Abstract
Human DNA polymerase δ is normally present in unstressed, non-dividing cells as a heterotetramer (Pol δ4). Its smallest subunit, p12, is transiently degraded in response to UV damage, as well as during the entry into S-phase, resulting in the conversion of Pol δ4 to a trimer (Pol δ3). In order to further understand the specific cellular roles of these two forms of Pol δ, the gene (POLD4) encoding p12 was disrupted by CRISPR/Cas9 to produce p12 knockout (p12KO) cells. Thus, Pol δ4 is absent in p12KO cells, leaving Pol δ3 as the sole source of Pol δ activity. GFP reporter assays revealed that the p12KO cells exhibited a defect in homologous recombination (HR) repair, indicating that Pol δ4, but not Pol δ3, is required for HR. Expression of Flag-tagged p12 in p12KO cells to restore Pol δ4 alleviated the HR defect. These results establish a specific requirement for Pol δ4 in HR repair. This leads to the prediction that p12KO cells should be more sensitive to chemotherapeutic agents, and should exhibit synthetic lethal killing by PARP inhibitors. These predictions were confirmed by clonogenic cell survival assays of p12KO cells treated with cisplatin and mitomycin C, and with the PARP inhibitors Olaparib, Talazoparib, Rucaparib, and Niraparib. The sensitivity to PARP inhibitors in H1299-p12KO cells was alleviated by expression of Flag-p12. These findings have clinical significance, as the expression levels of p12 could be a predictive biomarker of tumor response to PARP inhibitors. In addition, small cell lung cancers (SCLC) are known to exhibit a defect in p12 expression. Analysis of several SCLC cell lines showed that they exhibit hypersensitivity to PARP inhibitors, providing evidence that loss of p12 expression could represent a novel molecular basis for HR deficiency.
Keywords: DNA polymerase δ, p12 subunit, HR deficiency, PARP inhibitor, Pol δ4, Pol δ3
1. Introduction
Eukaryotic DNA replication depends on three DNA polymerases, Pol δ Pol ε and Pol α/primase. Pol α synthesizes the RNA/DNA primers that are extended by Pol δ and Pol ε. Biochemical analyses and reconstitution studies as well as genetic approaches (in yeast) support a current paradigm that Pol δ is primarily involved in DNA synthesis on the lagging strand, while Pol ε is primarily involved in synthesis of the leading strand [1,2]. Recent cryo-EM structural analysis of the yeast CMG helicase/Pol ε complex further supports this paradigm [3–5].
Human DNA Pol δ (Pol δ4) consists of a catalytic subunit (p125) and three smaller subunits (p50, p68 and p12) [6–8]. The p12 subunit has emerged as a critical factor in understanding the role of human Pol δ in both replication and repair [6–8]. The p12 subunit is transiently degraded in response to DNA damage by UV and alkylating agents, and also in response to replication stress, leading to the conversion of Pol δ4 to an active trimer, Pol δ3 [9]. Thus, there are two physiologically relevant forms of Pol δ4. Pol δ3 has distinct enzymatic properties from Pol δ4, and exhibits a greater proofreading activity and a decreased tendency for lesion bypass [10,11]. Pre-steady state kinetic analyses have revealed the basis for the differences in properties of Pol δ4 and Pol δ3 [6,7]. Pol δ3 has a decreased kpol, the rate constant for polymerization, and an increased kpol-exo, the rate constant for translocation of the primer terminus from the pol to the exo sites [10]. The decrease in kpol raises the kinetic barrier to polymerization, and the increase in the translocation rate act together to increase proofreading [12,13]. The generation of Pol δ3 in response to UV supports its role as a gap-filling enzyme in nucleotide excision repair [14], and its increased surveillance against translesion synthesis as an adaptation to alkylation damage [6,11].
Pol δ3 has also emerged as having a role in human DNA replication, as p12 is also transiently degraded during the G1/S transition, under the control of CRL4Cdt2. Pol δ3 is the major form of Pol δ during S phase [7,8,15,16]. Both Pol δ3 and Pol δ4 are able to cooperate with Fen1 and DNA ligase I to perform Okazaki fragment processing in reconstitution assays [17]. With Pol δ4 and Fen1, the processing of the blocking oligonucleotide occurs via creation of a flap by strand displacement, followed by Fen1 cleavage, in a manner previously described for the yeast system where short flaps are cleaved [18]. However, the main products with Pol δ3 are primarily single nucleotides. This difference emerged as a lack of strand displacement activity by Pol δ3, which can be understood by the same kinetic differences that govern its proofreading behavior [17].
Both Pol δ4 and Pol δ3 are active polymerases, so that one of the challenges is the assessment of their respective contributions to both DNA synthesis and DNA repair processes in vivo [8]. To address this question, we generated p12-knockout cells by CRISPR/Cas9 gene editing. In this study, we tested the hypothesis that Pol δ4, but not Pol δ3, is involved in HR DNA repair. This is based on observations that Pol δ3 does not perform strand displacement synthesis, so that it might not be expected to be proficient in the process of D-loop extension. Our studies show that p12-knockout cells exhibit a defect in HR.
HR defects have been critically important in cancer etiology, as exemplified by the roles of BRCA1/2 mutations in breast and ovarian cancer predisposition [19]. HR-deficient cells exhibit a higher sensitivity towards chemotherapeutic agents, including platinum derivatives and topoisomerase inhibitors, that cause DNA damage which require HR for repair. Major advances in treatment of ovarian cancers in patients with BRCA1/2 mutations have come from the use of poly (ADP-ribose) polymerase (PARP) inhibitors. These act via a synthetic lethal mechanism with HR defects [19–21]. PARP1 is a sensor for single stranded breaks and was initially shown to have a role in repair of single stranded breaks in a base excision repair pathway [22,23]. The basis for PARP inhibitor synthetic lethality in BRCA1/2 associated cancer was originally thought to be due to generation of excessive DSBs, but is now thought to occur via a trapping mechanism [24]. We have examined the effects of several PARP inhibitors on p12KO cells, and confirmed that they exhibit a greatly increased sensitivity to PARP inhibitors. In addition, there exists a known defect in p12 expression in small cell lung cancers (SCLC) [25]. This defect in p12 was also detected in a subgroup of non-small cell lung cancer patients that was correlated with poorer prognosis [25]. We have examined a group of SCLC derived cell lines. These cells exhibit a defect in p12 expression, and their sensitivities to PARP inhibitors are comparable or greater than those of p12KO cells.
2. Materials and Methods
2.1. Materials
Olaparib, Talazoparib (BMN 673), Niraparib (MK-4827) and Rucaparib were purchased from Medchem Express (MCE). Cisplatin and mitomycin C were obtained from Sigma-Aldrich.
2.1. Cells
Non-small cell lung cancer cell lines A549, H1299, and SCLC cell lines DMS114, H446, H1688 and H69AR were obtained from ATCC.
2.3. Western blot analysis
Western blotting for p12 and other proteins was performed as described previously [9]. Antibodies against individual Pol δ subunits were as previously described [14].
2.4. CRISPR/Cas9-mediated knockout of p12
A549 and H1299 cells in which the p12 subunit was knocked out were generated by CRISPR/Cas9 gene targeting of the p12 gene (POLD4). Plasmids and protocols were supplied by Santa Cruz Biotechnology. Cells were co-transfected with Human POLD4 CRISPR/Cas9 KO Plasmid and POLD4 HDR Plasmid. The first plasmid pool consisted of 3 plasmids with individual guide sequences as well as a gene for expression of the Cas9 nuclease. The second plasmid allows for HR guided repair of the sites targeted by the CRISPR/Cas9 plasmids with the insertion of a Red Fluorescent Protein gene and a puromycin resistance gene, flanked by LoxP sites. Puromycin resistant clones were transfected with Cre vector for the removal of genetic material flanked by LoxP sites. A control CRISPR/Cas9 plasmid containing a non-targeting 20 nt scrambled guide RNA (gRNA) was used as a negative control. Selected clones were analyzed by Western blotting for p12 to confirm disruption of the POLD4 gene.
H1299-KO cells into which Flag-tagged p12 was stably expressed were generated by the use of the mammalian expression vector pcDNA3 (Amersham Biosciences).
2.5. Homologous recombination assay using a GFP reporter system
The HR assay was performed essentially as described by Pierce et al. [26]. The DR-GFP plasmid [26], and the I-SceI expression plasmid, pCBASceI [27] were gifts from Maria Jasin and supplied by Addgene. The readout for the assay was the determination of the number of GFP positive cells by flow cytometry. Statistical analysis was performed with Graphpad Prism v 7.01 by either unpaired t-test or Chi-squared tests.
2.6. Cytotoxicity analysis by clonogenic cell survival assays
Clonogenic cell survival assays were performed by published protocols [28]. Cells were treated with PARP inhibitors at different concentrations for 24 h. The plates were washed and the cells cultured for 11 to 14 days. The plates were stained with crystal violet. Digital images of the colonies were counted using ImageJ. Plating efficiencies (number of colonies counted/number of cells seeded) were determined and used to convert colony counts to the Survival Fraction (colony count/cells seeded X plating efficiency) [28]. Experiments were performed in triplicate. Results were presented as means ± SD. All statistical tests and graphing were performed with the GraphPad Prism version 7.01. Cytotoxicity analysis for the chemotherapeutic agents cisplatin and mitomycin C were performed in a similar manner except that exposure to the drugs was for 3 h.
3. Results and Discussion
3.1. Generation of p12 knockout cells by CRISPR/Cas9 gene editing
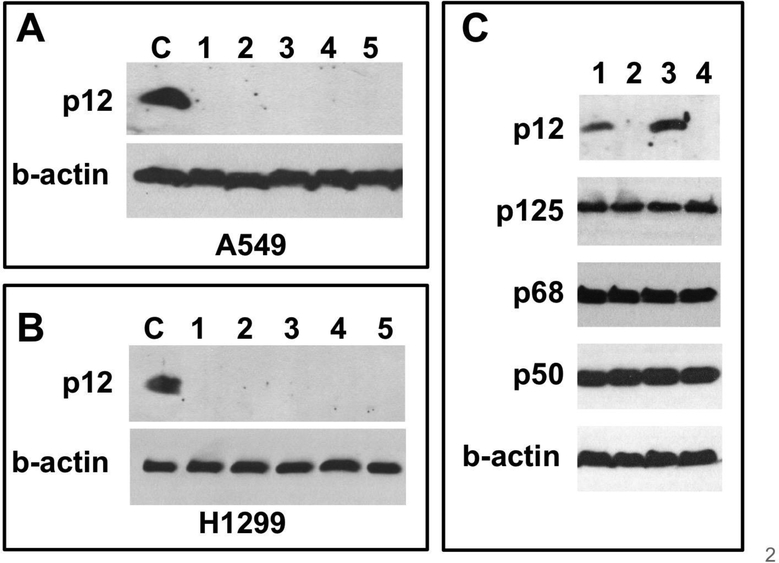
A549 and H1299 non-small lung cancer cell lines were used for these studies. We have previously studied the regulation of p12 levels in response to DNA damage in these cells [9,14,16,29]. The POLD4 gene that encodes the p12 subunit was disrupted by CRISPR/Cas9 gene editing (Materials and Methods). Screening was performed by Western blotting for p12. The results for a typical experiment are shown in Fig. 1A for A549 cells, and for H1299 cells in Fig. 1B. The A549 and H1299 knockout cells will be referred to as A549-p12KO and H1299-p12KO cells.
Fig. 1.
CRISPR/Cas9 ablation of p12 expression in A549 and H1299 cells. (A, B) Western blots for p12 in single cell isolates of A549 and H1299 cells after CRISPR/Cas9 disruption of the gene (POLD4) encoding the p12 subunit of Pol δ (C) Western blots for the p125, p50, p68 and p12 subunits of Pol δ: lanes 1–4: H1299, H1299-p12KO, A549 wt and A549-p12KO cells, respectively. B-actin was used as a loading control.
No significant differences were observed for levels of the p125, p50 and p68 subunits in H1299-p12KO and A549-p12KO and their wild type parental cells as determined by Western blotting (Fig. 1C, cf. lanes 1,2, and lanes 3,4 for the H1299 and A549 cells, respectively). The presence of the p125, p50 and p68 subunits confirms that Pol δ activity in the knockout cells is due solely to the Pol δ3 enzyme.
We encountered little difficulty in isolating p12KO knockout cells. These cells appear to grow normally compared to the wild type cells. The p12KO cells displayed no significant differences from the wild type cells in growth rates by MTT assays, in colony formation efficiency or colony size. Examination of A549 and A549-p12KO cells by standard flow cytometry did not reveal any major changes in cell cycle phase distributions (data not shown). The behavior of our p12KO cells contrasts with reports that siRNA depletion of p12 leads to decreased growth rates, cell cycle abnormalities as well as chromosomal abnormalities including polyploidy and aneuploidy [25,30]. These differences may be due to the different methodologies used, as siRNA methods may be less specific in targeting than the CRISPR/Cas9 gene editing methods.
Thus, these results show that Pol δ4 is not essential for cell survival. This is consistent with a view that Pol δ3 is the primary form of Pol δ involved in Okazaki fragment processing. Nevertheless, these findings do not exclude an ancillary role for Pol δ4 in DNA replication, where it might be involved in negotiating regions of secondary structure [7,8].
3.2. Loss of Pol δ4 leads to a defect in HR repair of double strand breaks
The central goal of this study was to assess the role of Pol δ4 in homologous recombination. D-loop extension is a key step in HR, in which the invading strand is extended by DNA polymerases. The requirement for strand displacement during extension of the invading strand is well recognized, and is somewhat analogous to the processes involved during Okazaki fragment processing [31]. In yeast, reconstitution and genetic studies have identified Pol δ as the primary polymerase involved in HR, noting that Pol ε does not have strand displacement activity [31–33]. In humans, in vitro studies have established that Pol δ4 is able to perform D-loop strand displacement synthesis [34,35]. Since Pol δ3 does not perform strand displacement [17,36], this provides a strong rationale for the view that only Pol δ4 but not Pol δ3 participates in D-loop strand displacement synthesis.
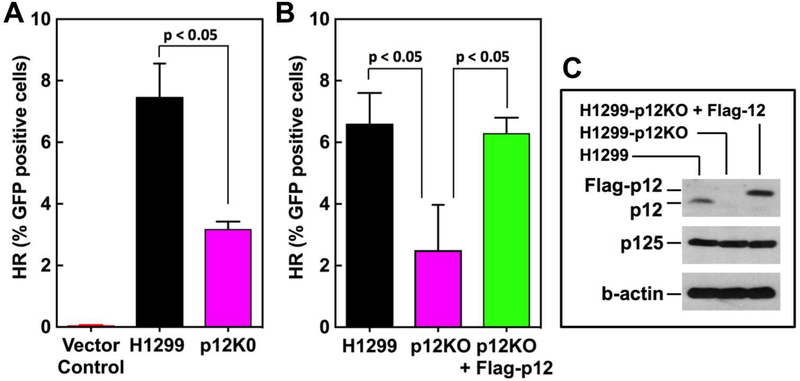
A well-established method based on the DR-GFP plasmid that relies on a GFP reporter gene [26] was used to assess HR repair in H1299 and H1299-p12KO cells. The H1299p12KO cells exhibited a significantly lower level of HR repair (57% decrease) compared to the parental cells (Fig. 2A).
Fig. 2.
GFP reporter assays for HR in p12KO cells. Cells were analyzed for HR activity using the DR-GFP based reporter assay (Material and Methods). Data show the HR activity expressed as % GFP positive cells (n=3). Statistical analysis was performed using Graphpad Prism software. (A) H1299 and H1299-p12KO cells. (B) H1299, H1299-p12KO and H1299-p12KO cells into which Flag-tagged p12 was stably expressed. (C) H1299, H1299-p12KO and H1299-p12KO+Flag-p12 cells were Western blotted for p12, Flag-p12 and the p125 subunit of Pol δ. B-actin was used as a loading control.
To confirm that the loss of HR efficiency is due to loss of p12, we used a H1299-p12KO cell line in which Flag-tagged-p12 was stably expressed (H1299-p12KO+Flag-p12). The results of this complementation experiment (Fig. 2B) show that HR was restored to the level of the wild type H1299 cells. This removes concerns that the HR deficiency observed is due an off-target effect of the CRISPR/Cas9 gene editing process. Validation of the introduction of Flag-p12 into the H1299-p12KO+Flag-p12 cells was demonstrated by Western blotting for p12 (Fig. 2C).
The above experiments, and those that follow in Sections 3.3 and 3.4, demonstrate that Pol δ4 is required for full expression of HR activity in human cells. This contrasts to the situation in budding yeast, which lacks a homolog of p12, so that yeast Pol δ is a three-subunit enzyme. Yeast Pol δ is able to perform strand displacement and is competent in HR [31–33]. Human Pol δ3 does not perform strand displacement and therefore would not be expected to perform displacement synthesis in D-loop extension.
3.3. Loss of Pol δ4 leads to an increased sensitivity to chemotherapeutic agents, cisplatin and mitomycin C
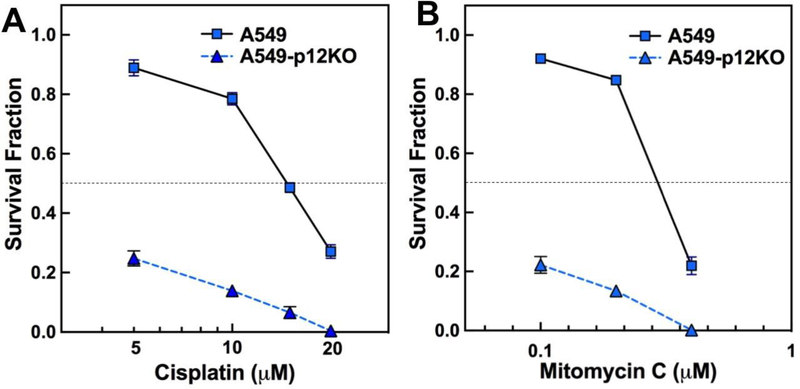
The presence of an HR-defective phenotype in the p12KO cells indicates that these cells should be more sensitive to genotoxic agents that can lead to DNA strand breaks. We examined the cytotoxic effects of two well-established chemotherapeutic agents, cisplatin and mitomycin C, which form intrastrand and interstrand adducts with DNA that lead to increased dependence on HR repair [37,38]. Cytotoxic effects of cisplatin and mitomycin C were analyzed by clonogenic cell survival assays (Materials and Methods). Survival was expressed as the survival fraction, after correction of the colony counts for plating efficiency [28]. The A549-p12KO cells exhibited markedly higher sensitivity to both cisplatin (Fig. 3A) and mitomycin C (Fig. 3B).
Fig. 3.
Cytotoxicity of cisplatin and mitomycin C toward A549 and A549-p12KO cells. The cytotoxicity of cisplatin and mitomycin C was determined by clonogenic survival assays. Cell survival was expressed as survival fraction (Materials and Methods). Cells were exposed to different concentrations of the drugs for 3 h, and analyzed after 11 days of culture. (A) Data for cisplatin. (B) Data for mitomycin C. Results are shown as mean ± SD (n=3). Data for A549 cells are shown as solid squares, solid line and for A549-p12KO cells as solid triangles, dotted line.
3.4. Loss of Pol δ4 leads to an increased sensitivity to PARP inhibitors
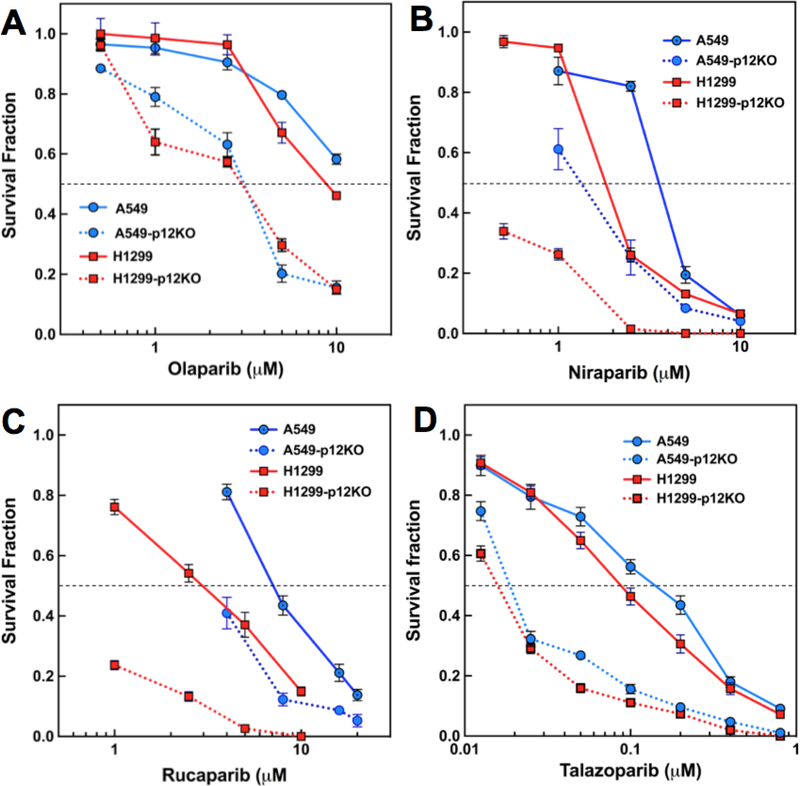
Since the GFP-based reporter assays demonstrated a defect in HR repair, it is predicted that the p12KO cells should exhibit an enhanced sensitivity to PARP inhibitors. The cytotoxicity of the PARP inhibitors Olaparib [39], Niraparib [40], and Rucaparib [41] and Talazoparib (BMN 673) [24] were determined. The first three are in clinical use while Talazoparib is in clinical trials [42]. The p12KO cell lines exhibited an increased sensitivity to the PARP inhibitors by comparison to the wild type A549 and H1299 cells for all four PARP inhibitors (Fig. 4). The increases in Olaparib, Niraparib and Rucaparib cytotoxicity were within a similar concentration range, but Talazoparib was cytotoxic over a ten-fold lower range. This is consistent with biochemical evidence for the greater potency of Talazoparib over Olaparib, Rucaparib and Niraparib as gauged by their relative IC50’s for inhibition of PARP1/2 [43].
Fig. 4.
Cytotoxicity of PARP inhibitors toward wt and p12KO cells. (A-D) Cytotoxicity of the PARP inhibitors Olaparib, Niraparib, Rucaparib and Talazoparib respectively, as determined by clonogenic survival assays (Materials and Methods). Data are shown as survival fraction (mean ± SD, n=3) and plotted against the log of the drug concentrations. Symbols and lines for A549 cells, solid blue circles, solid blue line; A549-p12KO cells, solid blue circles, dotted blue line; H1299 cells, solid red squares, solid red line; H1299-p12KO, solid red squares, dotted red line.
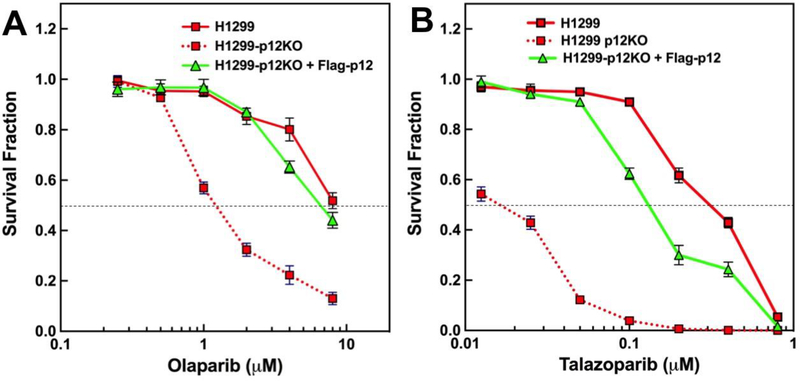
The findings that p12KO cells exhibit increased sensitivity to PARP inhibitors is fully consistent with their well established synthetic lethal effect on cells with HR defects. To provide additional confirmation that the synthetic lethal effects of PARP inhibitors were due to the loss of p12 and the consequent absence of Pol δ4, the H1299-p12KO+Flag-p12 cell line in which Flag-tagged 12 was stably expressed was examined for sensitivity to Olaparib and Talazoparib (Fig. 5). In both cases, the expression of Flag-p12 led to significant resistance to the PARP inhibitors. These findings provide further evidence for a significant HR defect when p12 is knocked out, as well as an affirmation that Pol δ4 plays a significant role in HR repair of chromosomal DNA in a cellular context.
Fig. 5.
Cytoxicity of Olaparib and Talazoparib toward H1299, H1299-p12KO and H1299-p12KO+Flag12 cells. Experiments were performed as in Fig. 4. Symbols for H1299 and H1299-p12KO are as in Fig. 4. Symbols for H1299-p12KO+Flagp12 cells are shown in solid green triangles and solid green lines.
3.5. Sensitivity of SCLC cell lines to PARP inhibitors
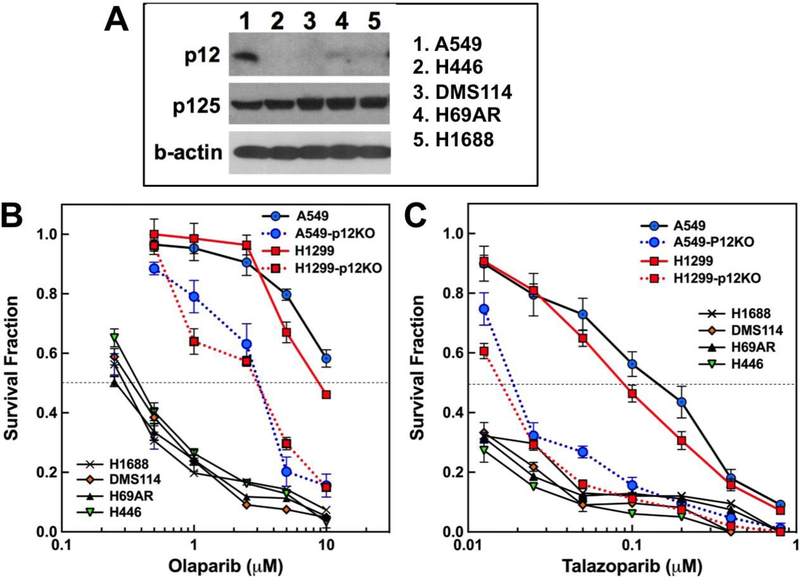
A defect in p12 expression at the mRNA and protein level in a sample of 9 small cell lung cancers has been reported [25]. p12 levels in these cancers are barely detectable by Western blotting [25]. In addition, this defect in p12 was also detected in a subgroup of non-small cell lung cancer patients that was correlated with poorer prognosis [25]. SCLC’s are highly aggressive and exhibit high rates of recurrence, despite the fact that they are initially responsive to treatment by cisplatin and etoposide, together with radiotherapy [44]. Cell lines derived from SCLC tumors would be expected to exhibit sensitivities to PARP inhibitors that are consistent with those we observed with the p12KO cells. We compared the sensitivities of four cell lines derived from small cell lung tumors to PARP inhibitors. These were H446, H1688, H69AR, and DMS114, obtained from ATCC. All four cell lines display significantly reduced levels of p12 expression in comparison to that of A549, but normal levels of the p125 subunit (Fig. 6A). It should be noted that these cells were not totally devoid of p12 expression, as this could be detected after much longer exposure of the Western blots.
Fig. 6.
Cytotoxicity of Olaparib and Talazoparib toward Small Cell Lung Cancer cell lines. Experimental procedures were as for Fig. 4. (A) Western blots for p12 and p125 levels in A549 and SCLC cell lines H446, DMS114, H69AR and H1688. (B, C) Cytotoxicity of Olaparib and Talazoparib, respectively. Symbols for H1688, x; DMS114, solid red diamonds; H69AR solid black triangle; H446, solid inverted green triangle. Symbols for A549, A549-p12KO, H1299-p12KO are as in Fig. 4.
The sensitivities of the SCLC cell lines to Olaparib and Talazoparib, together with those of A549, A549-p12KO, H1299 and H1299-p12KO cells were compared (Fig. 6B,C). All four SCLC cell lines exhibit comparable sensitivities to Olaparib. However, these were much more sensitive than the A549-p12KO and H1299-p12KO cell lines, by a factor of about 10 based on their approximate IC50’s (Fig. 6B). A similar behavior was observed with Talazoparib, where the SCLC cell lines exhibited similar sensitivities that were at least several fold more than those of the p12KO cells (Fig. 6C). These findings are significant as they are consistent with a hypothesis that SCLC lung cancer cells that exhibit low expression for p12 are likely to exhibit a HR defect.
4. Summary and Perspective
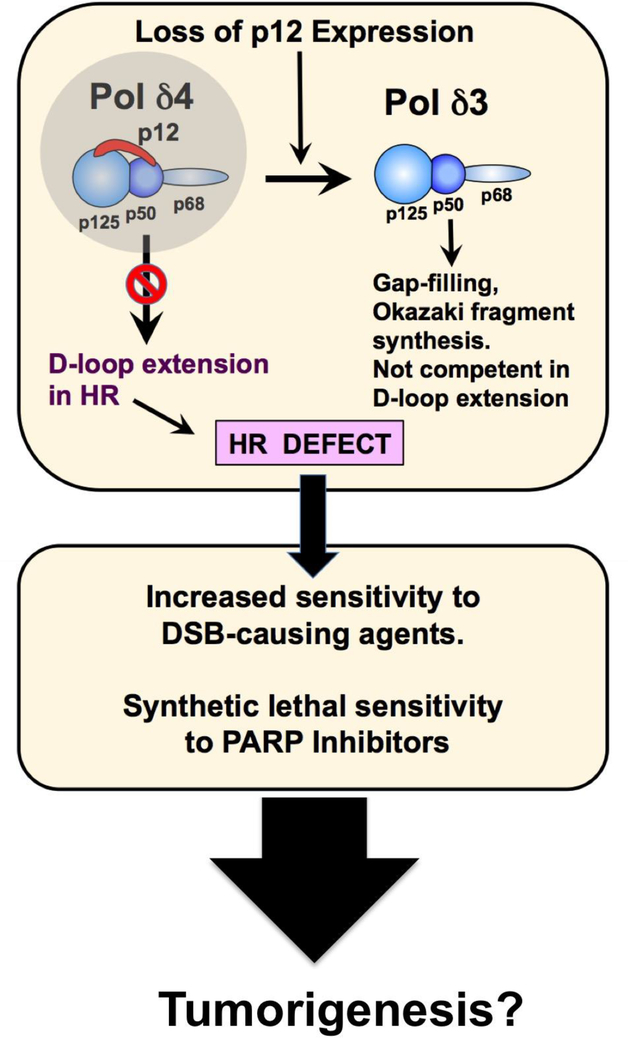
Our findings support a scenario that is shown diagrammatically in Fig. 7. Loss of p12 results in viable cells in which the only form of Pol δ activity is that of the Pol δ3 trimer. The cells remain viable because Pol δ3 is able to sustain Okazaki fragment synthesis at the lagging strand. In addition, Pol δ3 is able to maintain a role as a gap filling enzyme in nucleotide excision repair. HR is compromised because Pol δ4 has a significant role in D-loop strand displacement synthesis. While there are extensive studies on the involvement of other DNA polymerases in HR [31], these clearly cannot compensate for loss of Pol δ4. Thus the p12KO cells exhibit a HR-deficient phenotype, noting that this is possible because Pol δ4 is not essential for cell viability. Consequently, the p12KO cells exhibit increased sensitivity to chemotherapeutic agents and also synthetic lethal sensitivity to PARP inhibitors.
Fig. 7.
Consequences of the loss of expression of p12: generation of a HR deficient phenotype. The diagram shows the basis for the generation of the HR deficiency by loss of p12 and is discussed in the text.
These findings suggest that p12 expression is a potential biomarker for assessing the potential of tumor response to PARP inhibitors, an area that is of considerable current interest [45,46]. Given that SCLC tumors are characterized by low expression of p12, it may be speculated that these may be amenable to PARP inhibitor therapies. The observations that SCLC cell lines exhibit low expression levels of p12, and their sensitivity to PARP inhibitors, strongly suggests they may be HR deficient. This now appears a promising area of enquiry that in the future may reveal whether p12 defects play a role in the development of SCLC. Interestingly, Talazoparib has been shown to a potent radiosensitizer for SCLC [47].
Highlights.
p12-knockout generates a HR deficiency phenotype.
Pol δ4 is required for full expression of cellular HR activity
p12 knockout cells are sensitized to PARP inhibitors
p12 expression is a potential biomarker for the use of PARP inhibitors
p12 deficiency may have a role in SCLC tumorigenesis.
Acknowledgements
We thank Dr. Samuel H. Wilson for insightful suggestions.
Grant support
This work is supported by grants from the National Institute of Environmental Health Sciences (ES014737 to MYWTL and ZZ) and a Concept Award from USAMRAA (W81XWH-18–1-0353 to SZ).
Footnotes
Conflict of interest statement
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References.
- [1].Burgers PM, Kunkel TA, Eukaryotic DNA Replication Fork. Annu Rev Biochem, 86 (2017) 417–438. DOI: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kunkel TA, Burgers PM, Dividing the workload at a eukaryotic replication fork. Trends Cell Biol, 18 (2008) 521–7. DOI: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kurat CF, Yeeles JT, Patel H, Early A, Diffley JF, Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Mol Cell, 65 (2017) 117–130. DOI: 10.1016/j.molcel.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li H, O’Donnell ME, The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism. Bioessays, 40 (2018) DOI: 10.1002/bies.201700208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Georgescu R, Yuan Z, Bai L, de R Santos Luna Almeida, Sun J, Zhang D, Yurieva O, Li H et al. , Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci U S A, 114 (2017) E697–E706. DOI: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lee MY, Zhang S, Lin SH, Chea J, Wang X, LeRoy C, Wong A, Zhang Z et al. , Regulation of human DNA polymerase Delta in the cellular responses to DNA damage Environ Mol Mutagen, 53 (2012) 683–98. DOI: 10.1002/em.21743. [DOI] [PubMed] [Google Scholar]
- [7].Lee MY, Zhang S, Lin SH, Wang X, Darzynkiewicz Z, Zhang Z, Lee EY, The tail that wags the dog: p12, the smallest subunit of DNA polymerase delta, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression. Cell Cycle, 13 (2014) 23–31. DOI: 10.4161/cc.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee MYWT, Wang X, Zhang S, Zhang Z, Lee EYC, Regulation and Modulation of Human DNA Polymerase δ Activity and Function. Genes (Basel), 8 (2017) E190 DOI: 10.3390/genes8070190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY, A novel DNA damage response: rapid degradation of the p12 subunit of dna polymerase delta. J. Biol. Chem, 282 (2007) 15330–40. DOI: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- [10].Meng X, Zhou Y, Lee EY, Lee MY, Frick DN, The p12 subunit of human polymerase delta modulates the rate and fidelity of DNA synthesis. Biochemistry, 49 (2010) 3545–54. DOI: 10.1021/bi100042b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meng X, Zhou Y, Zhang S, Lee EY, Frick DN, Lee MY, DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res, 37 (2009) 647–57. DOI: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Khare V, Eckert KA, The proofreading 3’−5’ exonuclease activity of DNA polymerases: a kinetic barrier to translesion DNA synthesis. Mutat. Res, 510 (2002) 45–54. DOI: 10.1016/S0027-5107(02)00251-8. [DOI] [PubMed] [Google Scholar]
- [13].Johnson KA, Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem, 62 (1993) 685–713. DOI: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- [14].Chea J, Zhang S, Zhao H, Zhang Z, Lee EY, Darzynkiewicz Z, Lee MY, Spatiotemporal recruitment of human DNA polymerase delta to sites of UV damage. Cell Cycle, 11 (2012) 2885–95. DOI: 10.4161/cc.21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhao H, Zhang S, Xu D, Lee MY, Zhang Z, Lee EY, Darzynkiewicz Z, Expression of the p12 subunit of human DNA polymerase delta (Pol delta), CDK inhibitor p21(WAF1), Cdt1, cyclin A, PCNA and Ki-67 in relation to DNA replication in individual cells. Cell Cycle, 13 (2014) 3529–40. DOI: 10.4161/15384101.2014.958910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang S, Zhao H, Darzynkiewicz Z, Zhou P, Zhang Z, Lee EY, Lee MY, A novel function of CRL4Cdt2: regulation of the subunit structure of DNA polymerase delta in response to DNA damage and during the S phase. J Biol Chem, 288 (2013) 29950–61. DOI: 10.1074/jbc.M113.490466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lin SH, Wang X, Zhang S, Zhang Z, Lee EY, Lee MY, Dynamics of Enzymatic Interactions During Short Flap Human Okazaki Fragment Processing by Two forms of Human DNA Polymerase δ DNA Repair (Amst), 12 (2013) 922–35. DOI: 10.1016/j.dnarep.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Burgers PM, Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem, 284 (2009) 4041–5. DOI: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Walsh CS, Two decades beyond BRCA1/2: Homologous recombination, hereditary cancer risk and a target for ovarian cancer therapy. Gynecol Oncol, 137 (2015) 343–50. DOI: 10.1016/j.ygyno.2015.02.017. [DOI] [PubMed] [Google Scholar]
- [20].Lord CJ, Ashworth A, PARP inhibitors: Synthetic lethality in the clinic. Science, 355 (2017) 1152–1158. DOI: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Evans T, Matulonis U, PARP inhibitors in ovarian cancer: evidence, experience and clinical potential. Ther Adv Med Oncol, 9 (2017) 253–267. DOI: 10.1177/1758834016687254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH, Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem, 276 (2001) 25541–8. DOI: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- [23].Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH, DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem, 276 (2001) 32411–4. DOI: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- [24].Pommier Y, O’Connor MJ, de Bono J, Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med, 8 (2016) 362ps17 DOI: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- [25].Huang QM, Tomida S, Masuda Y, Arima C, Cao K, Kasahara TA, Osada H, Yatabe Y et al. , Regulation of DNA polymerase POLD4 influences genomic instability in lung cancer. Cancer Res, 70 (2010) 8407–16. DOI: 10.1158/0008-5472.CAN-10-0784. [DOI] [PubMed] [Google Scholar]
- [26].Pierce AJ, Johnson RD, Thompson LH, Jasin M, XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev, 13 (1999) 2633–8. PMCID: PMC317094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Richardson C, Moynahan ME, Jasin M, Homologous recombination between heterologs during repair of a double-strand break. Suppression of translocations in normal cells. Ann N Y Acad Sci, 886 (1999) 183–6. PMID: [DOI] [PubMed] [Google Scholar]
- [28].Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C, Clonogenic assay of cells in vitro. Nat Protoc, 1 (2006) 2315–9. DOI: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- [29].Zhang S, Zhou Y, Sarkeshik A, Yates JR, Thomson T, Zhang Z, Lee EY, Lee MY, Identification of RNF8 as a Ubiquitin Ligase Involved in Targeting the p12 Subunit of DNA Polymerase δ for Degradation in Response to DNA Damage J Biol Chem, 288 (2013) 2941–50. DOI: 10.1074/jbc.M112.423392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huang QM, Akashi T, Masuda Y, Kamiya K, Takahashi T, Suzuki M, Roles of POLD4, smallest subunit of DNA polymerase delta, in nuclear structures and genomic stability of human cells. Biochem Biophys Res Commun, 391 (2010) 542–6. DOI: 10.1016/j.bbrc.2009.11.094. [DOI] [PubMed] [Google Scholar]
- [31].McVey M, Khodaverdian VY, Meyer D, Cerqueira PG, Heyer WD, Eukaryotic DNA Polymerases in Homologous Recombination. Annu Rev Genet, 50 (2016) 393–421. DOI: 10.1146/annurev-genet-120215-035243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ganai RA, Zhang XP, Heyer WD, Johansson E, Strand displacement synthesis by yeast DNA polymerase epsilon. Nucleic Acids Res, 44 (2016) 822940 DOI: 10.1093/nar/gkw556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kwon Y, Daley JM, Sung P, Reconstituted System for the Examination of Repair DNA Synthesis in Homologous Recombination. Methods Enzymol, 591 (2017) 307–325. DOI: 10.1016/bs.mie.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sneeden JL, Grossi SM, Tappin I, Hurwitz J, Heyer WD, Reconstitution of recombination-associated DNA synthesis with human proteins. Nucleic Acids Res, 41 (2013) 4913–25. DOI: 10.1093/nar/gkt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sebesta M, Burkovics P, Haracska L, Krejci L, Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair (Amst), 10 (2011) 567–76. DOI: 10.1016/j.dnarep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang X, Zhang S, Zheng R, Yue F, Lin SH, Rahmeh AA, Lee EY, Zhang Z et al. , PDIP46 (DNA polymerase delta interacting protein 46) is an activating factor for human DNA polymerase delta. Oncotarget, 7 (2016) 6294–6313. DOI: 10.18632/oncotarget.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ito S, Murphy CG, Doubrovina E, Jasin M, Moynahan ME, PARP Inhibitors in Clinical Use Induce Genomic Instability in Normal Human Cells. PLoS One, 11 (2016) e0159341 DOI: 10.1371/journal.pone.0159341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dasari S, Tchounwou PB, Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol, 740 (2014) 364–78. DOI: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott CL, Meier W et al. , Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol, 15 (2014) 852–61. DOI: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- [40].Caruso D, Papa A, Tomao S, Vici P, Panici PB, Tomao F, Niraparib in ovarian cancer: results to date and clinical potential. Ther Adv Med Oncol, 9 (2017) 579–588. DOI: 10.1177/1758834017718775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Syed YY, Rucaparib: First Global Approval. Drugs, 77 (2017) 585–592. DOI: 10.1007/s40265-017-0716-2. [DOI] [PubMed] [Google Scholar]
- [42].Gunjur A, Talazoparib for BRCA-mutated advanced breast cancer. Lancet Oncol, 19 (2018) e511 DOI: 10.1016/S1470-2045(18)30650-8. [DOI] [PubMed] [Google Scholar]
- [43].Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, Wang B, Lord CJ et al. , BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res, 19 (2013) 5003–15. DOI: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mamdani H, Induru R, Jalal SI, Novel therapies in small cell lung cancer. Transl Lung Cancer Res, 4 (2015) 533–44. DOI: 10.3978/j.issn.2218-6751.2015.07.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ganguly B, Dolfi SC, Rodriguez-Rodriguez L, Ganesan S, Hirshfield KM, Role of Biomarkers in the Development of PARP Inhibitors. Biomark Cancer, 8 (2016) 15–25. DOI: 10.4137/BIC.S36679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Thomas A, Murai J, Pommier Y, The evolving landscape of predictive biomarkers of response to PARP inhibitors. J Clin Invest, 128 (2018) 1727–1730. DOI: 10.1172/JCI120388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Laird JH, Lok BH, Ma J, Bell A, de Stanchina E, Poirier JT, Rudin CM, Talazoparib is a Potent Radiosensitizer in Small Cell Lung Cancer Cell Lines and Xenografts. Clin Cancer Res, (2018) DOI: 10.1158/1078-0432.CCR-18-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]