMs [10, 11]. Third, the discovery by Bliss and Lomo in the early 1970s that long-term potentiation (LTP; i.e., enhanced efficacy of presynaptic transmission and post-synaptic excitability) may serve as a substrate for LTM [12]. Together, these three findings fortified a view that LTP, long-term depression (LTD) and spike-timing dependent plasticity [13] may provide a mechanism for acquiring and consolidating LTMs. While the in vivo roles of LTP and LTD in learning and memory have been debated [14-16], there is a general consensus that the consolidation of LTMs requires de novo mRNA transcription and protein synthesis [17], as evidenced by their requirement in the late-phase of LTP [18] for episodic (e.g., hippocampal) memories in rodents [19]. Indeed, we have learned a great deal about the cellular and molecular mechanisms of LTM consolidation at the level of the synapse. However, there are still a number of important questions about the cellular and molecular mechanisms of systems consolidation (for etymological considerations on synaptic and systems consolidation see [20] and Glossary).
In this opinion article, we first provide an overview of what is known about the molecular biology of synaptic consolidation. We then focus on candidate cellular mechanisms of systems consolidation that require further investigation. Finally, we highlight recent studies on the organization of LTM traces in the mammalian brain and consider how certain technological breakthroughs will help to elucidate the molecular biology of systems consolidation.
Molecular Mechanisms of Long-Term Memory Storage
Reductionist approaches in the mid-20th century employed a number of model systems to identify the molecular mechanisms of synaptic consolidation (for review see [1, 21, 22]). Research using the marine snail Aplysia californica was particularly helpful in providing insight into the differences between short- and long-term memories. These studies revealed that short-term memories require increased pre-synaptic glutamate release as well as changes in post-synaptic glutamatergic receptor activity [23, 24] mediated by the covalent modification of existing proteins at preexisting synapses [21]. In contrast, long-term memories require de novo gene transcription [25], new protein translation [26], and synaptic growth at pre- and post-synaptic terminals [27, 28]. Of critical importance were findings that revealed that in a number of instances both mitogen activated protein kinase (MAPK) and protein kinase A (PKA) act in combination on cAMP response element binding protein (CREB) in the nucleus to consolidate a LTM [22]. In short-term memories, PKA functions in the cytoplasm to alter synaptic transmission. By contrast, in long-term memory the catalytic subunit of PKA translocates to the nucleus to phosphorylate CREB-1, which then modulates the transcription of genes containing cAMP response elements [29]. This transcriptional mechanism recruits a host of additional genes including the immediate-early gene CCAAT box/enhancer-binding protein (C/EBP) which, via dimerization with an activating protein [30], drives the transcription of genes necessary for synaptic growth (e.g., elongation-factor 1α). Importantly, MAPK indirectly regulates CREB-1 via the removal of CREB-2 – a protein which represses CREB-1 activity in the basal state [31, 32]. Moreover, MAPK also guides the internalization and redistribution of neural cell adhesion molecules to sites of new synaptic growth [33]. These studies illuminated how a signal originating from an activated synapse triggers a specific intracellular signaling cascade to alter nuclear function and synaptic connectivity in order to consolidate a LTM.
The subsequent extension of these biological mechanisms of memory from Aplysia to mice was critical for elucidating (1) the cross-species preservation of these signaling cascades (i.e., from long-term facilitation in Aplysia to the late-phase of LTP in rodents) and (2) the precise role of these molecules in the storage of episodic or hippocampus-dependent memories. Research using Aplysia and mice helped bridge the postulate of Hebb with the findings of Milner, Bliss, and Lomo to provide a biological framework for the synaptic consolidation of LTMs. Studies have since elaborated on how these proteins and pathways interact and how they are intricately regulated [34-36]. This research has also identified the function of several immediate-early genes and other proteins as important regulators of different phases of LTP, as well as certain stages of LTM. Examples of such proteins include cellular feline osteosarcoma (c-Fos), Zif-268 [37], activity-regulated cytoskeleton protein (Arc), calcium-calmodulin dependent protein kinase II (CaMKII; [38]) and protein kinase C (PKC) isoforms (for review see [35, 39]). Notably, both c-Fos and Arc are associated with LTP, and increases in their expression are observed within minutes following an experience (for review see [39, 40]). For these reasons, much of the work examining the cellular organization of memory traces or engrams in rodents has leveraged these properties to selectively tag neuronal ensembles recruited to a memory trace (e.g., [41]).
Persistence of Long-Term Memories
An enduring focus in the study of the molecular biology of memory storage has been in the search for mechanisms which allow a LTM to persist. Many of the transcriptional events and post-translational modifications of proteins are short-lived (i.e., on the order of a few hours to days). This raises the challenging question: How is a LTM maintained in synaptic connections that are so dynamic [42] in a manner that allows a memory to persist throughout the life-span of an organism? In the early 1980s, it was hypothesized that the persistence of memories requires an “intramolecular autocatalytic” reaction [43-45], that is, a molecular mechanism that once activated persists in a self-sustaining manner. Due to the ability to autophosphorylate at a specific threonine residue, calcium calmodulin dependent protein kinase II (CAMKII) became an attractive candidate mechanism for the maintenance of LTMs [46]. Another protein-kinase, protein-kinase-M-zeta (PKMζ), an atypical isoform of PKC, has also been proposed as a necessary component for the maintenance of LTP and the persistence of LTMs given that only the catalytically active form of the gene is transcribed following stimulation [47, 48]. Despite controversy [49], PKMζ is a particularly interesting candidate in the persistence of LTMs in that its mRNA is transported to dendrites and locally translated upon induction of LTP. Moreover, PKMζ regulates the endocytosis of GLUA2-containing AMPA receptors in addition to possibly regulating the structure of dendritic spines [50].
Similar to PKMζ, mRNA of the immediate-early gene Arc is also transported to activated synapses [51] where it too regulates AMPA receptor endocytosis [52]. Emerging studies on the properties of Arc have found that its function may be more complex than previously thought in that the protein contains a structural likeness to group-specific antigen proteins expressed by viruses [53, 54]. Moreover, Arc protein can bind its own mRNA and transport it across the synapse through extracellular vesicle [53-55]. These studies are of considerable importance in that they suggest a non-canonical mechanism for the synaptic transmission of information that would constitute a novel mechanism for the molecular control and possible persistence of LTMs [53].
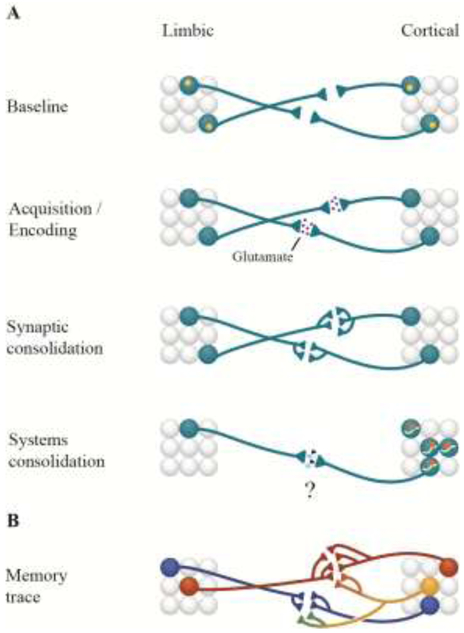
At the turn of the century, work from our laboratory identified functional prions as another molecular candidate for the persistence of long-term memories [56]. Like their pathogenic counterparts, functional prions contain Q/N rich “prion-like” domains that promote aggregation. However, functional prions are distinguished from pathogenic prions in that their aggregation is tightly regulated and serves a physiological function. These functional prions exist in a soluble conformation until activated, at which point they oligomerize, become self-sustaining, and contribute to the consolidation of LTMs [56]. One particular functional prion, the RNA-binding protein cytoplasmic polyadenylation binding protein-3 (CPEB-3), was identified by this laboratory as important for the maintenance of LTP and hippocampal-dependent memories in mice [57]. These studies, along with work on the function of Arc, raise several intriguing questions: Can a self-sustaining molecule transport RNA between the pre- and post-synaptic compartments of the activated synapses of a memory trace and modify the synaptic architecture of a LTM? Can RNA-binding proteins and epigenetic modifications to the RNA that they carry serve as a synaptic substrate for storing information in select neurons during systems consolidation, or under conditions of massive dynamic change (see Figure 1A; [58])?. Indeed, different types of RNA (e.g., mRNA, miRNA, snRNA, etc.) perform fundamentally different roles in LTMs [59, 60] and the synaptic transfer of exosomally-packaged RNA between neurons [61] may be an important mechanism that warrants further investigation.
Figure 1.
Molecular Mechanisms of the Memory Trace. (A) At baseline, increased levels of CREB (yellow circles) are thought to determine which neurons are preferentially incorporated into a memory trace (blue circles). Acquisition/encoding of a long-term memory involves increased glutamatergic transmission between neurons of a memory trace. Synaptic consolidation and the transition from short-term to long-term memory involves the transcription of new genes, new proteins, and synaptic growth. Systems consolidation involves a greater reliance on cortical areas with the passage of time, in addition to epigenetic changes (e.g., methylation; orange circles) in genes involved in learning and memory. An important future direction (question mark) is in understanding if/how RNA-binding proteins (light blue diamonds) and modifications to the RNA they carry as well as RNA packaged into exosomes (black triangles) contribute to systems consolidation. (B) Recent progress has dissected some of the principles of how individual memories are represented in the brain. Stronger memories (orange circles) involve greater initial synaptic connectivity between brain regions relative to weaker memories (blue circles). Moreover, similar memories acquired close in time recruit an overlapping ensemble of neurons (yellow circle). However, these neurons can represent individual memories in a synapse-specific fashion (light orange vs. green processes). An important direction for future research would be to identify the molecular mechanisms which control how similar or stronger memories within overlapping ensembles are preserved during systems consolidation.
Research examining the function of RNA-binding proteins and RNA trafficking between synapses of a memory trace at multiple time points following acquisition of LTMs will lend new insights into the self-sustaining molecular machinery which contribute to systems consolidation. To answer these questions, future research must blend activity-dependent tagging strategies (detailed below) with novel tools that offer temporally-precise molecular control over these molecules such as optically-controlled protein degradation [62], optically-controlled dominant-negative protein inhibition [63], and real-time molecular imaging as has recently been accomplished with Arc [64]. By combining these in vivo approaches with techniques in single-cell sequencing (e.g., patch-seq., act-seq, etc. [65, 66]) to isolate neurons of a memory trace at various time points, we will gain a deeper insight into how the molecular landscape changes during systems consolidation. These tools also offer the ability to determine the necessity and sufficiency of molecules such as ARC, CPEB-3, and others yet to be identified in systems consolidation [62, 63].
Beyond the traditional transcriptional and proteomic machinery of LTMs, recent studies have also identified the roles of a variety of post-translational epigenetic modifications in LTMs [6, 67, 68]. For example, methylation and acetylation of DNA as well as RNA [69] have a powerful influence over LTMs [70, 71]. The temporally-dependent emergence of epigenetic modifications to the DNA of key learning and memory proteins such as calcineurin following LTM acquisition has been suggested to contribute to systems consolidation and LTM maintenance [72]. The importance of site-specific epigenetic modifications may also extend to the transgenerational inheritance and persistence of specific LTMs [73]. However, much remains to be learned about the role of epigenetic modifications with regard to systems consolidation. Newer tools which provide high temporal control over RNA and epigenetic modifications such as transcription activator-like effectors (TALEs), chemo-optical modulation of epigenetically regulated transcription (COMET; [74]), and optically activated CRISPR/Cas9 [75] represent sophisticated new approaches for assessing the temporal necessity and sufficiency of epigenetic marks and synaptic RNA regulation. Moreover, integrating these tools with strategies to access select memory traces will enhance our understanding of which mechanisms modulate memory persistence and systems consolidation.
Memory Traces in mammalian systems
For nearly a century, the question of how long-term memories are stored within the mammalian brain has been an area of intense interest and debate [76-80]. Recent technological advances in tagging and optically controlling active neuronal ensembles have invigorated research into the cellular representation of discrete LTMs in mice [81]. This work has relied on viral delivery and antibiotic-dependent expression strategies [8, 82] to leverage the properties of immediate-early genes such as c-Fos and Arc to tag neuronal ensembles recruited to a particular memory.
The creation of transgenic mice that utilize a tetracycline-transactivator (tet) ON or OFF system or more recently a Cre-ERT2 fusion system [82] to achieve selective targeting of neurons activated during the acquisition of a LTM has been essential. In the tet-OFF system, a transgenic animal expresses the tTA gene (a fusion of the TetR repressor protein with the c-terminal domain of a herpes simplex virus protein) under the control of an immediate early gene promoter [8]. The activity of tTA can be inhibited by the antibiotic doxycycline. Removal of doxycycline from the diet enables tTA to bind to a Tet operator located within a tetracycline-response element (TRE) promoter to transcribe a target gene. By driving a viral transgene with a TRE promoter, regional and temporal expression of specific genes (e.g., opsins, designer receptors exclusively activated by designer drugs (DREADDs), fluorophores, toxins, etc.) can be achieved in a specific subset of recently activated neurons.
Using activity-dependent tagging approaches, research during the past decade has applied these and other novel technologies to dissect how neurons that are activated during the formation of a LTM can regulate distinct phases of hippocampal-dependent memories. For example, recent work has found that optogenetic reactivation of c-Fos+ neurons in the dentate gyrus (DG), a subregion of the hippocampal formation important for the acquisition and retrieval of contextual fear memories [83], tagged during the acquisition of a long-term fear memory is sufficient to elicit retrieval of a recently acquired LTM in a novel environment [84, 85]. Perhaps even more striking is the finding that when the hippocampal Cornu Ammonis field 1 (CA1) region or DG c-Fos+ neurons labeled in a neutral context are optogenetically stimulated during the acquisition of a long-term fear memory, these neurons are recruited to the fear memory trace, and fear behavior can be elicited in the once-neutral context [85, 86]. Conversely, optogenetic silencing of DG and CA3 ARC+ neurons or CA1 c-Fos+ neurons tagged during acquisition can inhibit the recall of a LTM [82, 87]. Consistent with the idea that fear memories require brain-wide networks [88], newer studies have demonstrated the functional contribution of neuronal ensembles in cortical areas such as the retrosplenial cortex [89]; and limbic areas such as the amygdala during the acquisition and recent retrieval of LTMs [90]. One particularly interesting discovery is that, if neurons labeled in a neutral context are re-activated during the retrieval of a fear memory, they can interfere with recall of the recently acquired LTM [85, 86]. These observations have been extended to show how activating neuronal ensembles associated with different types of memory (e.g., positive memories) can disrupt aversive LTMs and potentially serve as a therapeutic intervention [91]. These studies reveal the delicate spatial and temporal constraints under which neuronal ensembles represent discrete LTMs in the mammalian brain.
Much of this work has focused on recently acquired LTMs (i.e., a few days), but studies are now beginning to focus on remote LTMs (i.e., a few weeks old or older) to examine how memories evolve during systems consolidation (for review see [92]). This is important because with the passage of time, systems consolidation relies progressively more on cortical areas and less on the hippocampus in a process that may be mediated by hippocampal sharp-wave ripples (for review see [93, 94]; but also see recent evidence that cortical areas are recruited early-on for systems consolidation [89, 95-97]). Recent work has found that by tagging the subset of DG neurons that are still active during the remote recall of a fear memory, and subsequently activating those neurons during fear extinction, the extinction of an aversive memory at remote time-points is facilitated [98]. Moreover, optogenetic silencing of neurons initially recruited to the memory trace in the prefrontal cortex at remote, but not recent, time-points is sufficient to disrupt LTMs [97] – a finding which reinforces an early-role for the prefrontal cortex as well as the slow maturation of region- and gene-specific epigenetic marks in systems consolidation [72, 95, 99-101]. Together, these studies highlight how specific neuronal populations in different brain regions can have varying influence over a particular LTM with the passage of time. They also raise the important question: What are the molecular mechanisms which control how cortical cells mature to store information important for retrieving particular memories?
Studies are now beginning to examine the molecular and structural mechanisms which guide the allocation of particular neurons to a memory trace – a focus which will be critical to understanding how information is preserved during systems consolidation. This research has concentrated on answering questions such as: What are the molecular mechanisms that control the initial allocation of a neuronal ensemble to a particular memory trace? Or what differentiates stronger LTMs from weaker ones within discrete circuits? While it is generally thought that neurons with higher excitability are initially recruited to form a LTM trace [102], the molecular mechanisms which drive this allocation have been elusive. Key studies have found that in addition to its role in regulating transcription, elevated levels of CREB in neurons increases neuronal excitability and increases the likelihood that specific neurons (e.g., in the lateral amygdala) are recruited to a memory trace (see Figure 1A; [103-105]). Moreover, research has uncovered fundamental rules about neuronal allocation within a LTM trace. For example, similar, but non-identical, aversive memories (i.e., fear conditioning using different tones or contexts) that are acquired closely in time recruit an overlapping ensemble of neurons in the amygdala and CA1 [90, 106]. Furthermore, these ensembles can represent each memory in a synapse-specific manner (see Figure 1B; [105]). However, when the acquisition of similar LTMs has a greater temporal separation, the likelihood of this overlap diminishes. While neuronal ensembles of a particular memory are linked across brain regions, when similar memories differ in strength, stronger memories are initially differentiated by greater synaptic connectivity between CA3→CA1 neurons recruited to a memory trace relative to weaker memories [107]. It is still unclear how connectivity at the circuit-level for a particular memory evolves during systems consolidation given that synapses themselves are dynamic [42]. However, these studies are an important step forward in identifying the biological mechanisms of systems consolidation in mammals. They provide a foundation to interrogate, for the first time, the molecular mechanisms that control how distinct LTMs evolve and persist within shared neuronal ensembles (see Figure 1B).
The research on memory traces discussed above highlights a number of key organizing principles. First, levels of CREB may determine the excitability of neurons and guide which neurons are recruited to a LTM trace [103]. Second, a LTM trace is represented by a discrete subset of neurons, while activation of other neurons recruited to a different memory trace can interfere with recall. Third, and consistent with past work [27, 28], the initial strength of a LTM trace is represented by the number of pre- and post-synaptic connections between different brain regions (as shown using elegant virus-mediated tracing techniques [107]). Fourth, similar memories may recruit a shared neuronal ensemble, but this is constrained by the temporal separation of acquiring each memory in addition to a synapse-specific representation of each memory [105]. Fifth, with the passage of time, LTMs rely more heavily on cortical structures in a process that involves the delayed maturation of cortical neurons that were, in fact, initially recruited to the memory trace [95, 97, 99, 100]. While we have primarily focused on aversive learning and memory, these studies provide a framework for research to now examine the molecular biology of systems consolidation and for understanding how LTMs evolve over time within discrete neural circuits.
Concluding Remarks
In the coming decade, research can capitalize on new technologies to elucidate the molecular mechanisms (e.g., synaptic RNA, RNA-binding proteins, and epigenetic regulation of RNA) which govern systems consolidation and the persistence of LTMs (see Figure 1 and Outstanding Questions). By combining activity-dependent tagging strategies with neural-circuit targeting strategies and novel techniques for molecular interrogation, this research focus is poised to obtain important insights into how memories are stored and preserved across time. New techniques such as iDISCO [108], CLARITY [109], in vivo optical control of proteins [62, 63], optical control of epigenetic marks [74], next-generation sequencing [65, 66], optical control of gene editing [75], in vivo transcriptional imaging [64], and viral mediated-tracing tools [110, 111] will make a significant contribution to furthering our knowledge. Importantly, a focus on the molecular biology of systems consolidation will provide a critical framework for how memories are preserved in the face of biological and circuit dynamicity. The past half century has provided considerable insight into the molecular mechanisms of synaptic consolidation. Progress in the coming decades will provide the necessary scaffold for understanding the molecular biology of systems consolidation and how memories persist across our lifespan. This progress will be an essential component in the development of better-targeted treatments for disorders of memory such as Alzheimer’s disease, age-related memory loss, post-traumatic stress disorder, and many others.
Outstanding Questions.
Is the storage of long-term memory influenced by exosomally transported RNA and associated RNA-binding proteins? Can this transfer modify synaptic connections or store information in the face of dynamicity during systems consolidation?
How do epigenetic modifications to DNA and exosomally transported RNA evolve within a defined long-term memory trace? Can these epigenetic modifications contribute to long-term memory during systems consolidation?
What are the molecular mechanisms that preserve a long-term memory as neocortical areas undergo progressively greater recruitment over time?
Some memories are expected to initially recruit partly overlapping neuronal ensembles. What are the molecular mechanisms that regulate the systems-level consolidation of such memories? How are these overlapping representations preserved during systems consolidation?
How do highly-similar memories evolve within neuronal ensembles during systems consolidation?
Can overlapping ensembles represent similar memories that differ in strength? And if so, what are the molecular mechanisms which allow for these long-term memories to co-exist and persist during systems consolidation?
Highlights.
The molecular biology of Synaptic consolidation is relatively well-defined. By contrast, the molecular mechanisms of systems consolidation remain poorly understood.
Recent technological developments have helped advance our understanding of the cellular representation of memories in the brain.
One of the goals of future research is to clarify how non-canonical forms of synaptic transmission (e.g., exosomally transported RNA, associated RNA-binding proteins, and epigenetic modifications to the transported RNA) may contribute to systems consolidation.
Integrating tools for examining these non-canonical cellular and molecular mechanisms with tools for tagging select circuits of a memory trace will be highly informative for elucidating the molecular mechanisms of systems consolidation.
Acknowledgments
We are grateful for support from the Howard Hughes Medical Institute and from Cohen Veterans Biosciences to E.R.K in addition to the National Institute of Mental Health 1F32-MH114306 to A.A. We thank Sarah Mack and Mariah Widman for help with the figure.
Glossary
- Acquisition/encoding
the initial processing of information within a defined neuronal ensemble for a specific experience or event on times-scales lasting seconds or minutes.
- Extracellular vesicle
membranous compartment that is capable of packaging and transferring proteins, RNA, etc. from pre-synaptic to post-synaptic cells
- Long-term Memory (LTM)
the storage of information for a specific experience or event on a prolonged time scale such as days, weeks, months or years following acquisition.
- Molecular mechanisms of long-term memory
the transcriptomic, proteomic and epigenetic mechanisms that are involved in long-term memory consolidation.
- Memory trace/Engram
the cellular representation of a long-term memory by neuronal ensembles across different brain regions.
- Persistence
the preservation of a stored long-term across neuronal ensembles in the timeframe of days, weeks, months, or years following the initial storage.
- Storage/consolidation
the conversion of short-term memory into long-term memory within a defined neuronal ensemble in the timeframe of hours.
- Synaptic consolidation
the molecular mechanisms associated with the long-term potentiation of a synaptic connection and linked to a long-term memory.
- Systems consolidation
the maturation of a long-term memory to more heavily recruit cortical/neocortical brain regions.
- Recall/Retrieval
the reactivation of a neuronal ensemble which has stored a specific long-term memory.
- Recent LTM
a long-term memory acquired previously in the timeframe of days or less
- Remote LTM
a long-term memory acquired previously in the timeframe of weeks or longer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mayford M et al. (2012) Synapses and memory storage. Cold Spring Harb. Perspect. Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bliss TV and Collingridge GL (1993) A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361, 31–39. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y-S (2014) Genes and signaling pathways involved in memory enhancement in mutant mice. Mol. Brain 7, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarome TJ and Helmstetter FJ (2014) Protein degradation and protein synthesis in long-term memory formation. Front. Mol. Neurosci DOI: 10.3389/fnmol.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day JJ and Sweatt JD (2010) DNA methylation and memory formation. Nat. Neurosci 13, 1319–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gräff J and Tsai L-H (2013) Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci 14, 97–111. [DOI] [PubMed] [Google Scholar]
- 7.McGaugh JL (2000) Memory--a century of consolidation. Science 287, 248–251. [DOI] [PubMed] [Google Scholar]
- 8.Reijmers LG et al. (2007) Localization of a stable neural correlate of associative memory. Science 317, 1230–1233. [DOI] [PubMed] [Google Scholar]
- 9.Hebb DO (1949) The organization of behavior: A neurophysiological approach, Wiley JH. [Google Scholar]
- 10.Penfield W and Milner B (1958) Memory deficit produced by bilateral lesions in the hippocampal zone. AMA Arch. Neurol. Psychiatry 79, 475–497. [DOI] [PubMed] [Google Scholar]
- 11.Scoville WB and Milner B (1957) Loss of recent memory after bilateral hippocampal lesions. J. Neurol. Neurosurg. Psychiatry 20, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bliss TV and Lømo T (1973) Long- lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol 232, 331–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markram H et al. (2011) A history of spike-timing-dependent plasticity. Front. Synaptic. Neurosci DOI: 10.3389/fnsyn.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweatt JD (2009) Mechanisms of memory, Academic Press. [Google Scholar]
- 15.Malenka RC and Bear MF (2004) LTP and LTD: an embarrassment of riches. Neuron 44, 5–21. [DOI] [PubMed] [Google Scholar]
- 16.Nabavi S et al. (2014) Engineering a memory with LTD and LTP. Nature 511, 348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scharf MT et al. (2002) Protein synthesis is required for the enhancement of long-term potentiation and long-term memory by spaced training. J. Neurophysiol 87, 2770–2777. [DOI] [PubMed] [Google Scholar]
- 18.Nicoll RA (2017) A brief history of long-term potentiation. Neuron 93, 281–290. [DOI] [PubMed] [Google Scholar]
- 19.Whitlock JR et al. (2006) Learning induces long-term potentiation in the hippocampus. Science 313, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 20.Dudai Y (2004) The neurobiology of consolidations, or, how stable is the engram? Annu. Rev. Psychol 55, 51–86. [DOI] [PubMed] [Google Scholar]
- 21.Kandel ER (2001) The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030–1038. [DOI] [PubMed] [Google Scholar]
- 22.Kandel ER (2009) The biology of memory: a forty-year perspective. J. Neurosci 29, 12748–12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shuster M et al. (1985) Cyclic AMP-dependent protein kinase closes the serotonin-sensitive K+ channels of Aplysia sensory neurones in cell-free membrane patches. Nature 313, 392–395. [DOI] [PubMed] [Google Scholar]
- 24.Siegelbaum SA et al. (1982) Serotonin and cyclic AMP close single K+ channels in Aplysia sensory neurones. Nature 299, 413–417. [DOI] [PubMed] [Google Scholar]
- 25.Montarolo P et al. (1986) A critical period for macromolecular synthesis in long-term heterosynaptic facilitation in Aplysia. Science 234, 1249–1254. [DOI] [PubMed] [Google Scholar]
- 26.Schacher S et al. (1988) cAMP evokes long-term facilitation in Aplysia sensory neurons that requires new protein synthesis. Science 240, 1667–1669. [DOI] [PubMed] [Google Scholar]
- 27.Bailey CH and Chen M (1983) Morphological basis of long-term habituation and sensitization in Aplysia. Science 220, 91–93. [DOI] [PubMed] [Google Scholar]
- 28.Bailey CH and Chen M (1989) Structural plasticity at identified synapses during long-term memory in Aplysia. Dev. Neurobiol 20, 356–372. [DOI] [PubMed] [Google Scholar]
- 29.Dash PK et al. (1990) Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature 345, 718–721. [DOI] [PubMed] [Google Scholar]
- 30.Bartsch D et al. (2000) Enhancement of memory-related long-term facilitation by ApAF, a novel transcription factor that acts downstream from both CREB1 and CREB2. Cell 103, 595–608. [DOI] [PubMed] [Google Scholar]
- 31.Martin KC et al. (1997) MAP kinase translocates into the nucleus of the presynaptic cell and is required for long-term facilitation in Aplysia. Neuron 18, 899–912. [DOI] [PubMed] [Google Scholar]
- 32.Bartsch D et al. (1995) Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell 83, 979–992. [DOI] [PubMed] [Google Scholar]
- 33.Bailey CH et al. (1997) Mutation in the phosphorylation sites of MAP kinase blocks learning-related internalization of apCAM in Aplysia sensory neurons. Neuron 18, 913–924. [DOI] [PubMed] [Google Scholar]
- 34.Schmid RS et al. (1999) NCAM stimulates the ras- MAPK pathway and CREB phosphorylation in neuronal cells. Dev. Neurobiol 38, 542–558. [PubMed] [Google Scholar]
- 35.Giese KP and Mizuno K (2013) The roles of protein kinases in learning and memory. Learn. Mem 20, 540–552. [DOI] [PubMed] [Google Scholar]
- 36.Abel T and Lattal KM (2001) Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr. Opin. Neurobiol 11, 180–187. [DOI] [PubMed] [Google Scholar]
- 37.Ryan MM et al. (2012) Temporal profiling of gene networks associated with the late phase of long-term potentiation in vivo. PLoS One DOI: 10.1371/journal.pone.0040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayford M et al. (1996) Control of memory formation through regulated expression of a CaMKII transgene. Science 274, 1678–1683. [DOI] [PubMed] [Google Scholar]
- 39.Alberini CM (2009) Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev 89, 121–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korb E and Finkbeiner S (2011) Arc in synaptic plasticity: from gene to behavior. Trends Neurosci 34, 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reijmers L and Mayford M (2009) Genetic control of active neural circuits. Frontiers in molecular neuroscience 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choquet D and Triller A (2013) The dynamic synapse. Neuron 80, 691–703. [DOI] [PubMed] [Google Scholar]
- 43.Crick F (1984) Neurobiology: Memory and molecular turnover. Nature 312, 101. [DOI] [PubMed] [Google Scholar]
- 44.Lisman JE (1985) A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proc. Natl. Acad. Sci. U.S.A 82, 3055–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberson ED and Sweatt JD (1999) A biochemical blueprint for long-term memory. Learn. Mem 6, 381–388. [PMC free article] [PubMed] [Google Scholar]
- 46.Lisman J (1994) The CaM kinase II hypothesis for the storage of synaptic memory. Trends Neurosci 17, 406–412. [DOI] [PubMed] [Google Scholar]
- 47.Sacktor TC (2011) How does PKMζ maintain long-term memory? Nat. Rev. Neurosci 12, 9–15. [DOI] [PubMed] [Google Scholar]
- 48.Ling DS et al. (2002) Protein kinase Mζ is necessary and sufficient for LTP maintenance. Nat. Neurosci 5, 295–296. [DOI] [PubMed] [Google Scholar]
- 49.Kwapis JL and Helmstetter FJ (2014) Does PKM (zeta) maintain memory? Brain research bulletin 105, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glanzman DL (2013) PKM and the maintenance of memory. F1000 Biol. Rep DOI: 10.3410/B5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steward O and Worley PF (2001) Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron 30, 227–240. [DOI] [PubMed] [Google Scholar]
- 52.Chowdhury S et al. (2006) Arc/Arg3. 1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pastuzyn ED et al. (2018) The neuronal gene Arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell 172, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashley J et al. (2018) Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172, 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holm MM et al. (2018) Extracellular Vesicles: Multimodal Envoys in Neural Maintenance and Repair. Trends Neurosci 41, 360–372. [DOI] [PubMed] [Google Scholar]
- 56.Rayman JB and Kandel ER (2016) Functional prions in the brain. Cold Spring Harb. Perspect. Biol DOI: 10.1101/cshperspect.a023671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fioriti L et al. (2015) The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron 86, 1433–1448. [DOI] [PubMed] [Google Scholar]
- 58.Ziv Y et al. (2013) Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci 16, 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bédecarrats A et al. (2018) RNA from Trained Aplysia Can Induce an Epigenetic Engram for Long-Term Sensitization in Untrained Aplysia. eNeuro DOI: 10.1523/eneuro.0038-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajasethupathy P et al. (2012) A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. 149, 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chivet M et al. (2012) Emerging role of neuronal exosomes in the central nervous system. Front. Physiol DOI: 10.3389/fphys.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renicke C et al. (2013) A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol 20, 619–626. [DOI] [PubMed] [Google Scholar]
- 63.Ali AM et al. (2015) Optogenetic inhibitor of the transcription factor CREB. Chem. Biol 22, 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das S et al. (2018) A transgenic mouse for imaging activity-dependent dynamics of endogenous Arc mRNA in live neurons. Sci. Adv DOI: 10.1126/sciadv.aar3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu YE et al. (2017) Detecting activated cell populations using single-cell RNA-seq. Neuron 96, 313–329. [DOI] [PubMed] [Google Scholar]
- 66.Cadwell CR et al. (2016) Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol 34, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Landry CD et al. (2013) New mechanisms in memory storage: piRNAs and epigenetics. Trends Neurosci 36, 535–542. [DOI] [PubMed] [Google Scholar]
- 68.Blaze J and Roth TL (2013) Epigenetic mechanisms in learning and memory. Wiley Interdiscip. Rev. Cogn. Sci DOI: 10.1002/wcs.1205. [DOI] [PubMed] [Google Scholar]
- 69.Widagdo J et al. (2016) Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci 36, 6771–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chwang WB et al. (2006) ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn. Mem 13, 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller CA and Sweatt JD (2007) Covalent modification of DNA regulates memory formation. Neuron 53, 857–869. [DOI] [PubMed] [Google Scholar]
- 72.Miller CA et al. (2010) Cortical DNA methylation maintains remote memory. Nat. Neurosci 13, 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dias BG and Ressler KJ (2014) Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci 17, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reis SA et al. (2016) Light-controlled modulation of gene expression by chemical optoepigenetic probes. Nat. Chem. Biol 12, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nihongaki Y et al. (2015) Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol 33, 755–760. [DOI] [PubMed] [Google Scholar]
- 76.Semon RW (1911) Die Mneme als erhaltendes Prinzip im Wechsel des organischen Geschehens, Engelmann. [Google Scholar]
- 77.Tonegawa S et al. (2015) Memory engram cells have come of age. Neuron 87, 918–931. [DOI] [PubMed] [Google Scholar]
- 78.y Cajal SR (1894) The Croonian lecture: La fine structure des centres nerveux. Proc. R. Soc. Lond 55, 444–468. [Google Scholar]
- 79.Konorski J (1948) Conditioned reflexes and neuron organization, Cambridge University Press. [Google Scholar]
- 80.Josselyn SA et al. (2015) Finding the engram. Nat. Rev. Neurosci 16, 521–534. [DOI] [PubMed] [Google Scholar]
- 81.Lashley KS (1950) In search of the engram In Society for Experimental Biology, Physiological mechanisms in animal behavior, 454–482, Academic Press. [Google Scholar]
- 82.Denny CA et al. (2014) Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron 83, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bernier BE et al. (2017) Dentate gyrus contributes to retrieval as well as encoding: Evidence from context fear conditioning, recall, and extinction. J. Neurosci, 6359–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu X et al. (2012) Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garner AR et al. (2012) Generation of a synthetic memory trace. Science 335, 1513–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramirez S et al. (2013) Creating a false memory in the hippocampus. Science 341, 387–391. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka KZ et al. (2014) Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron 84, 347–354. [DOI] [PubMed] [Google Scholar]
- 88.Vetere G et al. (2017) Chemogenetic interrogation of a brain-wide fear memory network in mice. Neuron 94, 363–374. [DOI] [PubMed] [Google Scholar]
- 89.Cowansage KK et al. (2014) Direct reactivation of a coherent neocortical memory of context. Neuron 84, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rashid AJ et al. (2016) Competition between engrams influences fear memory formation and recall. Science 353, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramirez S et al. (2015) Activating positive memory engrams suppresses depression-like behaviour. Nature 522, 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frankland PW and Bontempi B (2005) The organization of recent and remote memories. Nat. Rev. Neurosci 6, 119–130. [DOI] [PubMed] [Google Scholar]
- 93.Tonegawa S et al. (2018) The role of engram cells in the systems consolidation of memory. Nat. Rev. Neurosci 19, 485–498. [DOI] [PubMed] [Google Scholar]
- 94.Squire LR and Alvarez P (1995) Retrograde amnesia and memory consolidation: a neurobiological perspective. Curr. Opin. Neurobiol 5, 169–177. [DOI] [PubMed] [Google Scholar]
- 95.Lesburguères E et al. (2011) Early tagging of cortical networks is required for the formation of enduring associative memory. Science 331, 924–928. [DOI] [PubMed] [Google Scholar]
- 96.Bero AW et al. (2014) Early remodeling of the neocortex upon episodic memory encoding. Proc. Natl. Acad. Sci. U.S.A 111, 11852–11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kitamura T et al. (2017) Engrams and circuits crucial for systems consolidation of a memory. Science 356, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khalaf O et al. (2018) Reactivation of recall-induced neurons contributes to remote fear memory attenuation. Science 360, 1239–1242. [DOI] [PubMed] [Google Scholar]
- 99.Vetere G et al. (2011) Spine growth in the anterior cingulate cortex is necessary for the consolidation of contextual fear memory. Proc. Natl. Acad. Sci. U.S.A 108, 8456–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takehara-Nishiuchi K et al. (2006) Systems consolidation requires postlearning activation of NMDA receptors in the medial prefrontal cortex in trace eyeblink conditioning. J. Neurosci 26, 5049–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao M-G et al. (2005) Roles of NMDA NR2B subtype receptor in prefrontal long-term potentiation and contextual fear memory. Neuron 47, 859–872. [DOI] [PubMed] [Google Scholar]
- 102.Bittner KC et al. (2015) Conjunctive input processing drives feature selectivity in hippocampal CA1 neurons. Nat. Neurosci 18, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Josselyn SA and Frankland PW (2018) Memory Allocation: Mechanisms and Function. Annu. Rev. Neurosci 41, 389–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Han JH et al. (2007) Neuronal competition and selection during memory formation. Science 316, 457–460. [DOI] [PubMed] [Google Scholar]
- 105.Abdou K et al. (2018) Synapse-specific representation of the identity of overlapping memory engrams. Science 360, 1227–1231. [DOI] [PubMed] [Google Scholar]
- 106.Cai DJ et al. (2016) A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Choi JH et al. (2018) Interregional synaptic maps among engram cells underlie memory formation. Science 360, 430–435. [DOI] [PubMed] [Google Scholar]
- 108.Pavlova IP et al. (2018) Optimization of immunolabeling and clearing techniques for indelibly- labeled memory traces. Hippocampus 28, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chung K and Deisseroth K (2013) CLARITY for mapping the nervous system. Nat. Methods 10, 508–513. [DOI] [PubMed] [Google Scholar]
- 110.Nassi JJ et al. (2015) Neuroanatomy goes viral! Front. Neuroanat DOI: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim J et al. (2012) mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat. Methods 9, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]