Abstract
Platelets are highly specialized cells that continuously patrol the vasculature to ensure its integrity (hemostasis). At sites of vascular injury, they are able to respond to trace amounts of agonists and to rapidly transition from an anti-adhesive/patrolling to an adhesive state (integrin inside-out activation) required for hemostatic plug formation. Pathological conditions that disturb the balance in the underlying signaling processes can lead to unwanted platelet activation (thrombosis) or to an increased bleeding risk. The small GTPases of the RAP subfamily, highly expressed in platelets, are critical regulators of cell adhesion, cytoskeleton remodelling and MAP kinase signaling. Studies by our group and others demonstrate that RAP GTPases, in particular RAP1A and RAP1B, are the key molecular switches that turn on platelet activation/adhesiveness at sites of injury. In this review, we will summarize major findings on the role of RAP GTPases in platelet biology with a focus on the signaling pathways leading to the conversion of integrins to a high-affinity state.
Keywords: small GTPases, RAP1, integrin activation, signal transduction, hemostasis
RAS/RAP GTPases expressed in platelets
Small GTPases are binary on/off switches that alternate between a GDP-bound off state and a GTP-bound on state, the latter evoking cellular responses by binding to downstream effectors. High-throughput profiling studies[1–4] identified among the most abundant signaling proteins in platelets (Table 1), the Ras-related protein (RAP) GTPases, a subset (5 members in mammals: RAP1A, RAP1B, RAP2A, RAP2B, RAP2C) of the larger RAS family that includes key regulators of cell adhesion, cytoskeleton remodelling and MAP kinase signaling[5–7]. These functions are critical for platelets that, in order to maintain the integrity of the vascular system, must be able to rapidly shift from an anti-adhesive/patrolling state to a pro-adhesive state, required to form a hemostatic plug and to prevent bleeding[8]. Indeed, studies by our group and others identified RAP GTPases as the critical molecular switches, which drive platelet activation at sites of vascular injury by switching on multiple platelet responses, including integrin inside-out activation, secretion, and eicosanoid mediator formation[9–13].
Table 1. Expression in platelets of RAS/RAP GTPases and their upstream regulators.
Estimated protein (protein copies/platelet) and mRNA (RPKM, reads per kilobase of exon model per million mapped reads) levels based on quantitative proteomic and transcriptomic analysis of human [1, 3] and mouse [2, 3] platelets.
| PROTEIN NAME (Gene name) |
Human Proteome[1] proteins/platelet |
Mouse Proteome[2] proteins/platelet |
Human Transcriptome[3] mRNA abundance (RPKM) |
Mouse Transcriptome[3] mRNA abundance (RPKM) |
|---|---|---|---|---|
| RAP1A (Rap1a) | 125,000 | 19,000 | 57 | 89 |
| RAP1B (Rap1b) | 297,000 | 210,000 | 186 | 1611 |
| RAP2A (Rap2a) | 2,900 | 4,000 | 11 | 41 |
| RAP2B (Rap2b) | 6,200 | 6,900 | 39 | 10 |
| RAP2C (Rap2c) | 3,100 | 1,600 | - | - |
| K-RAS (Kras) | 6,600 | 72 | 1 | 0 |
| N-RAS (Nras) | 6,600 | 1,400 | 5 | 40 |
| H-RAS (Hras) | 100 | 449 | 0 | 1 |
| R-RAS (Rras) | 2,600 | 3,400 | 19 | 11 |
| R-RAS2/TC21 (Rras2) | 100 | 0 | 0 | 0 |
| CALDAG-GEFI (Rasgrp2) | 9,100 | 32,000 | 136 | 766 |
| PDZ-GEF1 (Rapgef2) | 870 | 362 | 2 | 0 |
| PDZ-GEF2 (Rapgef6) | 590 | 51 | 0 | 1 |
| SOS1 (Sos1) | 780 | 98 | 2 | 0 |
| p120gap (RASA1) | 2,900 | 1,700 | 1 | 0 |
| RASA3 (Rasa3) | 8,300 | 27,000 | 153 | 210 |
| RAP1GAP2 (Rap1gap2) | 2,300 | 0 | 5 | 0 |
| RASAL3 (Rasal3) | 570 | 0 | 3 | 0 |
In contrast, the founding members of the RAS superfamily are either undetectable (H-RAS, R-RAS2) or present in very limited amounts in platelets (K-RAS, N-RAS, R-RAS)[1–4] (Table 1). Like RAP, RAS GTPases can be activated in vitro upon platelet stimulation with various agonists[14,15]. However, it is unclear whether they play a role in platelet biology, especially as their activation is not coupled to the activation of ERK MAP kinase[15], a common downstream target of RAS in other cells. Notably guanine nucleotide exchange factors (GEFs), the enzymes that catalyse GTP loading (activation), can discriminate between RAS and RAP proteins,[6,16] and high-throughput profiling studies[1–4] showed that the most highly expressed GEFs present in platelets (Table 1) preferentially activate RAP over RAS GTPases.
RAP1 activation drives platelet adhesion
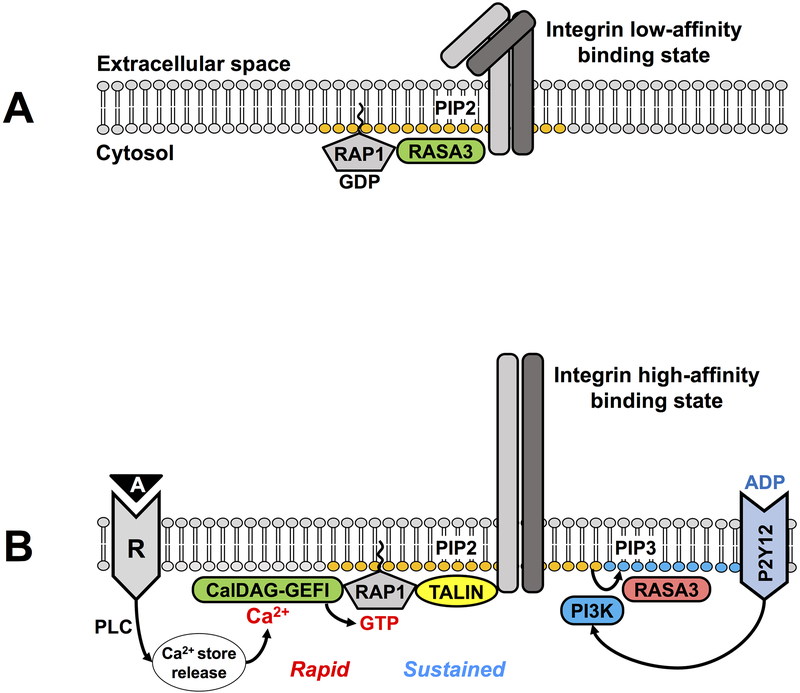
Tight regulation of platelet adhesiveness is essential to prevent clinical complications such as bleeding or thrombosis. Our studies demonstrated that the level of RAP1 activation directly correlates with the degree of platelet adhesiveness[17–20]. RAP1 becomes activated in response to all platelet agonists tested so far, including those for G-protein coupled receptors (GPCRs), immunoreceptor tyrosine-based activation motif (ITAM)-coupled receptors[18] and integrin receptors[21]. The upstream regulators of RAP1 most highly expressed in platelets are the guanine nucleotide exchange factor CALDAG-GEFI[11] and the GTPase-activating protein RASA3[22] (Figure 1). Both enzymes are multi-domain proteins, which can bind specific second messengers to integrate the signals triggered by engagement of surface agonist receptors and fine-tune RAP activation in time and space[23].
Figure 1. Working model or RAP1-dependent integrin affinity regulation.
A) In circulating/patrolling platelets the GTPase-activating protein RASA3 is active at the plasma membrane to counteract any spurious RAP1 activation and maintain integrins in a low affinity binding state for their ligands. B) Upon injury, platelet receptors (R) immediately respond to agonists (A) produced at the site of damage by mobilizing calcium ions (Ca2+) from the intracellular stores. The Ca2+-sensor CALDAG-GEFI, mediates the rapid activation of RAP1. Sustained RAP1 activation is achieved by turning off RASA3, following stimulation of the ADP (adenosine diphosphate) receptor P2Y12, and generation of PIP3 (phosphatidylinositol 3,4,5-trisphosphate) by the lipid kinase PI3K (phosphatidylinositol-4,5-bisphosphate 3-kinase). Once active, RAP1-GTP recruits TALIN to PIP2 (phosphatidylinositol 4,5-bisphosphate)-rich areas of the plasma membrane in proximity of the integrins. TALIN binding to the integrin cytoplasmic tail triggers the conversion of integrins to a high affinity binding state for their ligands, which is required for platelet adhesion and aggregation at sites of injury.
CALDAG-GEFI, also known as RASGRP2, consists of an N-terminal catalytic GEF domain and a C-terminal regulatory domain. The GEF domain is a CDC25 homology domain with high selectivity towards all RAP proteins[16,24]. In vitro studies suggest that CALDAG-GEFI can also weakly promote GTP-loading of R-RAS and R-RAS2/TC21[25], both of which have been implicated in the regulation of adhesion processes[26]. However, such in vitro data must be taken with caution because, although R-RAS is considered more similar to RAPs from a functional standpoint, it shares with RAS proteins the three residues (54, 61, 70) identified as key discriminators of GEF selectivity between RAP and RAS[16]. Moreover R-RAS has an intrinsic exchange rate that is approximately 10 times higher than that of RAP1, so the in vivo relevance of CALDAG-GEFI weak activity toward R-RAS awaits further investigation.
The C-terminal regulatory domain of CALDAG-GEFI[24] includes a pair of highly sensitive calcium ion (Ca2+)-binding EF hand domains (KD~80nM)[27] and an atypical C1 domain. The EF hand domains provide remarkable sensitivity towards minor changes in the cytoplasmic Ca2+ concentration[28], which in resting platelets was measured at ~20–50nM. The C1 domain is considered atypical because, unlike the prototypical C1 domain of protein kinase C (PKC), it has weak affinity for the second messenger diacylglycerol (DAG)[29]. Accordingly, DAG does not affect the subcellular localization or the activity of CALDAG-GEFI[30,31] and platelets lacking functional CALDAG-GEFI in humans or mice respond normally to stimulation with DAG mimetics[11,32]. However, the C1 domain is important for optimal CALDAG-GEFI function in platelets in vivo[20] and our ongoing studies suggest that this domain is important for the association of CalDAG-GEFI to the plasma membrane (unpublished data). When platelets encounter an injury, Ca2+ mobilization from the endoplasmic reticulum is the most rapid intracellular response to agonist receptor engagement by either exposed components of the extracellular matrix or by locally generated soluble agonists. Our current working model is that Ca2+ binding to the EF hands ensures the near-immediate activation of CALDAG-GEFI catalytic activity. The C1 domain, while it does not affect the catalytic activity per se, enhances the GEF efficiency of CALDAG-GEFI[20] by driving its localization to the inner leaflet of the plasma membrane (unpublished data), where RAP is enriched due to its post-translational modifications (see below for details). Both these regulatory mechanisms contribute to the efficient and very rapid activation of RAP.
Consistent with this model, platelets lacking functional CALDAG-GEFI show markedly reduced and delayed RAP1/2 and integrin activation in response to agonist stimulation, both in humans and mice[20,33]. CALDAG-GEFI deficiency inhibits the ability of platelets to form three-dimensional aggregates under conditions of flow, in particularly at high shear rates. In vivo this results in a marked protection against arterial thrombosis[11,34]. Moreover CALDAG-GEFI deficiency protects from atherosclerotic plaque development in hypercholesterolemic LDL receptor knockout mice, as it reduces platelet-leukocyte interactions at the site of lesion[35]. Based on this preclinical data and on its limited expression in tissues other than platelets[11,24], CALDAG-GEFI has been suggested as a new target for anti-platelet therapy. However, patients with mutations in Rasgrp2 present with a moderate or severe bleeding phenotype[32,33,36–39], suggesting that full inhibition of CALDAG-GEFI may not be a viable strategy to prevent arterial thrombosis. Interestingly, however, mice expressing low levels of CALDAG-GEFI were protected from arterial thrombosis, with minor impact on hemostasis[40]. Thus, partial inhibition of CALDAG-GEFI function, for instance by targeting a regulatory domain, such as the C1 domain[20], may be a safe but powerful approach to prevent platelet-driven thrombosis.
RAP1 inhibition limits platelet activation in circulation and at sites of vascular injury
Various mechanisms are in place to restrain platelet RAP activity in circulation and at sites of vascular injury[41]. For example, in circulating platelets, CALDAG-GEFI is negatively regulated by the endothelial-derived antagonist prostaglandin (PG)I2, which stimulates adenylyl cyclase to generate cyclic adenosine monophosphate (cAMP) and activates the cAMP-regulated protein kinase (PKA). PKA phosphorylates CALDAG-GEFI on serine587, serine116 and serine117 and thereby inhibits Ca2+/CalDAG-GEFI-dependent RAP1 activation and platelet aggregation[42,43]. However, at sites of injury the inhibitory effect of PGI2 is bypassed since its concentrations drop where the endothelial lining is disrupted, while activated platelets release agonists such as ADP that counteract PGI2 by lowering cAMP levels[44]. Another important regulatory mechanism to reduce RAP-GTP levels is through the action of GTPase-activating proteins (GAPs), which catalyse GTP hydrolysis. Human, but not mouse, platelets express low but significant amounts of RAP1GAP2[45] (Table 1), a GAP that is positively regulated by endothelial-derived inhibitors[46], in a reciprocal manner compared to CalDAG-GEFI. However, the main RAP-GAP that controls RAP1-GTP levels in platelets and antagonizes CALDAG-GEFI is RASA3 (also known as GAP1IP4BP)[22]. Since CALDAG-GEFI is extremely sensitive and unwanted RAP1 activation may occur as a consequence of shear stress or spurious platelet stimulation, platelets lacking functional RASA3 circulate in a pre-activated state, with elevated basal levels of RAP1-GTP and high-affinity integrin αIIbβ3. These hyperactive platelets undergo a more rapid clearance thus causing thrombocytopenia in mice. Platelets lacking RASA3 are more prone to stimulation by agonists and form more stable hemostatic plugs at sites of vascular injury. Thus, RASA3 is critical to maintain patrolling platelets in a quiescent/non-adhesive state and to limit platelet adhesion at sites of injury. Firm platelet adhesion and hemostatic plug formation, however, require downmodulation of RASA3 activity, a process mediated mainly by signaling through the purinergic receptor for ADP, P2Y12. Consistently, early studies have shown that ADP/P2Y12 signaling is critical for sustained RAP1 activation and our recent work demonstrates that platelets lacking functional RASA3 are insensitive to inhibitors of P2Y12 in vitro and in vivo[22]. The effect of P2Y12 signaling on RAP1 activity is believed to be mediated by the lipid product of PI3K, PIP3 [47,48]. Consistently, RASA3 contains a unique pleckstrin homology domain (PH/Btk), which binds PIP3 with high affinity (KD=0.5±0.2 μM), and platelets lacking functional RASA3 are insensitive to inhibitors of PI3K[22,49]. It is unclear, however, how RASA3 activity is regulated by PIP3, especially since RASA3 also binds PIP2 with similar affinity (KD=0.8±0.5 μM). Although in vitro studies show that RASA3 is sensitive to agonist-induced PIP3 generation in cells[50] and in platelets[49], only a small fraction of RASA3 seems to undergo agonist-induced translocation to the cytosol. Other more likely explanations that may be investigated in the future are whether PIP3 affects RASA3 by directly inhibiting its catalytic activity or by sequestering it in membrane microdomains[51] without RAP1.
RAP1 functions: understanding how different isoforms work in concert to activate platelets at sites of injury
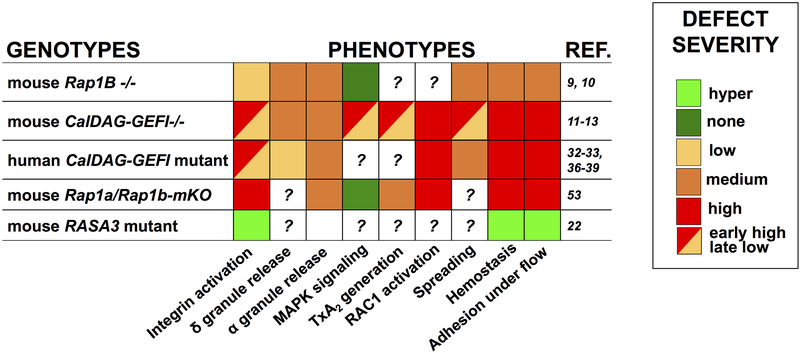
Most of what we know on RAP function comes from studies in mice lacking key upstream regulators (see above). Platelets lacking functional CALDAG-GEFI and/or P2Y12 exhibit defects in various platelet responses, including integrin activation, MAPK signaling, thromboxane A2 production, RAC1 activation and cytoskeleton-regulated responses such as secretion, spreading and clot retraction [11–13]. RAP1B is the only RAP isoform that has been examined for its pathophysiological role in hemostasis and thrombosis in mice. It is the most abundant RAP isoform in platelets, and germline deletion of the Rap1b gene in mice causes elevated embryonic lethality[52] and bleeding. However, the platelet defects in the surviving Rap1b−/− mice[9] are milder than those in mice lacking Caldaggef1[11] (Figure 2), as Rap1b-deficient platelets have decreased, but not abolished, agonist-induced fibrinogen binding and aggregation[9], partially impaired secretion and spreading, and no defect in MAPK signaling[10]. Thus, suggesting that there are RAP-independent mechanisms regulating platelet adhesion, or that other RAP isoforms have important roles in platelet activation and hemostasis. Our preliminary observations in mice lacking Rap1a and/or Rap1b in the platelet/megakaryocytic lineage support the latter conclusion, as combined deficiency of RAP1A and RAP1B leads to a severe integrin activation defect almost comparable to that observed in Talin1-deficient mice[53]. Additionally, we observe that RAP1A and RAP1B redundantly control TxA2 generation, while RAP1B alone affects RAC1 activation and granule secretion[53]. Conversely, in line with what was reported earlier for Rap1b−/− mice[10], we find that neither of the two RAP1 isoforms mediates ERK activation, opening the possibility that this pathway is controlled by RAP2 GTPases (see below).
Figure 2. Comparison of the platelet phenotype in the absence of functional RAP1 proteins or of their upstream regulators.
Table showing the effect of Rap1b deficiency (mouse Rap1b−/−), Caldaggef1 deficiency (mouse CalDAG-GEFI −/−), mutations in CalDAG-GEFI identified in human patients (human CalDAG-GEFI mutant), combined deficiency of Rap1a and Rap1b in the platelet-megakaryocytic lineage (mouse Rap1a/Rap1b-mKO) and expression of a Rasa3 mutant (H794L, mouse RASA3 mutant) on integrin activation, RAC1 activation, spreading, MAPK signaling, alpha (α) and dense (δ) granule secretion, TxA2 (thromboxane A2) production, hemostasis and adhesion under flow. The color code reflects the severity of the defect according to the legend shown on the left. Light green indicates hyperactivation. The question mark indicates ‘not known’.
RAP1 effectors: an old question takes a new turn
Cell-type-specific expression of RAP effectors is likely to explain the diverse actions of RAP proteins in specialized cells such as platelets. However, the identity of RAP downstream effectors in platelets remains elusive. The most well-established RAP effectors have been identified using cell models not related to the platelet/megakaryocytic lineage[7], and very few of these proteins are expressed at high enough levels in platelets[1–4]. Activated GTP-bound RAP1 binds preferentially to downstream effectors containing a RAS-binding-domain (RBD) or a RAS-association (RA) domain. Screening of the platelet proteome[1] for proteins containing the canonical RA domain revealed that platelets express at least two Rho-GAPs (ARAP1, Myosin IXb) that could potentially bind RAP1 and mediate RAP1-dependent cytoskeletal dynamics. However, a more systematic effort is required to clarify the signaling network downstream of these nodal signaling molecules.
Arguably the most important role for RAP1 in platelets is its ability to mediate integrin inside-out activation. Elegant studies conducted in heterologous cell systems (e.g. CHO or HEK293 cells) stably expressing αIIbβ3 suggested that, also in platelets, RAP1 promoted TALIN recruitment to the plasma membrane[54] through the formation of a ternary complex with the RAP1 effector RIAM/APBB1IP (RAP1-GTP-interacting adapter molecule)[55,56]. However, while RAP1 and TALIN are exceptionally abundant in platelets and megakaryocytes, RIAM is not[1–4]. Consistently, recent in vivo studies employing two independently generated knockout mouse models demonstrated that RIAM is dispensable for β1 and β3 integrin activation in platelets, while it is necessary for integrin β2 activation in leukocytes and neutrophils[57,58]. Interestingly, while RBD and RA domains do not share primary sequence similarity, they share the topology of the ubiquitin superfold[59,60], a structural element found in the N-terminal head domain of TALIN[61]. Thus, an alternative line of research has emerged, which investigates the hypothesis that RAP1-GTP can directly and specifically bind the F0 domain of TALIN[61–63]. At first this finding was not pursued since the affinity of the interaction was found to be very low (Kd~0.14mM)[61]. More recently, however, it was reported that membrane-anchored RAP1B-GTP binds the TALIN head domain with affinity two orders of magnitude greater than that measured without membranes and that site-directed mutations of critical residues at the RAP1/TALIN interface impairs integrin activation, adhesion and spreading of cultured cells[63]. In the amoeba Dictyostelium discoideum this interaction is biologically relevant as it is necessary for processes that require strong adhesion forces such as morphogenesis[62]. Future studies need to address the importance of the TALIN(F0)-RAP1 interaction for platelet function in vivo.
RAP1 subcellular localization: insights into new RAP1 functions
The ability of RAP proteins to relay signals to their downstream targets is also controlled by changes in their cellular distribution[64]. All small GTPases contain a highly-conserved GDP/GTP binding domain and a C-terminal hypervariable region, which is post-translationally modified by lipid anchors required for the docking to specific cellular membranes. Except RAP2A, which is farnesylated, RAP proteins are post-translationally modified by addition of a geranylgeranyl group. In addition, the C-terminal end of RAP1 proteins is rich of positively charged residues (lysine and arginine) that favor the recruitment to negatively charged membrane regions e.g. rich in PIP2 and PIP3[65]. It is very likely that these phospholipids spatially regulate RAP signaling by coordinating the assembly of the integrin activation complex consisting of RAP1, TALIN[66], and KINDLIN3[67]. In the near future, novel technologies such as super-resolution imaging, might give us insight on how these signaling molecules interact dynamically throughout the platelet activation process.
Nevertheless, it is clear that the C-terminal positively charged residues are crucial to position RAP1 in proximity of the integrin where TALIN needs to be recruited. Importantly within this stretch of acidic residues lies a serine (serine180 in RAP1A, serine179 in RAP1B) that is phosphorylated by PKA. While cAMP-directed phosphorylation of CALDAG-GEFI[42,43] and RAP1GAP2[46] is very rapid and produces the net effect of reducing RAP1-GTP levels, the direct phosphorylation of RAP1 occurs with a much slower kinetic and does not seem to affect GTP-loading[68]. Nonetheless it affects RAP1 function by promoting its translocation from the plasma membrane to the cytosol[69], thus terminating integrin activation. In cells other than platelets, it was shown that PKA phosphorylation of RAP1 is essential for the reversible regulation of integrin inside-out activation, which has to occur to enable cyclical cell detachment during migration[69]. Interestingly a recent study, using single-cell resolution analysis in vivo, observed for the first time that a fraction of adherent platelets at the periphery of an injury where able to migrate[70]. Since the periphery of the injury is where you might find higher levels of the endothelial-derived PKA-activator PGI2, it is possible that this signaling mechanism is in place also in platelets.
There is also increasing evidence that phosphorylation of RAP1 may also be creating new binding sites for downstream effectors[71,72]. Interestingly, 6 of the 9 residues in which RAP1A and RAP1B differ, lie within the C-terminal hypervariable region. Thus, differences in the localization and in the interactions with effectors could explain the functional differences between these isoforms and also reveal new ones[73].
RAP2: the mysterious sibling
Although RAP2 GTPases have been cloned more than 30 years ago[74], their functional role in platelets remains elusive. Despite sharing a high degree of homology (60%) and being activated by the same GEFs, mounting evidence from cells other than platelets indicates that RAP1 and RAP2 have distinct signaling functions. Due to a single replacement of serine39 by phenylalanine in the effector region, RAP2, but not RAP1, proteins interact with a subgroup of Ste20 kinases (TNIK, MINK and MAP4K4)[75–77], which are involved in many diverse signaling pathways by regulating MAPK signaling. The C-terminal region of RAP2 also contains two cysteine residues that undergo palmitoylation. In leukocytes[78], RAP2 GTPases are targeted to the recycling endosomal compartment[79] and regulate the recycling of integrins during migration, in a palmitoylation-dependent manner. In platelets, this modification was shown to target RAP2, but not RAP1, to lipid rafts[80]; however, the functional consequences of this localization are not clear yet.
Redundancy between the three RAP2 isoforms and a lack of mouse models have made it difficult to evaluate the role of RAP2 in platelet biology. In our previous work, we have shown that the activity of both RAP1 and RAP2 is regulated by CALDAG-GEFI and P2Y12/RASA3[13]. Thus, comparing platelet activation in Rap1a/b−/− and Caldaggef1/P2y12−/− mice provides indirect information on how RAP2 contributes to murine platelet function. Our preliminary studies suggest that RAP2 is not implicated in integrin affinity regulation, while it may play a role in MAPK signaling in platelets. In line with these conclusions, deficiency of the RAP2-specific effector MINK impairs platelet MAPK signaling, spreading and secretion in response to low doses of agonists[81]. Further insights into RAP2 function may be achieved by studying the role of other RAP2 effectors expressed in platelets, such as TNIK and MAP4K4.
Conclusions
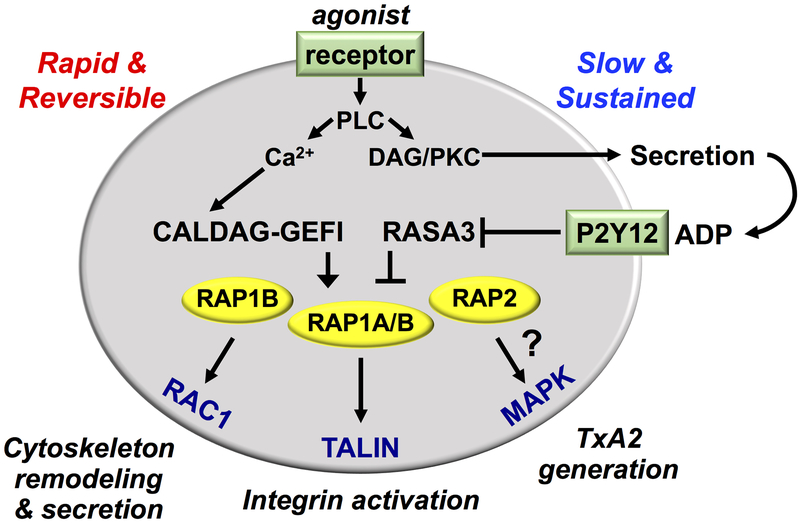
In recent years our understanding of how circulating platelets rapidly shift from an anti-adhesive to a pro-adhesive state has made huge progress. We and others established RAP1 GTPases as a central signaling node in this process, while the role of the closely related RAP2 proteins remains elusive (Figure 3). Their upstream regulators, CalDAG-GEFI and RASA3 are critical signal integrators for second messengers generated in response to agonist receptor engagement at sites of injury. While CALDAG-GEFI is critical for rapid RAP1 activation in response to trace amounts of agonists, RASA3 is necessary to limit platelet activation in circulation and needs to be turned off by ADP-mediated P2Y12 signaling to ensure sustained platelet activation and form stable hemostatic plugs. Once active, RAP1 is critical for the efficient formation of the integrin activation complex as it recruits TALIN in proximity of the integrin. Interestingly, recent studies have finally shed new light in understanding how this happens, suggesting that RAP1-GTP may be anchoring TALIN to the inner leaflet of the plasma membrane without the need of an intermediate effector. Moreover, we have strong indications that RAP1 activation and the formation of the integrin activation complex is coordinated by the localized synthesis of PIP2 and PIP3, although the details need to be clarified. In summary, we have now in-depth knowledge of the critical role of RAP1 GTPases in the molecular processes that go from injury recognition to platelet adhesion, which may lead to a better understanding of the variability in platelet reactivity, improved diagnosis of rare bleeding disorders and to the exploration of safer strategies for the treatment of thrombosis.
Figure 3. Schematic summary of RAP signaling in platelets.
RAP GTPases are a central signaling node regulating platelet activation. Their upstream regulators, CalDAG-GEFI and RASA3 are critical signal integrators for second messengers generated in response to agonist receptor engagement at sites of injury. While CALDAG-GEFI is critical for rapid RAP activation in response to trace amounts of agonists, sustained RAP activation is ensured by ADP-mediated P2Y12 signaling that turns off the RAP inhibitor RASA3. Once active, RAP GTPases drive platelet activation at sites of vascular injury by switching on multiple platelet responses. Our ongoing studies indicate that both RAP1A and RAP1B control integrin inside-out activation by recruiting TALIN, while RAP1B alone regulates granule secretion, probably though its effect on RAC1 activation. Furthermore, indirect observations suggest that RAP2 GTPases may be regulating MAPK signaling and MAPK-dependent responses such as TxA2 generation.
Acknowledgements
This work was supported by NIH grants R01 HL121650 and P01 HL120846 (W.B.) and the Ministero della Istruzione e della Ricerca Young Researchers Program Rita Levi Montalcini (L.S.).
Footnotes
Conflict of interest
The authors report no declarations of interest.
References
- 1.Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, Geiger J, Sickmann A, Zahedi RP. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood 2012; 120: e73–82. [DOI] [PubMed] [Google Scholar]
- 2.Zeiler M, Moser M, Mann M. Copy Number Analysis of the Murine Platelet Proteome Spanning the Complete Abundance Range. Molecular & Cellular Proteomics 2014; 13: 3435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood 2011; 118: e101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon LM, Edelstein LC, Nagalla S, Woodley AB, Chen ES, Kong X, Ma L, Fortina P, Kunapuli S, Holinstat M, McKenzie SE, Dong J-F, Shaw CA, Bray PF. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood 2014; 123: e37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron E Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. Journal of Cell Science 2003; 116: 435–40. [DOI] [PubMed] [Google Scholar]
- 6.Raaijmakers JH, Bos JL. Specificity in Ras and Rap Signaling. J Biol Chem 2009; 284: 10995–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos JL. Linking Rap to cell adhesion. Current Opinion in Cell Biology 2005; 17: 123–8. [DOI] [PubMed] [Google Scholar]
- 8.Bergmeier W, Stefanini L. Platelets at the vascular interface. Res Pract Thromb Haemost 2017; 16: 58–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC. Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest 2005; 115: 680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, Xiang B, Ye S, Chrzanowska-Wodnicka M, Morris AJ, Gartner TK, Whiteheart SW, White GC, Smyth SS, Li Z. Distinct roles for Rap1b protein in platelet secretion and integrin αIIbβ3 outside-in signaling. J Biol Chem 2011; 286: 39466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crittenden JR, Bergmeier W, Zhang Y, Piffath CL, Liang Y, Wagner DD, Housman DE, Graybiel AM. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat Med 2004; 10: 982–6. [DOI] [PubMed] [Google Scholar]
- 12.Stefanini L, Roden RC, Bergmeier W. CalDAG-GEFI is at the nexus of calcium-dependent platelet activation. Blood 2009; 114: 2506–14. [DOI] [PubMed] [Google Scholar]
- 13.Stefanini L, Boulaftali Y, Ouellette TD, Holinstat M, Désiré L, Leblond B, Andre P, Conley PB, Bergmeier W. Rap1-Rac1 circuits potentiate platelet activation. Arterioscler Thromb Vasc Biol 2012; 32: 434–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shock DD, He K, Wencel-Drake JD, Parise LV. Ras activation in platelets after stimulation of the thrombin receptor, thromboxane A2 receptor or protein kinase C. Biochem J 1997; 321: 525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tulasne D, Bori T, Watson SP. Regulation of RAS in human platelets. Evidence that activation of RAS is not sufficient to lead to ERK1–2 phosphorylation. Eur J Biochem 2002; 269: 1511–7. [DOI] [PubMed] [Google Scholar]
- 16.Popovic M, Leeuw MR-D, Rehmann H. Selectivity of CDC25 Homology Domain-Containing Guanine Nucleotide Exchange Factors. Journal of Molecular Biology Elsevier Ltd; 2013; 425: 2782–94. [DOI] [PubMed] [Google Scholar]
- 17.Franke B, van Triest M, de Bruijn KM, van Willigen G, Nieuwenhuis HK, Negrier C, Akkerman JW, Bos JL. Sequential regulation of the small GTPase Rap1 in human platelets. Molecular and Cellular Biology 2000; 20: 779–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke B, Akkerman JW, Bos JL. Rapid Ca2+-mediated activation of Rap1 in human platelets. The EMBO Journal 1997; 16: 252–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cifuni SM, Wagner DD, Bergmeier W. CalDAG-GEFI and protein kinase C represent alternative pathways leading to activation of integrin alphaIIbbeta3 in platelets. Blood 2008; 112: 1696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stolla M, Stefanini L, Roden RC, Chavez M, Hirsch J, Greene T, Ouellette TD, Maloney SF, Diamond SL, Poncz M, Woulfe DS, Bergmeier W. The kinetics of αIIbβ3 activation determines the size and stability of thrombi in mice: implications for antiplatelet therapy. Blood 2011; 117: 1005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernardi B, Guidetti GF, Campus F, Crittenden JR, Graybiel AM, Balduini C, Torti M. The small GTPase Rap1b regulates the cross talk between platelet integrin alpha2beta1 and integrin alphaIIbbeta3. Blood 2006; 107: 2728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefanini L, Paul DS, Robledo RF, Chan ER, Getz TM, Campbell RA, Kechele DO, Casari C, Piatt R, Caron KM, Mackman N, Weyrich AS, Parrott MC, Boulaftali Y, Adams MD, Peters LL, Bergmeier W. RASA3 is a critical inhibitor of RAP1-dependent platelet activation. J Clin Invest 2015; 125: 1419–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical Elements in the Control of Small G Proteins. Cell 2007; 129: 865–77. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, Matsuda M, Housman DE, Graybiel AM. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci USA 1998; 95: 13278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohba Y, Mochizuki N, Yamashita S, Chan AM, Schrader JW, Hattori S, Nagashima K, Matsuda M. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J Biol Chem; 2000; 275: 20020–6. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Vuori K, Wang H, Reed JC, Ruoslahti E. Integrin activation by R-ras. Cell 1996; 85: 61–9. [DOI] [PubMed] [Google Scholar]
- 27.Iwig JS, Vercoulen Y, Das R, Barros T, Limnander A, Che Y, Pelton JG, Wemmer DE, Roose JP, Kuriyan J. Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1. eLife 2013; 2: e00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook AA, Deng W, Ren J, Li R, Sondek J, Bergmeier W. Calcium-induced structural rearrangements release autoinhibition in the Rap-GEF, CalDAG-GEFI. Journal of Biological Chemistry 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Czikora A, Lundberg DJ, Abramovitz A, Lewin NE, Kedei N, Peach ML, Zhou X, Merritt RC Jr., Craft EA, Braun DC, Blumberg PM. Structural Basis for the Failure of the C1 Domain of Ras Guanine Nucleotide Releasing Protein 2 (RasGRP2) to Bind Phorbol Ester with High Affinity. J Biol Chem 2016; 291: 11133–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irie K, Masuda A, Shindo M, Nakagawa Y, Ohigashi H. Tumor promoter binding of the protein kinase C C1 homology domain peptides of RasGRPs, chimaerins, and Unc13s. Bioorganic & Medicinal Chemistry 2004; 12: 4575–83. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JE, Goulding RE, Ding Z, Partovi A, Anthony KV, Beaulieu N, Tazmini G, Cornell RB, Kay RJ. Differential membrane binding and diacylglycerol recognition by C1 domains of RasGRPs. Biochem J 2007; 406: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canault M, Ghalloussi D, Grosdidier C, Guinier M, Perret C, Chelghoum N, Germain M, Raslova H, Peiretti F, Morange PE, Saut N, Pillois X, Nurden AT, Cambien F, Pierres A, van den Berg TK, Kuijpers TW, Alessi M-C, Tregouet D-A. Human CalDAG-GEFI gene (RASGRP2) mutation affects platelet function and causes severe bleeding. J Exp Med 2014; 211: 1349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H, Nakazawa Y, Kurokawa Y, Kashiwagi H, Morikawa Y, Morita D, Banno F, Honda S, Kanakura Y, Tomiyama Y. Human CalDAG-GEFI deficiency increases bleeding and delays αIIbβ3 activation. Blood 2016; 128: 2729–33. [DOI] [PubMed] [Google Scholar]
- 34.Stolla M, Stefanini L, André P, Ouellette TD, Reilly MP, McKenzie SE, Bergmeier W. CalDAG-GEFI deficiency protects mice in a novel model of FcγRIIA-mediated thrombosis and thrombocytopenia. Blood 2011; 118: 1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boulaftali Y, Owens AP, Beale A, Piatt R, Casari C, Lee RH, Conley PB, Paul DS, Mackman N, Bergmeier W. CalDAG-GEFI Deficiency Reduces Atherosclerotic Lesion Development in Mice. Arterioscler Thromb Vasc Biol 2016; 36: 792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bermejo E, Alberto MF, Paul DS, Cook AA, Nurden P, Sanchez Luceros A, Nurden AT, Bergmeier W. Marked bleeding diathesis in patients with platelet dysfunction due to a novel mutation in RASGRP2, encoding CalDAG-GEFI (p.Gly305Asp). Platelets 2018; 29: 84–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westbury SK, Canault M, Greene D, Bermejo E, Hanlon K, Lambert MP, Millar CM, Nurden P, Obaji SG, Revel-Vilk S, Van Geet C, Downes K, Papadia S, Tuna S, Watt C, NIHR BioResource–Rare Diseases Consortium, Freson K, Laffan MA, Ouwehand WH, Alessi M-C, et al. Expanded repertoire of RASGRP2 variants responsible for platelet dysfunction and severe bleeding. Blood; 2017; 130: 1026–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sevivas T, Bastida JM, Paul DS, Caparros E, Palma-Barqueros V, Coucelo M, Marques D, Ferrer-Marin F, González-Porras JR, Vicente V, Hernández-Rivas JM, Watson SP, Lozano ML, Bergmeier W, Rivera J. Identification of two novel mutations in RASGRP2 affecting platelet CalDAG-GEFI expression and function in patients with bleeding diathesis. Platelets. 2018. March;29(2):192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozano ML, Cook A, Bastida JM, Paul DS, Iruin G, Cid AR, Adan-Pedroso R, Ramón González-Porras J, Hernández-Rivas JM, Fletcher SJ, Johnson B, Morgan N, Ferrer-Marin F, Vicente V, Sondek J, Watson SP, Bergmeier W, Rivera J. Novel mutations in RASGRP2, which encodes CalDAG-GEFI, abrogate Rap1 activation, causing platelet dysfunction. Blood; 2016; 128: 1282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piatt R, Paul DS, Lee RH, McKenzie SE, Parise LV, Cowley DO, Cooley BC, Bergmeier W. Mice Expressing Low Levels of CalDAG-GEFI Exhibit Markedly Impaired Platelet Activation With Minor Impact on Hemostasis. Arterioscler Thromb Vasc Biol; 2016; 36: 1838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefanini L, Bergmeier W. Negative regulators of platelet activation and adhesion. J Thromb Haemost 2017; 8: 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian H, Zahedi RP, Sickmann A, Walter U, Gambaryan S. Phosphorylation of CalDAG-GEFI by protein kinase A prevents Rap1b activation. Journal of Thrombosis and Haemostasis 2013; 11: 1574–82. [DOI] [PubMed] [Google Scholar]
- 43.Guidetti GF, Manganaro D, Consonni A, Canobbio I, Balduini C, Torti M. Phosphorylation of the guanine-nucleotide-exchange factor CalDAG-GEFI by protein kinase A regulates Ca 2+-dependent activation of platelet Rap1b GTPase. Biochem J 2013; 453: 115–23. [DOI] [PubMed] [Google Scholar]
- 44.Beck F, Geiger J, Gambaryan S, Solari FA, Dell’Aica M, Loroch S, Mattheij NJ, Mindukshev I, Pötz O, Jurk K, Burkhart JM, Fufezan C, Heemskerk JWM, Walter U, Zahedi RP, Sickmann A. Temporal quantitative phosphoproteomics of ADP stimulation reveals novel central nodes in platelet activation and inhibition. Blood; 2017; 129: e1–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultess J, Danielewski O, Smolenski AP. Rap1GAP2 is a new GTPase-activating protein of Rap1 expressed in human platelets. Blood 2005; 105: 3185–92. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmeister M, Riha P, Neumüller O, Danielewski O, Schultess J, Smolenski AP. Cyclic Nucleotide-dependent Protein Kinases Inhibit Binding of 14-3-3 to the GTPase-activating Protein Rap1GAP2 in Platelets. J Biol Chem 2008; 283: 2297–306. [DOI] [PubMed] [Google Scholar]
- 47.Cozier GE, Bouyoucef D, Cullen PJ. Engineering the phosphoinositide-binding profile of a class I pleckstrin homology domain. J Biol Chem 2003; 278: 39489–96. [DOI] [PubMed] [Google Scholar]
- 48.Cozier GE, Lockyer PJ, Reynolds JS, Kupzig S, Bottomley JR, Millard TH, Banting G, Cullen PJ. GAP1IP4BP contains a novel group I pleckstrin homology domain that directs constitutive plasma membrane association. J Biol Chem 2000; 275: 28261–8. [DOI] [PubMed] [Google Scholar]
- 49.Battram AM, Durrant TN, Agbani EO, Heesom KJ, Paul DS, Piatt R, Poole AW, Cullen PJ, Bergmeier W, Moore SF, Hers I. The Phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3) Binder Rasa3 Regulates Phosphoinositide 3-kinase (PI3K)-dependent Integrin αIIbβ3 Outside-in Signaling. J Biol Chem 2017; 292: 1691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammond GRV, Sim Y, Lagnado L, Irvine RF. Reversible binding and rapid diffusion of proteins in complex with inositol lipids serves to coordinate free movement with spatial information. The Journal of Cell Biology 2009; 184: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Richards DA. Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biology Open 2012; 1: 857–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chrzanowska-Wodnicka M, White GC, Quilliam LA, Whitehead KJ. Small GTPase Rap1 Is Essential for Mouse Development and Formation of Functional Vasculature. Boggon TJ, editor. PLoS ONE; 2015; 10: e0145689–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stefanini L, Paul DS, OShaughnessy E, Jones CI, Piatt R, Lee RH, Petrich BG, Cooley B, Hahn K, Gibbins J, Bergmeier W. Rap1A and Rap1B Functional Redundancy in Platelets and Megakaryocytes. Res Pract Thromb Haemost 2017; 1: 226.(OC06.1). [Google Scholar]
- 54.Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Current Biology 2006; 16: 1796–806. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe N, Bodin L, Pandey M, Krause M, Coughlin S, Boussiotis VA, Ginsberg MH, Shattil SJ. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. The Journal of Cell Biology 2008; 181: 1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee H-S, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J Biol Chem 2009; 284: 5119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stritt S, Wolf K, Lorenz V, Vögtle T, Gupta S, Bösl MR, Nieswandt B. Rap1-GTP-interacting adaptor molecule (RIAM) is dispensable for platelet integrin activation and function in mice. Blood 2015; 125: 219–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su W, Wynne J, Pinheiro EM, Strazza M, Mor A, Montenont E, Berger J, Paul DS, Bergmeier W, Gertler FB, Philips MR. Rap1 and its effector RIAM are required for lymphocyte trafficking. Blood; 2015; 126: 2695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, Serrano L. Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. Journal of Molecular Biology 2005; 348: 759–75. [DOI] [PubMed] [Google Scholar]
- 60.Wohlgemuth S, Kiel C, Krämer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. Journal of Molecular Biology 2005; 348: 741–58. [DOI] [PubMed] [Google Scholar]
- 61.Goult BT, Bouaouina M, Elliott PR, Bate N, Patel B, Gingras AR, Grossmann JG, Roberts GCK, Calderwood DA, Critchley DR, Barsukov IL. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. The EMBO Journal 2010; 29: 1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plak K, Pots H, Van Haastert PJM, Kortholt A. Direct Interaction between TalinB and Rap1 is necessary for adhesion of Dictyostelium cells. BMC Cell Biology; 2016; 17:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu L, Yang J, Bromberger T, Holly A, Lu F, Liu H, Sun K, Klapproth S, Hirbawi J, Byzova TV, Plow EF, Moser M, Qin J. Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation. Nat Commun US; 2017; 8: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wittchen ES, Aghajanian A, Burridge K. Isoform-specific differences between Rap1A and Rap1B GTPases in the formation of endothelial cell junctions. Small Gtpases 2011; 2: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science; 2006; 314: 1458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Legate KR, Takahashi S, Bonakdar N, Fabry B, Boettiger D, Zent R, Fässler R. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. The EMBO Journal 2011; 30: 4539–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J, Fukuda K, Xu Z, Ma Y-Q, Hirbawi J, Mao X, Wu C, Plow EF, Qin J. Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation. J Biol Chem; 2011; 286: 43334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hata Y, Kaibuchi K, Kawamura S, Hiroyoshi M, Shirataki H, Takai Y. Enhancement of the actions of smg p21 GDP/GTP exchange protein by the protein kinase A-catalyzed phosphorylation of smg p21. J Biol Chem 1991; 266: 6571–7. [PubMed] [Google Scholar]
- 69.Takahashi M, Dillon TJ, Liu C, Kariya Y, Wang Z, Stork PJS. Protein Kinase A-dependent Phosphorylation of Rap1 Regulates Its Membrane Localization and Cell Migration. J Biol Chem 2013; 288: 27712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaertner F, Ahmad Z, Rosenberger G, Fan S, Nicolai L, Busch B, Yavuz G, Luckner M, Ishikawa-Ankerhold H, Hennel R, Benechet A, Lorenz M, Chandraratne S, Schubert I, Helmer S, Striednig B, Stark K, Janko M, Böttcher RT, Verschoor A, et al. Migrating Platelets Are Mechano-scavengers that Collect and Bundle Bacteria. Cell; 2017; 171: 1368–1382.e23. [DOI] [PubMed] [Google Scholar]
- 71.Riou P, Kjær S, Garg R, Purkiss A, George R, Cain RJ, Bineva G, Reymond N, McColl B, Thompson AJ, O’Reilly N, McDonald NQ, Parker PJ, Ridley AJ. 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell 2013; 153: 640–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahashi M, Li Y, Dillon TJ, Stork PJS. Phosphorylation of Rap1 by cAMP-dependent Protein Kinase (PKA) Creates a Binding Site for KSR to Sustain ERK Activation by cAMP. J Biol Chem 2017; 292: 1449–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson JM, Prokop JW, Lorimer E, Ntantie E, Williams CL. Differences in the Phosphorylation-Dependent Regulation of Prenylation of Rap1A and Rap1B. Journal of Molecular Biology 2016; 428: 4929–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pizon V, Chardin P, Lerosey I, Olofsson B, Tavitian A. Human cDNAs rap1 and rap2 homologous to the Drosophila gene Dras3 encode proteins closely related to ras in the “effector” region. Oncogene 1988; 3: 201–4. [PubMed] [Google Scholar]
- 75.Taira K, Umikawa M, Takei K, Myagmar B-E, Shinzato M, Machida N, Uezato H, Nonaka S, Kariya K-I. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem 2004; 279: 49488–96. [DOI] [PubMed] [Google Scholar]
- 76.Machida N, Umikawa M, Takei K, Sakima N, Myagmar B-E, Taira K, Uezato H, Ogawa Y, Kariya K-I. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J Biol Chem 2004; 279: 15711–4. [DOI] [PubMed] [Google Scholar]
- 77.Nonaka H, Takei K, Umikawa M, Oshiro M, Kuninaka K, Bayarjargal M, Asato T, Yamashiro Y, Uechi Y, Endo S, Suzuki T, Kariya K-I. MINK is a Rap2 effector for phosphorylation of the postsynaptic scaffold protein TANC1. Biochem Biophys Res Commun 2008; 377: 573–8. [DOI] [PubMed] [Google Scholar]
- 78.Stanley P, Tooze S, Hogg N. A role for Rap2 in recycling the extended conformation of LFA-1 during T cell migration. Biology Open 2012; 1: 1161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uechi Y, Bayarjargal M, Umikawa M, Oshiro M, Takei K, Yamashiro Y, Asato T, Endo S, Misaki R, Taguchi T, Kariya K-I. Rap2 function requires palmitoylation and recycling endosome localization. Biochem Biophys Res Commun 2009; 378: 732–7. [DOI] [PubMed] [Google Scholar]
- 80.Canobbio I, Trionfini P, Guidetti GF, Balduini C, Torti M. Targeting of the small GTPase Rap2b, but not Rap1b, to lipid rafts is promoted by palmitoylation at Cys176 and Cys177 and is required for efficient protein activation in human platelets. Cell Signal 2008; 20: 1662–70. [DOI] [PubMed] [Google Scholar]
- 81.Yue M, Luo D, Yu S, Liu P, Zhou Q, Hu M, Liu Y, Wang S, Huang Q, Niu Y, Lu L, Hu H. Misshapen/NIK-related kinase (MINK1) is involved in platelet function, hemostasis, and thrombus formation. Blood; 2016; 127: 927–37. [DOI] [PubMed] [Google Scholar]