Introduction
Significant inter-individual variability in medication response can result in adverse drug reactions (ADRs) and increased health care costs. Based on ADR prevalence and mounting evidence linking genetics and pharmacokinetic variability, Cincinnati Children’s Hospital Medical Center (CCHMC) launched the Genetic Pharmacology Service (GPS) in 2004 and has since performed >25,000 tests. Herein, we describe how the service developed, launched, and has been updated along with how it is currently utilized, and key lessons learned.
Development
The impetus for developing the GPS started with a single patient. In 2001, at 18 months old, this previously healthy male was diagnosed with language-regression autism and developed significant side effects when prescribed selective serotonin reuptake inhibitors (SSRIs). This prompted his clinicians to examine the impact of genetics on adverse drug reactions for this patient and in general. A multi-disciplinary team was assembled including a psychiatrist, neurologist, informaticist, clinical pharmacologist, clinical molecular geneticist and licensed advanced practice nurse. The group met weekly for nine months to discuss potential genetic variants to test, testing methods, interpretation of results, result presentation options, ethical implications, test costs, and provider education. A donor provided funding to purchase laboratory instruments needed for pharmacogenetic testing while hospital leadership provided initial funding to cover other costs involved with launching the new service.
Launch
In 2004, the GPS was launched as a clinical service available to any practitioner at our institution; within months it was expanded to external prescribers. The clinical service included genotyping, clinical interpretation, and consultation. Genotyping was available for CYP2D6 (*3, *4, *5), CYP2C19 (*2), CYP2C9 (*2, *3), and TPMT (*2, *3A, *3B, *3C). Genotyping analyses were designed and validated based on CLIA and CAP standards. The wild type genotypes were inferred from the absence of the tested alleles. At that time, most alleles causing altered activity could be accurately identified by genotyping these alleles. The choice of alleles tested was also influenced by consideration of turnaround time (goal was 2 days) and cost to the patients (goal was $300). Initially, genotyping was performed using DNA extracted from peripheral blood obtained at the time other labs were drawn from the patient. Testing was by either TaqMan allelic discrimination system on an ABI-7500 real-time PCR system (Applied Biosystem, Forest City, CA) or long-range PCR, which was used to detect the CYP2D6 *5 deletion and duplication.
The service was designed for practicing clinicians’ point of care use. Tests were orderable by single drug. CCHMC’s internally developed electronic ordering system was modified to notify clinicians of available testing or existing results when certain drugs were ordered. A subsequent migration to Epic retained these alerts (and remains unchanged). When no prior results are recorded, the provider receives an alert indicating a pharmacogenetic test is available for a medication. When a result exists for an alternative test, the provider can request a reinterpretation without collecting additional specimens. In all cases, the provider is given an alert with guidance to the results. Report templates were created with therapeutic recommendations for each drug-gene pair.
Education
A variety of instructional strategies were used to provide education about both the impact of genetics on drug response and the service itself to multiple groups of health care professionals. The education process began six months prior to launching the service with short lectures and discussion sessions at divisional staff meetings. In addition to targeting prescribers, education was provided to unit-based education specialists, staff nurses and clinical pharmacists. Patient education handouts, written at the 7th grade level, were created and distributed to each unit. The handouts were later translated to Spanish, and both versions were posted on the GPS website. After GPS was launched in 2004, periodic education updates have been given to physician, nurse and pharmacist groups by GPS team members.
Early Adoption by Psychiatry
Leadership in the Division of Psychiatry quickly realized the potential benefits of using pharmacogenetic information during medication dosing for hospitalized patients. It was well known that patient response to psychotropic medications varied, with 30–75% achieving the desired therapeutic benefit and 65–85% experiencing adverse reactions (1). CYP2D6 and CYP2C19 genotype-guided dosing recommendations had been published for many psychotropic medications (2). However, the process of ordering by drug was cumbersome and impractical for psychiatrists’ purposes. In response, the GPS developed a psychiatry panel including both CYP2D6 and CYP2C19 and the report provided predicted metabolizing phenotypes, potential dose adjustments for 34 different psychotropic medications as a percentage of the recommended usual dose, and related information on drug-drug interactions. Beginning January 1, 2005, patients admitted to the inpatient psychiatry service at CCHMC received GPS testing as part of routine care.
Early Clinical Experience
While Psychiatry was the primary clinical service ordering GPS tests, there were psychiatrists who rarely used the service as they believed the time to receive test results unnecessarily delayed drug therapy initiation. Infrequent users were not accustomed to considering GPS results from a previous admission when children were readmitted to a psychiatry unit. In response, a clinical pharmacist was hired to attend daily psychiatry rounds, alert physicians to existing GPS results and therapeutic recommendations, notify prescribers about newly identified poor and ultra-rapid metabolizers, and be available for consultation. Avoidance of blood draws in young patients was another reason for not ordering a GPS test. In response, in 2006, the GPS began offering buccal swabs as an alternative to blood for collecting DNA.
Evolution
Psychiatry Panel
In 2012, the GPS decided sufficient new evidence existed to update its psychiatry panel (Figure 1) (3). New alleles of CYP2D6 and CYP2C19 had been discovered and new medications had been approved for pediatric use. An Expanded Psychiatry Panel was developed that included many of the new CYP2D6 and CYP2C19 variants and updated the medications that were included on the report. When the new test was implemented, the interpretation for CYP2D6 genotype to phenotype was changed to follow Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines (4). Whereas, with the first version of the panel, patients who carried one functional and one non-functional allele were intermediate metabolizers, after the update they were considered extensive (normal) metabolizers, and intermediate metabolizers are individuals who have one non-functional allele and one reduced function allele. When the Expanded Psychiatry Panel was implemented, rather than update historical reports, the group decided to re-test patients that had previous results including *1 alleles, since it was possible they carried one of the other alleles that weren’t previously tested. The new test was implemented in September 2013 and is currently being used. Rather than place the burden on prescribers to identify who should be retested, all patients admitted to a Psychiatry unit had a buccal swab or blood sample sent to the laboratory for the new panel test. Samples were analyzed with the new panel of genetic variants unless a patient was a known poor metabolizer, for whom the laboratory canceled the order but generated a new report with the updated slate of medications.
Figure 1: Implementation steps for a new gene-drug pair.

GPS, Genetic Pharmacology Service. SOP, standard operating procedure. BPA, best practice alert.
We have tested more than 8,700 patients with the Expanded Psychiatry Panel since 2013. Only about a third of our patients are CYP2D6 and CYP2C19 normal (extensive) metabolizers (Figure 2). With the large size of our population tested, we have seen all possible genotype-predicted phenotype combinations. The frequency of each metabolizer status does not seem to be enriched for extreme metabolizers.
Figure 2: Frequencies of CYP2D6 and CYP2C19 metabolizers tested with the Expanded Psychiatry Panel from September 2013 to June 2016.
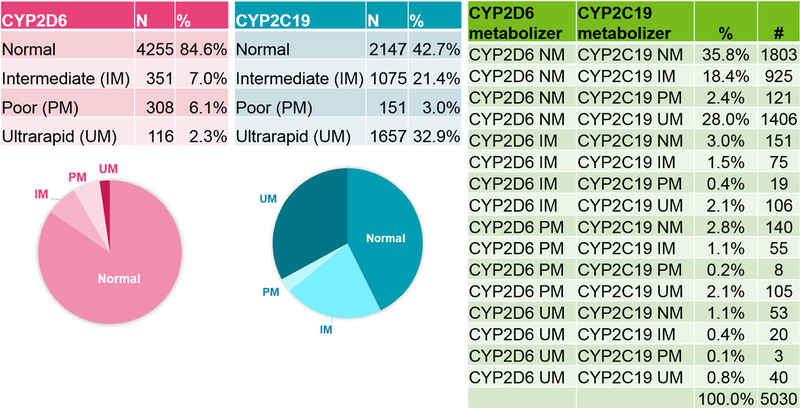
Individual genes are shown at left and combinations of the two genes are shown at right. 6147 patients were tested, with 451 with a result of “Indeterminate” for either gene and 666 genotyping failures not shown. NM, normal metabolizer; PM, poor metabolizer; IM, intermediate metabolizer; UM, ultra-rapid metabolizer. The 20 CYP2D6 alleles tested: *2A, *3, *4, *5, *6, *7, *8, *9, *10, *11, *14, *15, *17, *18, *19, *20, *40, *41, *42, *44 and the long-range PCR is used for the duplication. There are 8 CYP2C19 alleles tested: *2, *3, *4, *5, *6, *7, *8, and *17. *1 is inferred by the absence of any of these alleles.
Opioid panel
A GPS test for codeine was activated beginning in 2004, testing CYP2D6. An opioid panel was developed in 2013 that tests the 21 variants in CYP2D6 included on the Expanded Psychiatry Panel. Therapeutic recommendations consistent with the CPIC guidelines (4, 5) are included for codeine, tramadol, hydrocodone and oxycodone. Recently, the pain management team and some surgeons have begun ordering it prior to procedures for patients who are likely to receive oxycodone. It is also ordered when patients have adverse effects or are non-responsive to recommended usual doses of opioids.
Other updates
The TPMT and CYP2C9 tests have not changed since implementation in 2004, and VKORC1 testing was added in 2007. On average, we now perform approximately 1900 Expanded Psychiatry Panel tests, 100 Opioid Panel tests, 120 TPMT tests, 120 CYP2C19 tests, and 15 warfarin tests per year. We have recently updated and expanded the number of patient education sheets to be specific for each gene and metabolism phenotype. These are available to the patient at the time they receive their results or through our website (www.cincinnatichildrens.org/gpsinfo).
Lessons learned
Multiple factors impacted the success of the GPS. First, hospital leadership financial support was critical to purchase necessary infrastructure and hire personnel. Clinician feedback shaped the format and focus of the genetic reports and the services provided. A physician champion, especially in a leadership role, accelerated uptake and implementation of GPS testing in specific domains like psychiatry. Our ordering and reporting systems allowed for rapid identification of a panel of drugs impacted by specific genetic variants; this enhanced clinical adoption of GPS testing. A team approach with specific clinical pharmacology input was crucial for the integration of GPS results into clinical care at the point of care. Our ability to develop clinically impactful pharmacogenetics algorithms resulted from a close integration between biomedical informatics and clinical expertise.
Future Improvements
We are currently working to add discrete data results into the electronic health record system to improve our decision support capabilities so that the alerts include dosing recommendations and highlight actionable results. As advances in pharmacogenetics are rapidly identifying more associations, we will implement additional gene-drug pairs with high levels of evidence (occasionally beyond CPIC guidelines), including an update to the psychiatry panel that will include pharmacodynamic genes. We anticipate that including additional genes with high levels of evidence will improve outcomes, although there will be challenges implementing the informatics required for medications with multiple associated genes. Since many patients are on medications metabolized by CYP2D6 and CYP2C19 that are not included in our opioid or psychiatry panel, we will add alerts and reports for medications such as proton pump inhibitors, beta blockers, and anti-coagulants when we are able. We are conducting studies to better understand long-term impact on patients that have already been tested, including how they responded to the medication and dose prescribed, and whether inclusion of additional genes could have improved response. We plan to expand our education for clinicians through presentations and discussions at departmental or specialty group meetings. Finally, we plan to launch a hospital-wide consultative service where clinicians can contact us with questions about patients with unexpected drug responses such as those experienced by the young boy who inspired this innovative effort back in 2001.
Acknowledgments
Funding: Initiation of the opioid CYP2D6 pharmacogenetics panel was part of a single site eMERGE II network project funded by the NHGRI through grant U01HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital).
Footnotes
Conflict of interest:
As an Associate Editor for Clinical Pharmacology & Therapeutics, Alexander A. Vinks was not involved in the review or decision process for this paper. The authors have no conflicts of interest to declare.
References
- (1).Kirchheiner J et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. (Nature Publishing Group, 2004). [DOI] [PubMed] [Google Scholar]
- (2).Roots I et al. Pharmacogenetics‐based new therapeutic concepts. Drug metabolism reviews 36, 617–38 (2004). [DOI] [PubMed] [Google Scholar]
- (3).Stingl JC, Brockmoller J & Viviani R Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol Psychiatry 18, 273–87 (2013). [DOI] [PubMed] [Google Scholar]
- (4).Crews K et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clinical pharmacology and therapeutics 91, 321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clinical Pharmacology & Therapeutics 95, 376–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]


