Abstract
The accessory optic system (AOS) detects retinal image slip and reports it to the oculomotor system for reflexive image stabilization. Here, we characterize two Cre lines that permit genetic access to AOS circuits responding to vertical motion. The first (Pcdh9-Cre) labels only one of the four subtypes of ON direction-selective retinal ganglion cells (ON-DS RGCs), those preferring ventral retinal motion. Their axons diverge from the optic tract just behind the chiasm and selectively innervate the medial terminal nucleus (MTN) of the AOS. Unlike most RGC subtypes examined, they survive after optic nerve crush. The second Cre-driver line (Pdzk1ip1-Cre) labels postsynaptic neurons in the MTN. These project predominantly to the other major terminal nucleus of the AOS, the nucleus of the optic tract (NOT). We find that the transmembrane protein semaphorin 6A (Sema6A) is required for the formation of axonal projections from the MTN to the NOT, just as it is for the retinal innervation of the MTN. These new tools permit manipulation of specific circuits in the AOS and show that Sema6A is required for establishing AOS connections in multiple locations.
Keywords: Retinal ganglion cells, Medial terminal nucleus, Accessory optic system, Sema6A
Graphical Abstract
1. Introduction
Retinal ganglion cells (RGCs) transmit information from the eye to the brain, allowing for central processing of visual stimuli. Retinal ganglion cells can be classified into ~30 subtypes distinguished by visual response properties, morphology, synaptic connectivity, gene expression profiles and response to optic nerve injury (Duan et al., 2015; Sanes & Masland, 2015). The visual response properties encoded by retinal ganglion cells include intrinsic photosensitivity, color opponency, contrast sensitivity, looming object detection, and direction selectivity (Duan, Krishnaswamy, la Huerta, & Sanes, 2014; Ecker et al., 2010; Huberman et al., 2009; Joesch & Meister, 2016; K. P. Johnson, Zhao, & Kerschensteiner, 2018; Kim, Zhang, Yamagata, Meister, & Sanes, 2008; Krishnaswamy, Yamagata, Duan, Hong, & Sanes, 2015; Sabbah, Berg, Papendorp, Briggman, & Berson, 2017a; Y. Zhang, Kim, Sanes, & Meister, 2012). Directionally selective retinal ganglion cells (DS RGCs) increase their firing rate when movement in a preferred direction occurs within their receptive fields (Mauss, Vlasits, Borst, & Feller, 2017). One type of DS RGC (ON-DS RGC) responds to slow movement of bright objects (Barlow & Levick, 1965). The ON-DS RGCs project to the accessory optic system (AOS) in the brain and are believed to be responsible for stabilizing images on the retina during animal self-motion (Dhande et al., 2013; Simpson, 1984; Yonehara et al., 2009). This image-stabilization mechanism is highly conserved and thought to be essential for maintaining image clarity (Oyster, Takahashi, & Collewijn, 1972). Retinal direction selectivity depends critically on selective inhibitory input onto ON-DS RGC dendrites from pre-synaptic starburst amacrine cells (SACs) (Mauss et al., 2017). The dendrites of ON-DS RGCs that contribute to AOS image stabilization responses stratify mostly in the S4 sublamina of the retinal inner plexiform layer (IPL), and there form synaptic connections with ON SAC dendrites (Yonehara et al., 2010). The axons of ON-DS RGCs project to three subnuclei in the AOS: the MTN, the dorsal terminal nucleus (DTN) and the NOT (Dhande et al., 2013; Sabbah, et al., 2017b; Zhang, Kolodkin, Wong, & James, 2017). ON-DS RGCs that prefer dorsal and ventral motion project to the MTN, whereas those that detect forward and reverse movements mainly target the NOT and DTN (Dhande et al., 2013; Sabbah, Gemmer, Bhatia-Lin, Manoff, Castro, Siegel, et al., 2017b; Yonehara et al., 2009; 2008). The AOS nuclei themselves are interconnected; MTN-resident neurons project to the NOT, and NOT-resident neurons project to the MTN (reviewed in (Giolli, Blanks, & Lui, 2006)). How specific RGC subtypes target their appropriate brain regions, and how these brain regions are interconnected to allow proper visual system function, are questions that remain to be fully addressed.
Several molecular pathways have been described that direct RGC targeting to AOS nuclei. Signaling between the transmembrane protein Sema6A, which is expressed on the surface of AOS-targeting RGCs and functions as a receptor, and PlexA2/PlexA4 expressed in the MTN functioning as ligands, is required for ON-DS RGC innervation of the MTN and also AOS image stabilization (Sun, et al., 2015a). In addition, the immunoglobulin superfamily member contactin-4 (CNTN4) and its binding partner amyloid precursor protein (APP) are required in ON-DS RGCs for innervation of the NOT (Osterhout, Stafford, Nguyen, Yoshihara, & Huberman, 2015). It is unknown whether these molecular cues are also required for establishing connections between AOS nuclei. Furthermore, the AOS cell types residing in these distinct brain nuclei are not well described and their functions are unclear.
Another feature that distinguishes RGC subtypes is their differential abilities to survive and re-extend axons after sustaining an injury to the optic nerve. After optic nerve injury, only about 20% of total RGCs survive, and the survivors are predominantly α RGCs and M1-intrinsically photosensitive RGCs (M1-ipRGCs) (Duan et al., 2015; Park et al., 2008). All other RGC subtypes examined to date, including M2-ipRGCs and also ON-OFF DS RGCs that project to the superior colliculus (SC) and the dorsal lateral geniculate nucleus (dLGN), undergo axotomy-induced apoptosis (Duan et al., 2015). One hypothesis to explain why certain RGC subtypes survive after axotomy focuses on mTOR signaling, α RGCs have high endogenous levels of mTOR activity and the mTOR signaling effector osteopontin, and activation of mTOR signaling in all RGCs selectively promotes axonal regeneration of a RGCs, but not ON-OFF DS RGCs or W3-RGCs (Duan et al., 2015). Additionally, M1-ipRGCs, which express the GPCR melanopsin at high levels, show increased mTOR activation after melanopsin activation, and this in turn stimulates axon regeneration (S. Li et al., 2016). Overexpression of melanopsin in all RGCs selectively promotes axonal regeneration of a RGCs but not CART+ ON-OFF DS RGCs or M1-ipRGCs (S. Li et al., 2016). Whether the RGCs that project to the AOS survive after axotomy and re-extend their axons remains to be determined.
Here we characterize two mouse lines that allow investigation into AOS development, function, and response to injury. The Pcdh9-Cre line labels a type of ON-DS RGC that prefers ventral motion and projects to the MTN via the inferior fasciculus of the accessory optic tract. The Pdzk1ip1-Cre line labels retinorecipient MTN neurons that project to the NOT. Using these lines we demonstrate that Sema6A is required to form MTN-NOT projections, suggesting that the molecular cues directing RGC targeting to visual system nuclei are also used to establish interconnectivity between visual system nuclei. Finally, we demonstrate that the ON-DS RGCs that project to the MTN survive after optic nerve crush, providing a paradigm for utilizing regeneration-promoting strategies to re-establish AOS function after nerve damage.
2. Methods
2.1. Animals
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocol was approved by the Animal Care and Use Committees of the Johns Hopkins University School of Medicine (protocol number M017M26) and of Brown University. The mouse strains HB9-GFP (Tg(Hlxb9-GFP)1Tmj; RRID:IMSR_JAX:005029), Sema6A (Sema6aGt(KST069)Bys; RRID:MGI:3037891), AH4 (Gt(ROSA)26Sortm14(CAT-tdTomato)Hze; RRID: IMSR_JAX:007914) have been described previously (Kerjan et al., 2005; Madisen et al., 2010; Wichterle, Lieberam, Porter, & Jessell, 2002). Cryopreserved sperm samples from strain Pcdh9-Cre (Tg(Pcdh9-cre)NP276GsatMmucd; RRID:MMRRC_036084-UCD) and Pdzk1ip1-Cre (Tg(Pdzk1ip1-cre)KD31GsatMmucd; RRID:MMRRC_030851-UCD) were obtained from the Mutant Mouse Resource and Research Center and were used for in vitro fertilization according to established methods. Progeny were genotyped with PCR using primers for Cre and were then bred to C57/B6J for at least 5 generations.
2.2. Surgical procedures
Intravitreal injections were performed with animals under isoflurane anesthesia as previously described (Sun et al., 2015a). Briefly, a 30 Gauge needle was used to make an incision at the limbus. A 33 G Hamilton syringe was filled and inserted into the hole and solution of CTB or AAV was slowly injected. Following needle removal, the eyelids were forced closed and the animal was placed on a heated pad until it recovered.
For stereotaxic injection, animals were anesthetized by induction with isoflurane (3% in Oxygen) and were transferred to a stereotactic frame (Knopf) with a heating pad and a nose piece for inhaled isoflurane anesthesia (2% in Oxygen). The skull was shaved and an incision was made along the midline exposing the skull. The coordinates of Bregma and Lambda were identified, and a 0.7 mm burr drill bit was used to drill a hole in the skull at the appropriate location. Stereotactic coordinates were derived from Paxinos and Watson. Coordinates for MTN injection (relative to Bregma in mm): 30 degree anterior angle, AP +0.05, ML +0.82, DV −5.36. Injections into the NOT were made using coordinates (from bregma): AP +2.8, ML +1.1, DV −1.1. Alexa dye conjugated Cholera toxin subunit B (1 mg/ml, 200 nl) or AAV vectors (200 nl) were injected at a rate of 10nL/sec using a Neuros Hamilton Syringe controlled by a syringe pump. Following injection, the needle was left in place for 5 min and was then withdrawn slowly. Wound clips were used to close the incision in the scalp, and the animals were given post-operative analgesia and were monitored closely until they recovered.
Optic nerve crush was performed with animals under isoflurane anesthesia. Two weeks after intravitreal viral injection, the optic nerve was exposed intraorbitally and crushed with jeweler’s forceps (Dumont #5, FST) for 10 seconds approximately 1 mm behind the optic disc (Leon, Yin, Nguyen, Irwin, & Benowitz, 2000). Optic nerve crush was verified by intravitreal injection of CTB one day before tissue harvest.
High titer (>1012 genome copies per ml) aliquots of AAV2-CAG-FLEX-GFP and AAV2-CAG-FLEX- tdTomato were obtained from the University of North Carolina Viral Vector Core. The AAV2retro-CAG- FLEX-tdTomato virus was obtained from the Howard Hughes Medical Institute Janelia Farms Viral Vector Core (Tervo et al., 2016). Viral vectors were diluted to 1×1012 GC/ml prior to intravitreal injection, and undiluted aliquots were used for injection into the brain.
2.3. Tissue preparation, Immunohistochemistry and Antibodies
Animals were transcardially perfused with 20 ml of ice-cold Ringer solution followed by 25 ml of 4% w/v Paraformaldehyde (PFA). Brains were removed and were post-fixed in 4% PFA for 16 hours followed by extensive washing in PBS. Brains were sectioned on a vibrating microtome (Leica VT1000) at 50–100 microns and sections were collected in PBS. Tissue sections and whole retinas were incubated free floating in blocking buffer (PBS with 10% Normal goat or donkey serum, 0.3% Triton X-100) for 2 hours, and were then incubated for 48–72 hours at 4°C with primary antibody diluted in blocking buffer. Sections were washed in PBS and incubated for 2 hours at room temperature secondary antibodies diluted in blocking buffer. Following additional washes, sections were incubated with Neurotrace 647 (Invitrogen) and mounted with Vectashield (Vector Laboratories).
The following antibodies were used for immunohistochemistry: chicken anti-GFP (Aves Labs, GFP-1020, 1:1000; RRID:AB_10000240), rabbit anti-RFP (Rockland, cat # 600–401-379, 1:1000; RRID:AB_2209751), goat anti-ChAT (Millipore, cat #AB144, 1:200; RRID: AB_2079751), guinea pig anti- RBPMS (PhosphoSolutions 1832-RBPMS, 1:500; RRID:AB_2492226). All antibodies have been characterized previously and used extensively in numerous published reports. Dye-conjugated secondary antibodies were from Invitrogen or Jackson Immunoresearch. Brain clearing and immunohistochemical staining for GFP was performed as described (Chi et al., 2018).
2.4. Tissue imaging
Confocal microscopy was performed with a Zeiss LSM 510 with 20× 0.8 NA and 40× 1.3 NA oil immersion objectives as described (Sun et al., 2015a). Confocal z stacks were collected with a 2 and 0.5 micron step size for the 20X and 40X objectives, respectively. Imaging of whole brain sections was done using a Keyence microscope with a 5× objective and filters for TRITC and Cy5. Tiled images covering the entire brain section were stitched together and exported as tiff files for processing in Adobe Photoshop. Cleared brains were prepared by the method described (Chi et al., 2018). Cleared brains were imaged by Light Sheet Fluorescence Microscopy (LSFM) using a LaVision BioTEC light sheet fluorescence microscope (Chi et al., 2018).
2.5. Image analysis
ImageJ was used for analysis of confocal image stacks and for quantification of dendritic field area and soma size. Analysis of Pcdh9-Cre+ RGC mosaic spacing was performed by collecting images from Pcdh9-Cre retinas infected with AAV2-FLEX-tdTomato and immunostained for RFP and RBPMS to visualize RGCs. Flat mounted retinas from three independently injected animals were imaged at 20X with 4–5 fields collected per retina. The positions of RBPMS+/tdTomato+ cells were measured in each field (640 × 640 microns) and were exported to the WinDRP program to generate a density recovery profile (Rockhill, Euler, & Masland, 2000; Rodieck, 1991). The mean ± SEM (n=3 animals) of the relative density as a function of distance from the reference cell was plotted. Stitching of 3D image stacks was done using the Stitching plugin in Fiji (Preibisch, Saalfeld, & Tomancak, 2009). For examination of dendritic stratification, retinal whole mounts stained for tdTomato and ChAT were imaged at 40X and image stacks were stitched together to generate a stack that contained a cell with its full dendritic arbor. Z sections containing the S2 and S4 ChAT sublaminae were processed separately and a maximum intensity projection was made of each. Quantification of dendritic stratification was performed by creating orthogonal views of stitched z-stacks in ImageJ using the 3D project function. The Mean value was projected for the tdTomato and ChAT signals separately and images were adjusted to normalize brightness. Pixel density as a function of IPL depth was generated for both sets of images using Matlab. Imaris software was used to visualize data captured with LSFM.
2.6. Tissue harvest and retinal dissection for patch recording and dye filling of RGCs
Right eye was removed and immersed in oxygenated Ames medium (95% O2, 5% CO2; Sigma-Aldrich; supplemented with 23 mM NaHCO3 and 10 mM D-glucose). Under dim red light, the globe was cut along the ora serrata, and cornea, lens and vitreous removed. Four radial relieving cuts were made in the eyecup, the largest centered on the insertions of the lateral and medial recti, useful later as a reference axis. The other two were deliberately asymmetric (roughly dorsotemporal and ventral) to disambiguate retinal orientation. Using gentle suction, the retina was flat-mounted on a custom-machined hydrophilic polytetrafluoroethylene membrane (cell culture inserts, Millicell) and secured in a chamber on the microscope stage. Retinas were continuously superfused with oxygenated Ames’ medium (32–34°C).
Whole-cell patch-clamp current-clamp and voltage-clamp recordings of isolated flat-mount retinas were performed as described (Sabbah, Berg, Papendorp, Briggman, & Berson, 2017a; Sabbah, et al., 2017b), using a Multiclamp 700B amplifier, Digidata 1550 digitizer, and pClamp 10.5 data acquisition software (Molecular Devices; 10 kHz sampling). Pipettes were pulled from thick-walled borosilicate tubing (P-97; Sutter Instruments); tip resistances were 5–6 MΩ when filled with internal solution, which, for current- clamp recordings, contained (in mM): 120 K-gluconate, 5 NaCl, 4 KCl, 2 EGTA, 10 HEPES, 4 ATP-Mg, 7 phosphocreatine-Tris, and 0.3 GTP-Tris, pH 7.3, 270–280 mOsm). Red fluorescent dye (Alexa Fluor 568; Invitrogen) was added to the pipette for visual guidance under two-photon imaging and dye-filling for later morphological characterization.
Whole-cell recordings were conducted on a multiphoton Olympus FV1200MPE BASIC (BX-61WI) microscope equipped with a 25×, 1.05 NA water-immersion objective (XLPL25XWMP, Olympus) and an ultrafast pulsed laser (Mai Tai DeepSee HP, Spectra-Physics) tuned to 910 nm. Epifluorescence emission was separated into “green” and “red” channels with a 570 nm dichroic mirror and a 525/50 bandpass filter (FF03–525/50–32, Semrock, green channel) and 575–630 nm bandpass filter (BA575–630, Olympus, red channel), respectively. The microscope system was controlled by FluoView software (FV10-ASW v.4.1).
2.7. Visual stimulation
Patterned visual stimuli, synthesized by custom software using Psychophysics Toolbox under Matlab (The MathWorks), were projected (AX325AA, HP) and focused onto photoreceptor outer segments through the microscope’s condenser (Borghuis, Marvin, Looger, & Demb, 2013; Sabbah, et al., 2017b). The projected display covered 1.5 × 1.5 mm; each pixel was 5χ5 /x m. The video projector was modified to use a single ultraviolet LED lamp (NC4U134A, Nichia). The LED’s peak wavelength (385 nm) shifted to 395 nm after transmission through a 440 nm short-pass dichroic filter (FF01–440/SP, Semrock), a dichroic mirror (T425lpxr, Chroma), and various reflective neutral density filters (Edmund Optics).
Photoisomerization rates were derived from the stimulus spectrum (measured using an absolute-irradiance-calibrated spectrometer [USB4000-UV-VIS-ES, Ocean Optics]), estimated rod (0.85 μ m2) and cone (1 μ m2) collecting areas (Naarendorp et al., 2010); and spectral absorbances of mouse rod and cone pigments (Govardovskii, Calvert, & Arshavsky, 2000). Rates were very similar among rods and cones [~104 photoisomerizations/s (R*/photoreceptor/s)], independent of their relative expression of S- and M-cone pigments (Szel et al., 1992).
To probe ON and OFF responses, we used a bright baron a dark background (barwidth=1500 μm, interstimulus duration=5 sec) drifting perpendicular to the bars long axis in 8 randomized directions (45° interval, speed=300 pm/sec, 4 repetitions). To assess directional tuning, we used a sinusoidal grating spanning two spatial periods (spatial frequency=0.132 cycle/degree, Michelson contrast=0.95, stimulus duration=3.65 sec, inter-stimulus duration=5 sec at uniform mean grating luminance) drifted in 8 randomized directions (45° interval, drift speed=4.5 degree/sec, 4 repetitions). Grating and bar parameters were optimized for ON-DSGCs (Dhande et al., 2013). For all stimuli, frames of the stimulus movie were brief but frequent, appearing for 50 μs during the short 185 μs interval between successive sweeps of the imaging laser. Response persistence allowed us to observe the evoked light responses even though no stimulus was present during the interval of laser scanning and associated imaging (300 μs/sweep). The very rapid stimulus flicker (>2000 Hz) was well above critical fusion frequency in mice (Dai et al., 2015).
2.8. Electrophysiology data analysis
Prior to analysis, voltage traces were down-sampled to 0.1 KHz, and peristimulus-time histograms (PSTHs, bin width = 0.155 sec) were calculated. The preferred direction of a cell was estimated as the angle of the vector sum following (Kim et al., 2008):
| (1) |
where r is the response amplitude to stimuli moving at direction φ (0, 45,...,315). The direction selectivity index (DSI) ranges between 0 (no direction selectivity) and 1 (maximal direction selectivity). It was calculated as:
| (2) |
The response amplitude represented the mean firing rate over the stimulus duration and the subsequent 2-sec period, account for the ON and OFF responses. All data were analyzed using custom Matlab scripts.
3. Results
3.1. Pcdh9-Cre labels a subtype of RGC that projects to the MTN
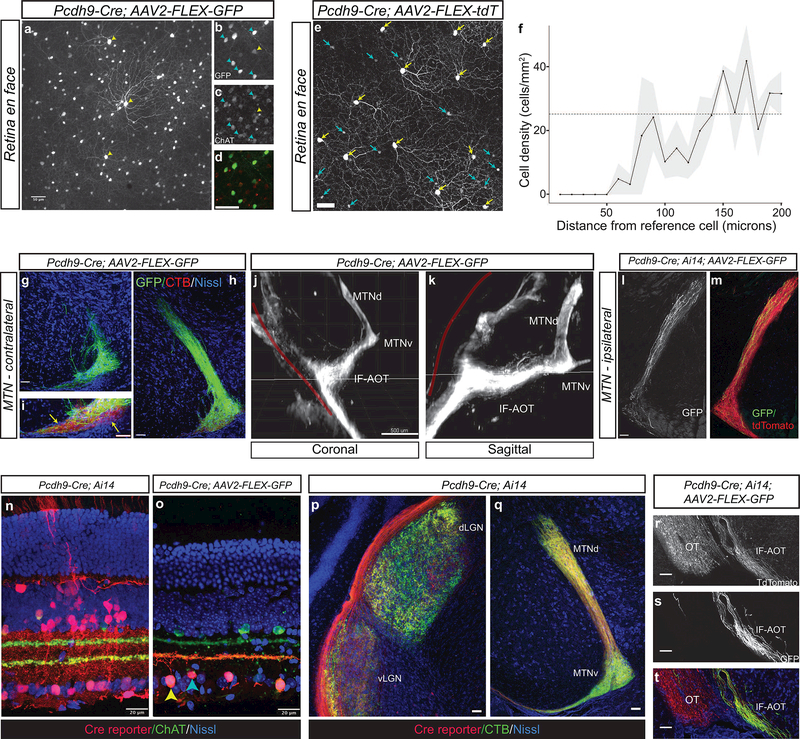
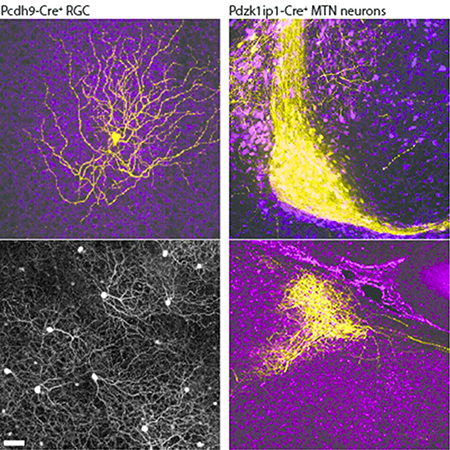
To identify tools that permit genetic access to the neurons within the AOS, we examined images from the GENSAT and Allen Brain Atlas collections and found two mouse lines (Pcdh9-Cre and Pdzk1ip1-Cre) that exhibited expression in the MTN, the AOS nucleus that receives inputs from RGCs preferring dorsal and ventral motion (Simpson, Leonard, & Soodak, 1988). We initially confirmed previous results with the Pcdh9-Cre line (Martersteck et al., 2017) by performing intravitreal infection of adult Pcdh9-Cre animals with AAV2 expressing Cre-dependent fluorophores. When injected into the eyes of Pcdh9-Cre animals, no difference was observed in the patterns of labeling between the AAV2-FLEX-GFP or AAV2-FLEX- tdTomato viruses. We observed expression of Cre in two types of cells in the ganglion cell layer: large diameter RGCs with elaborate dendritic arbors and smaller SACs that expressed choline acetyltransferase (ChAT) (Figure 1a-d). Infected Pcdh9-Cre+ cells that express RBPMS, a pan-RGC marker (Rodriguez, de Sevilla Muller, & Brecha, 2014), had an average density of 23.3± 0.533 cells/mm (mean ± SEM, n=180 cells from 3 independently injected animals). Analysis of the spatial distribution of Pcdh9-Cre+ RGCs revealed non-random spacing in a mosaic pattern, with a clear exclusion zone around each cell (Figure 1e, f). This density and spacing pattern suggest that Pcdh9-Cre+ RGCs represent an RGC subtype.
Figure 1: The Pcdh9-Cre line labels starburst amacrine cells and retinal ganglion cells that project to the MTN.
(a-d) En face view of retina following intravitreal injection of AAV2-CAG-FLEX-GFP into adult Pcdh9-cre animals shows labeling of large diameter RGCs (yellow arrows) and ChAT+ SACs (cyan arrows, c,d). (e) Intravitreal injection of AAV2-FLEX-tdTomato reveals Pcdh9-Cre+ RGCs. Yellow arrows denote RGCs identified by RBPMS immunopositivity. RGC somas form a regular mosaic. Cyan arrows denote ON starburst amacrine cells. (f) Density recovery profile plot of Pcdh9-Cre+ RGCs showing a prominent exclusion zone (averages from 4–5 fields per retina from three different mice; line denotes mean and grey ±SEM). The dashed line shows the expected profile if the cells were randomly distributed at the same density. (g, h) Projections of Pcdh9-Cre+ RGCs to the contralateral medial terminal nucleus (MTN). Extensive GFP+ axon arborization (green) is seen in the MTN. g and h are 100 microns apart. (i) Pcdh9- Cre+ RGC arbors do not fully innervate the more ventrolateral aspect of the MTN (arrows). Pcdh9-Cre+ terminals (green) occupy a fraction of the total area of RGC terminals labeled with CTB (red) in this area. (j, k) Axon tracts from Pcdh9-Cre+ RGCs labeled with AAV2-FLEX-GFP were visualized by brain clearing followed by LSFM. Axons target the if-AOT and avoid the main OT (red line) on their way to the MTN. (l, m) Pcdh9-Cre+ RGC innervation of the ipsilateral MTN after unilateral intravitreal injection of AAV2-FLEX- GFP (GFP, l) into Pcdh9-Cre; Ai14 which labels large numbers of RGCs with tdTomato (red, m). (n, o) Retinal cross sections of adult Pcdh9-Cre; Ai14 animals (n) and Pcdh9-Cre animals intravitreally injected with AAV2-FLEX-GFP (o) reveals that Pcdh9-Cre expresses in a wide range of retinal cells types early in development, but is restricted to ON SACs (cyan arrow) and RGCs (yellow arrow) at early postnatal times. (p, q) RGCs from adult Pcdh9-Cre; Ai14 animals target many retinorecipient areas later including the dLGN and vLGN (p) and the MTN (q). (r-t) Intravitreal injection of AAV2-FLEX-GFP into Pcdh9-Cre; Ai14 animals showing that Pcdh9-Cre expression becomes restricted to the MTN projecting RGCs that traverse the if-AOT (r, s), whereas other RGCs project throughout both the main optic tract as well as the if-AOT (r, t). Scale bars: a-e, l-m, 50 microns; g-i, 100 microns; j, k, 500 microns; n, o, 20 microns; p-t, 25 microns.
We then examined the axonal projections of Pcdh9-Cre+ RGCs labeled with AAV2-FLEX-GFP and found that the vast majority targeted the contralateral MTN (Figure 1 g-i), similar to previous observations (Martersteck et al., 2017). A very small number of axons (<1%) targeted the ventral and dorsal lateral geniculate nuclei (vLGN and dLGN), or superior colliculus (SC), thus demonstrating the nearly exclusive targeting of the MTN by Pcdh9-Cre+ RGCs. Within the MTN, Pcdh9-Cre+ RGC axon arbors filled the MTN, with the more ventrolateral aspect being less densely innervated relative to RGC axons labeled by intravitreal injection of Alexa-555 conjugated CTB (Figure 1i). Light sheet fluorescence microscopy imaging of cleared brains from adult Pcdh9-Cre animals confirmed that Pcdh9-Cre+ RGCs labeled with AAV2-FLEX-GFP largely project to the MTN via the inferior fasciculus of the AOT (if-AOT), the route taken by SPIG1-GFP+ RGCs and ON-DS RGCs with a preference for ventral motion on the retina (Dhande et al., 2013; Sabbah et al.,, 2017b; Yonehara et al., 2008; 2009) (Figure 1j, k). Monocular viral injections also revealed a small number of labeled axons within the ipsilateral MTN (Figure 1l, m) consistent with previous results showing that a minority of MTN-targeting RGCs project ipsilaterally (Dhande et al., 2013). Thus, Cre-mediated expression using an AAV reporter in adult animals confirms that Pcdh9-Cre labels a subtype of RGC with a unique projection pattern in the brain.
When the Pcdh9-Cre line was crossed to the Ai14 Cre reporter (Madisen et al., 2010) located in the ROSA26 locus (Pcdh9-Cre; Ai14), the retinas from these animals had large numbers of cells labeled including cones, horizontal cells, Muller glia, amacrine cells and many subtypes of RGCs (Figure 1n). All retinorecipient areas were targeted by the axons of Pcdh9-Cre; Ai14 RGCs (Figures 1 p, q and 2d, e). This pattern of expression suggests that Pcdh9-Cre is expressed early during development in retinal progenitors, but that postnatally expression becomes restricted to a subset of MTN projecting RGCs and ON-SACs. This difference in expression pattern is illustrated dramatically when Pcdh9-Cre; Ai14 adult animals are infected intravitreally with AAV2-FLEX-GFP to label Cre-expressing retinal cells (Figure 10). In these doubly labeled animals, GFP+ axons almost exclusively target the if-AOT and avoid the main OT, whereas tdTomato+ axons are present throughout all branches of the optic tract (Figure 1 r-t). Selective labeling of Pcdh9-Cre+ RGCs therefore requires viral, or other, methods that can be switched on after early postnatal periods when Pcdh9-Cre becomes restricted to MTN projecting RGCs.
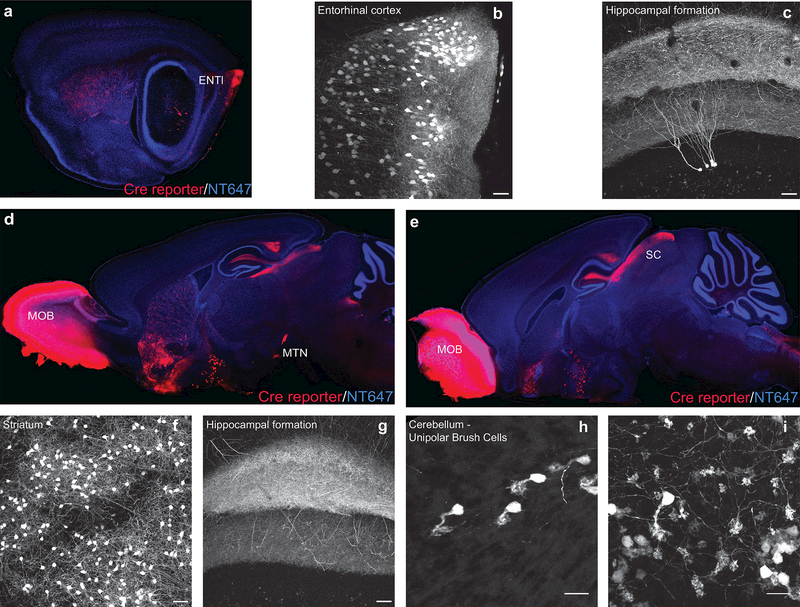
Figure 2: Pcdh9-Cre is expressed in many brain areas.
The brains of adult Pcdh9-Cre; Ai14 animals show strong tdTomato expression in the entorhinal cortex (a), the striatum (a, d), the olfactory bulb (d, e) as well as retinorecipient areas (Superior Colliculus, SC, e). Closeup images in (b, c, f-i) show Pcdh9-Cre+ cells in the entorhinal cortex (b), hippocampal formation (c, g), striatum (f) and cerebellum (unipolar brush cells: h, soma/dendritic brush; i, mossy terminals).Scale bars: a-c, 500 microns, d-i, 100 microns.
Examination of brains from Pcdh9-Cre; Ai14 animals revealed Cre expression in several specific areas of the brain in addition to sporadic expression in isolated cells. Strongly labeled areas included the main olfactory bulb, the caudatoputamen, and the entorhinal cortex (ENTI) including projections from ENTI to the hippocampus (Figure 2a-g). Additional expression was observed in unipolar brush cells, a type of local excitatory interneuron in the cerebellum (van Dorp & De Zeeuw, 2015), and so the Pcdh9-Cre line may prove to be a useful tool for examining the function of this relatively poorly characterized cell type (Figure 2h, i).
3.2. Pcdh9-Cre+ RGCs display features of ON-DSGCs
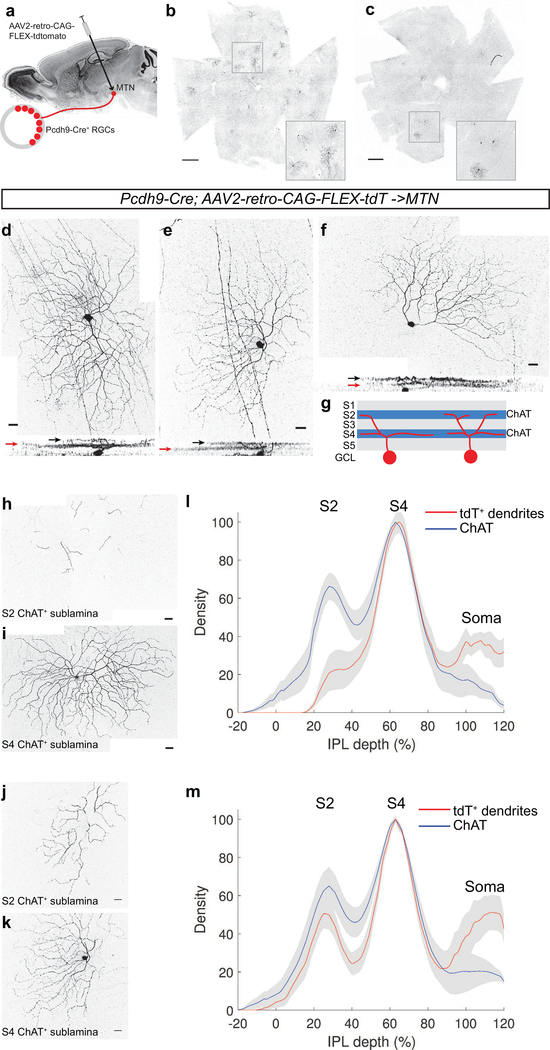
The expression of Pcdh9-Cre in ON-SACs limited our ability to visualize the morphological features of Pcdh9-Cre+ RGCs using traditional AAV2. Therefore, we took advantage of the newly developed AAV2- retro virus that infects cells at nerve terminals and is retrogradely transported to the cell bodies of infected cells (Tervo et al., 2016). We stereotaxically injected AAV2-retro-CAG-FLEX-tdTomato into the MTN of adult Pcdh9-Cre animals and examined tdTomato expression in the retina two weeks later (Figure 3a). Small numbers of RGCs were efficiently labeled in Pcdh9-Cre animals but not in lines that do not express Cre in RGCs (Figure 3b, c and data not shown). Retrogradely labeled Pcdh9-Cre+ RGCs were present throughout the retina and did not exhibit clustering in a specific retinal quadrant. The labeled RGCs had large soma diameters (18.58 ± 0.63 microns, mean ± SEM, n=19 cells from 4 independently injected animals) and dendritic field areas (78,532 ± 3370 μm2, mean ± SEM, n=19 cells), consistent with their putative identity as ON-DS RGCs known to project to the MTN (Yonehara et al., 2009).
Figure 3: Dendritic architecture of Pcdh9-Cre+ RGCs.
(a) Schematic of retrograde infection with AAV2-retro. (b,c) En face view of retinal whole mounts of Pcdh9-Cre animals injected with AAV2-retro into the MTN. Many tdTomato+ RGCs are found throughout the retinas. Insets show higher magnification view of selected regions. (d-f) Individually labeled Pcdh9- Cre+ RGCs retrogradely labeled from the MTN with AAV2-retro-CAG-FLEX-tdTomato. Horizontal rotations of the confocal image stacks are shown below. 9 of 17 cells exhibit a primarily S4 dendritic lamination (example in d), and 8/17 are more bistratified (examples in e, f). Black and red arrows denote lamination with the ChAT+ plexuses in the S2 and S4 sublaminae, respectively. (g) Schematic of sublaminae in the inner plexiform layer showing locations of ChAT plexuses and Pcdh9-Cre+ RGC dendrites (h-k) Dendrites of retrogradely labeled Pcdh9-Cre+ RGCs cofasciculating with S2 (h, j) and S4 (I, k) ChAT plexuses for exemplary monostratified (d) and bistratified (e, f) cells. (l, m) Dendritic stratification analysis of monostratifed (l) and bistratified (m) cells dendrites compared with position of ChAT as a function of IPL depth. Red and blue lines denote mean with grey showing ±SEM, n=9 monostratified, n=8 bistratified. Scale bars: b,c, 500 microns, d-k, 25 microns.
All isolated Pcdh9-Cre+ RGCs labeled by AAV2-retro-CAG-FLEX-tdTomato had the majority of their dendritic arbors in the S4 sublamina, where they cofasciculated with the dendrites of ON SACs (Figure 3d-g, l, m). In 9 of 17 cells we observed occasional dendritic sprouts that extended up to the S2 sublamina, overlapping with the OFF SAC plexus (Figure 3d,h, I, l). This type of dendritic stratification is similar to what has been reported for ON-DS RGCs that project to the MTN (Dhande et al., 2013; Sun, Deng, Levick, & He, 2006). The other 8 labeled cells that we examined had more of their dendritic arbors in the OFF ChAT plexus in the S2 sublamina of the IPL (Figure 3e, f, j, k, m). This feature of Pcdh9-Cre+ RGCs is in contrast to SPIG1-GFP+ RGCs that have dendrites confined to the S4 sublamina (Yonehara et al., 2008). Bi-stratified ON-DS cells have been reported in the rat (Dann & Buhl, 1987) and in the mouse (Dhande et al., 2013), where there are ranges of bistratification levels. Pcdh9-Cre+ RGCs may account for much of the diversity in dendritic stratification reported for the ON-DS cells labeled in the Hoxd10-GFP line (Dhande et al., 2013). In addition, 8 of 17 retrogradely labeled Pcdh9-Cre+ RGCs had asymmetric dendritic arbors (Figure 3f) reminiscent of the HB9-GFP+ RGCs (Trenholm, Johnson, Li, Smith, & Awatramani, 2011). These cells did not display a consistent orientation of their dendrites in relation to a particular region of the retina (Kim et al., 2008). It will be interesting to determine whether this asymmetry aligns with the preferred direction selectivity of RGCs that prefer ventral motion (V cells) depending on their overall position within the retina (Sabbah et al., 2017b), or whether this asymmetry is independent of directional preference, as in the case of the asymmetric W9 ON-OFF DSGCs (Kay et al., 2011). Thus, Pcdh9-Cre+ RGCs have features of previously characterized ON-DS RGCs that respond to vertical motion, but a subset of these cells exhibits more extensive dendritic stratification in the S2 sublamina that may alter their responses to moving stimuli.
3.3. Pcdh9-Cre+ RGCs are ON type DS cells that prefer ventral motion
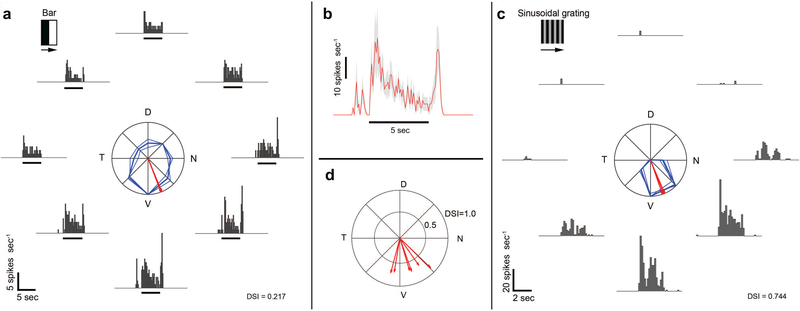
We used electrophysiology to explore the response polarity and direction selectivity of Pcdh9-Cre+ RGCs. In the central retina, identified Pcdh9-Cre+ RGCs labeled with AAV2-FLEX-GFP were shown a bright bar on a dark background drifting perpendicular to the bar’s long axis in 8 randomized directions. Figure 4a summarizes the responses of one of these cells. The firing rate varied across drifting directions, but sustained firing was observed in response to the leading edge of the bright bar (ON response) in all 8 directions. In some directions, typically around the cell’s preferred direction, a transient firing burst in response to the trailing edge of the bright bar was observed (OFF response). This observed OFF response likely represents input from OFF bipolar cells to the S2 sublamina dendrites of the Pcdh9-Cre+ RGC (Figure 3). The firing rate in response to bars drifting in the preferred direction, averaged across all examined cells (n = 6), illustrates the transient pre-ON response, sustained ON response, and transient OFF response of Pcdh9-Cre+ RGCs (Figure 4b). To determine the preferred direction of Pcdh9-Cre+ RGCs, we used sinusoidal gratings drifting in 8 randomized directions. Figure 4c summarizes the responses of one of the examined cells. Firing rate varied dramatically across drifting directions, with the responses averaged over the stimulus duration peaking at a single general direction [direction selectivity index (DSI) = 0.744]. The preferred direction of all examined cells (n = 6) pointed ventrally on the retina, and thus preferred stimuli drift dorsally in visual space (Figure 4d). Directional tuning revealed using drifting gratings was significantly higher than that determined using drifting bars (paired t-test, t = 3.83, d.f. = 5, p = 0.012; DSIgrating = 0.745 ± 0.074, DSIbar = 0.309 ± 0.231; average ± standard deviation). In summary, ganglion cells marked in central retina of the Pcdh9-Cre line are direction selective (DS) cells of the ON type, preferring ventral retinal motion.
Figure 4: Pcdh9-Cre+ RGCs are ON-DS cells that prefer ventral motion.
(a) Directional tuning of Pcdh9-Cre+ RGCs, targeted based on their labeling after ocular injections of AAV2-FLEX-GFP. Firing rate of a representative RGC in response to bright bars drifting in eight directions at 45° intervals, shown as peristimulus time histograms (PSTHs). The black marker indicates the estimated time when the stimulus bar was within the cell’s receptive field. The polar plot shows response amplitude (normalized to maximum) for each direction (bold curves = mean of four trials; thin = single trial). Red vectors show preferred direction (bold, mean; thin, single trials; N, nasal; D, dorsal; T, temporal; V, ventral). The direction-selectivity index (DSI) is indicated on the bottom-right. (b) Average ± SEM firing rate in response to a bright bar drifting in the preferred direction (n = 6 cells). Pcdh9-Cre+ cells exhibit (1) a transient ON response preceding the entry of the bright bar into the cell’s receptive field, (2) a sustained ON response evoked by the bright bar, and (3) a transient OFF response evoked by the trailing edge of the bright bar. (c) Firing rate of a representative Pcdh9-Cre+ RGC in response to sinusoidal gratings drifting in eight directions at 45□ intervals. Conventions are similar to panel (a). (d) Preferred directions among cells examined (n = 6). Direction and length of vectors indicate the cell’s preferred direction and DSI, respectively. Preferred directions were determined from responses to drifting gratings.
3.4. Pcdh9-Cre+ RGCs survive after optic nerve injury
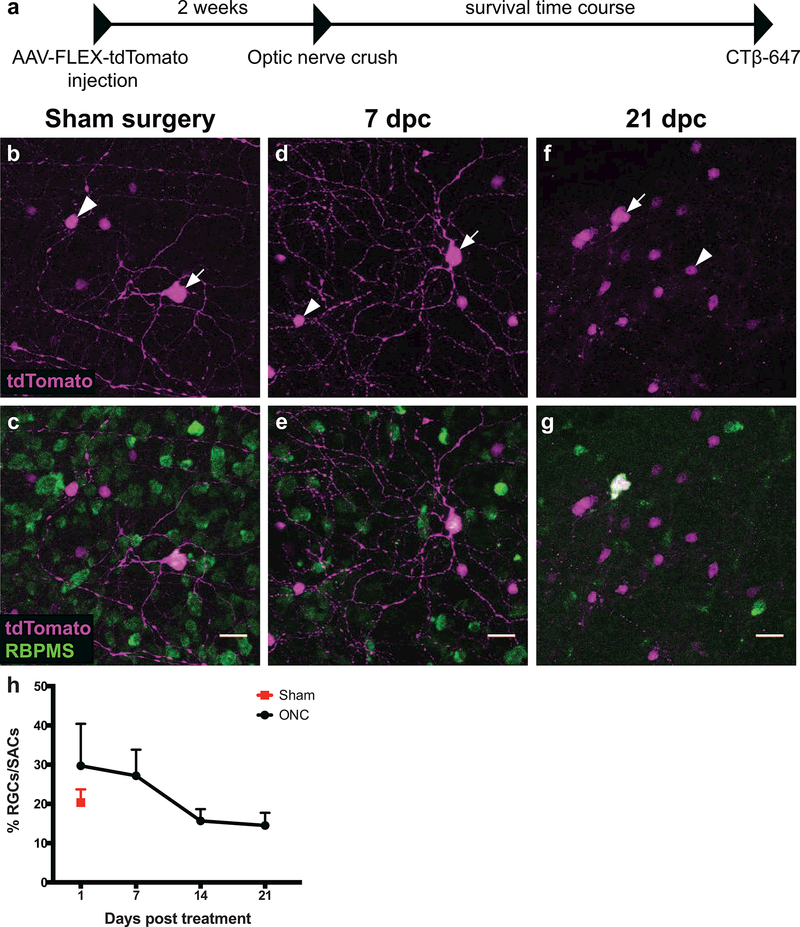
Another feature that varies among RGC subtypes is the ability to survive and regenerate axons after injury to the optic nerve. The ON-alpha and M1 intrinsically photosensitive RGC subtypes are fairly resistant to injury-induced apoptosis following optic nerve crush (ONC), whereas nearly all other subtypes previously described are not (Duan et al., 2015). We examined whether Pcdh9-Cre+ RGCs displayed an intrinsic ability to survive following injury by first labeling Pcdh9-Cre+ RGCs with unilateral intravitreal injection of AAV2-FLEX-tdTomato, and then crushing the optic nerves and assessing RGC survival 1,7, 14, and 21 days post-crush (dpc) (Figure 5a). We labeled all RGCs using the anti-RBPMS antibody (Rodriguez et al., 2014), which allowed us to distinguish between Pcdh9-Cre+ RGCs and other retinal cells labeled in the Pcdh9-Cre line. To control for variation in infection efficiency, we compared the ratio of RBPMS+/tdTomato+ RGCs to RBPMS−/tdTomato+ SACs before and after ONC since SAC cell survival is not affected by ONC (Nadal-Nicolas, Sobrado-Calvo, Jimenez-Lopez, Vidal-Sanz, & Agudo-Barriuso, 2015). At 1 and 7 dpc, Pcdh9-Cre+ RGCs were present in normal ratios compared to Pcdh9-Cre+ SACs and had prominent dendritic arbors (Figure 5b-e, h). By 14 and 21 dpc, the ratio of Pcdh9-Cre+ RGCs to SACs remained nearly constant (20.3% infected RGCs/SACs in sham compared to 15.7% at 14 dpc and 14.5% at 21 dpc; one-way ANOVA, p=0.2081; Figure 5h). The effectiveness of the crush was confirmed by the dramatic decrease in the number of RBPMS+ RGCs (Sham RGC density: 1365.9 ± 38.9 RGCs mm−2; 21 dpc RGC density: 357.5 ± 109.5 RGCs mm−2; n=2 animals mean±SEM; see also Figure 5c,e, g). Interestingly, the Pcdh9-Cre+ RGCs that remained at 21 dpc had no discernible dendritic arbors (Figure 5f-g), indicating that surviving RGCs may have initiated apoptosis. Our results demonstrate that some Pcdh9-Cre+ RGCs can survive up to 21 dpc, whereas nearly all other types of DS RGCs are lost by 14 dpc (Duan et al., 2015). Future studies will focus on identifying strategies to promote long-term survival and axon regeneration of the remaining Pcdh9-Cre+ RGCs.
Figure 5: Pcdh9-Cre+ RGCs can survive up to 21 days after optic nerve injury.
(a) Timeline of optic nerve crush experiments. (b-f) Representative images of Pcdh9-Cre+ retinas infected with AAV2-CAG-FLEX-tdTomato (magenta) and labeled with anti-RBPMS antibody (green). White arrows mark RBPMS-positive RGCs and white arrowheads mark RBPMS-negative starburst amacrine cells (SACs). (b-c) In the sham surgery condition, the percentage of infected RGCs to SACs is 20.3%, and the RGCs have dendritic arbors (n=2 retinas). (d-e) This percentage at 7 dpc is similar to the sham condition and dendritic arbors are clearly present (n=2 retinas). (f-g) By 21 dpc, the percentage of infected RGCs to SACs is 14.5% and dendrites are absent (n=3 retinas). (h) Percentage of infected RGCs to SACs at different time points following optic nerve crush (Black circles) or from Sham treated animals (red square). The number of Pcdh9-Cre+ RGCs relative to Pcdh9-Cre+ SACs decreases around 14 dpc but shows no further reduction by 21 dpc; these differences are not statistically significant (One-way ANOVA, p = 0.2081). Error bars denote mean and SEM. Numbers of retinas and RGCs examined for each time point were: Sham: N=2 retinas, 104 RGCs; 1 dpc: N=3 retinas, 136 RGCs; 7 dpc: 2 retinas, 69 RGCs; 14 dpc: 4 retinas, 129 RGCs; 21 dpc: 3 retinas, 128 RGCs. Scale bars: b-g, 20 microns.
3.5. The Pdzk1ip1-Cre line labels MTN neurons that project to the NOT
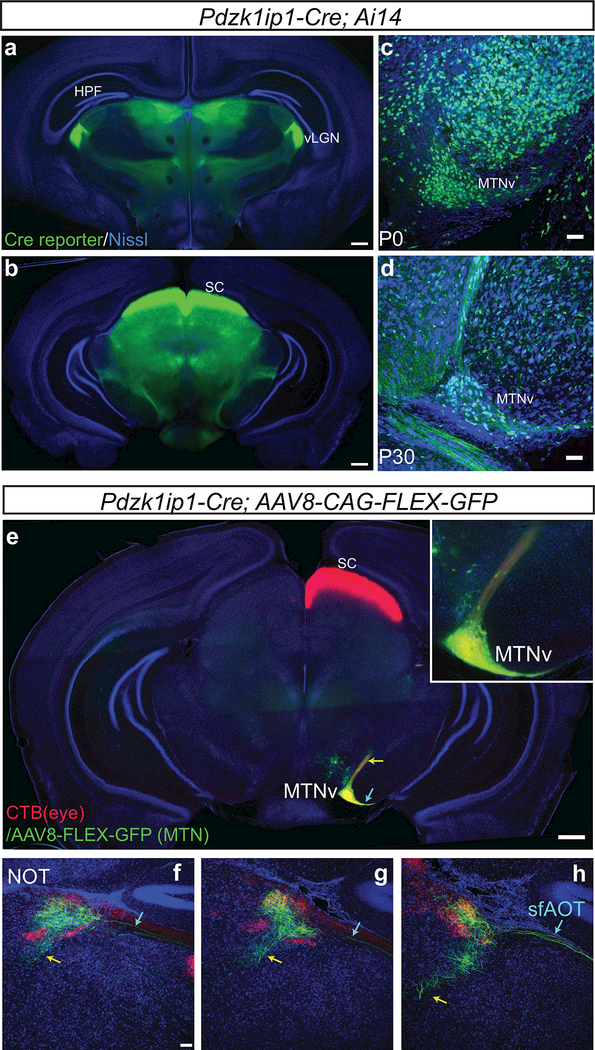
After characterizing the Pcdh9-Cre line that selectively labels a subtype of MTN projecting ON-DS RGCs, we sought to identify mouse lines that would allow us to assess the post-synaptic targets of these RGCs in the MTN. Screening through Cre-expressing lines in the GENSAT database revealed that the Pdzk1ip1-Cre line drives strong expression in the midbrain, including focal expression in the ventral MTN. Pdzk1ip1-Cre; Ai14 animals showed strong expression in the midbrain including the SC, vLGN and also in the MTN as early as P0 (Figure 6a-d). Both cell bodies and axons of midbrain neurons were labeled, however, within the retina no neuronal cells were labeled (data not shown).
Figure 6: Pdzk1ip1-Cre is expressed in MTN resident neurons that project to NOT.
(a-b) Coronal sections of midbrain from Pdzk1ip1-Cre; Ai14 animals at the level of the vLGN (a) and the superior colliculus (b), show strong expression of Pdzk1ip1-Cre in the midbrain. (c-d) Closeup view of the MTN at P0 (c) and P30 (d) shows Cre expressing cells in the MTNv. (e) The MTN of Pdzk1ip1-Cre animals was infected with AAV8-CAG-FLEX-GFP, and the contralateral eye was subsequently injected with CTB-555. Coronal section of the midbrain at the level of the MTN shows limited infection of neuronal somata in the ventral MTN with prominent GFP labeling. Neuronal processes extend both laterally (cyan arrow) and dorsally (yellow arrow) from the labeled MTNv neurons. Inset shows magnified area of MTNv with labeled somata. (f-h) GFP+ axons from Pdzk1ip1-Cre+ MTN neurons reach the NOT (labeled with CTB-555 from the right eye) via a direct, internal tract (yellow arrow) and a tract that overlaps with the sf- AOT (cyan arrow) shown in sections taken at 100 micron intervals from rostral (f) to caudal (h). Scale bars: a, b, e, 500 microns; c, d, f-h, 50 microns.
To examine the projections of Pdzk1ip1-Cre expressing MTN neurons, we stereotaxically injected AAV8- CAG-FLEX-GFP into the MTN. Two weeks after infection, we observed strong labeling of MTN neurons without significant expression in surrounding brain areas or in RGCs (Figure 6e and data not shown). The labeled MTN neurons largely resided in the more ventral region (Figure 6e, inset). The axons of these cells take two distinct routes to target cells both in the MTNd as well as the NOT, a projection that has been described previously (Giolli, Blanks, & Torigoe, 1984). Some axons travel internally through the midbrain tegmentum, coursing dorsally and rostrally from the MTN to reach the NOT (Figure 6f-h). Other processes course laterally out of the MTNv and follow the sf-AOT to the NOT. Both the internal and the sf-AOT output pathways are exclusively ipsilateral. Their terminals appear restricted to a retinal terminal zone corresponding to the NOT. Thus, Pdzk1ip1-Cre+ MTN neurons project to the NOT via two distinct routes.
3.6. Sema6A is required for the assembly of the MTN-NOT circuit
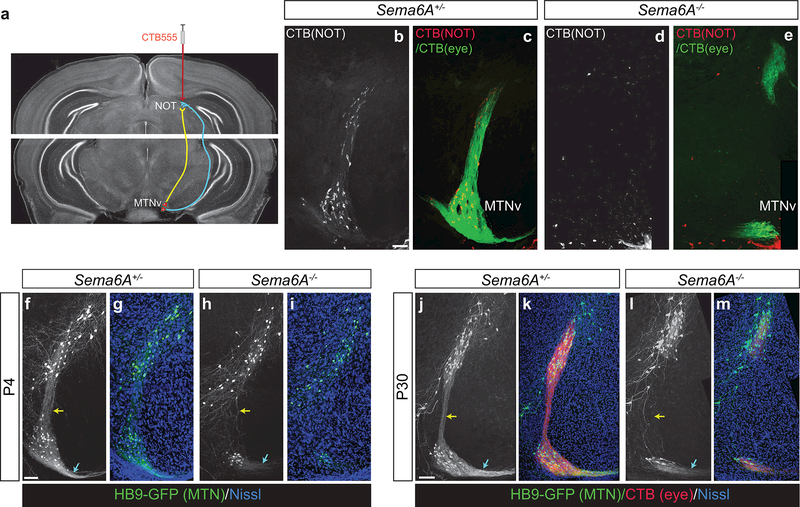
We next examined whether there are molecular factors that might control assembly of the midbrain MTN- NOT circuitry revealed by the Cre lines that we have characterized. Previous studies demonstrated a role for the transmembrane protein Sema6A in establishing direction selective circuits in the retina and in the targeting of the MTN by SPIG1-GFP+ RGCs (Sun et al., 2015a; Sun et al., 2013). In addition, Sema6A is expressed in both the MTN and also in the NOT (Sun et al., 2015a), suggesting a role in assembly of midbrain circuits. To ask whether Sema6A is required for formation of the MTN-NOT connection described above, we retrogradely labeled MTN-NOT projection neurons by injecting CTB into the NOT of control and Sema6A−/− animals (Giolli, Blanks, Torigoe, & Williams, 1985) (Figure 7a). In control animals, many cells throughout the MTN were retrogradely labeled (Figure 7b, c). In contrast, we were only able to detect retrogradely labeled MTN neurons in very rare instances in Sema6A−/− mutants (Figure 7d, e). Therefore, the loss of Sema6A function disrupts connections between the MTN and NOT.
Figure 7: Formation of AOS midbrain circuitry requires Sema6A.
(a) Schematic of retrograde labeling of MTN neurons by injection of CTB into the NOT. (b-e) Retrograde labeling of MTN-NOT projections neurons in control (b, c) and Sema6A−/− (d, e) animals with CTB-555 reveals a loss of MTN-NOT connection in the Sema6A mutants (n=3 animals per genotype). CTB-488 injected into the contralateral eye defines the extent of RGC MTN innervation (green, c, e). (f-m) Examination of MTN neurons labeled in the HB9-GFP line reveals that at P4 (f-i) and P30 (j-m), MTN neurons are reduced and GFP+ processes extending between the MTNv and MTNd (yellow arrows) and from the MTNv into the sf-AOT (cyan arrows) are absent in Sema6A mutants. Residual CTB-555 from the eye co-localizes only with the remaining HB9-GFP+ MTN neurons (n=3 animals per genotype). Scale bars: f-q, 50 microns.
To understand the basis for this loss of connectivity between the MTN and NOT, we took advantage of the observation that the HB9-GFP mouse line, known for its labeling of spinal motor neurons and also a subpopulation of ON-OFF DS RGCs (Trenholm et al., 2011; Wichterle et al., 2002), also labels MTN neurons. We observed GFP expression in the cells of the MTNv and MTNd in HB9-GFP animals as early as P4, and this expression persisted into adulthood (Figure 7f, j). Prominent GFP+ fibers were observed emanating from the MTNv dorsally into the MTNd and laterally into the sf-AOT, at P4 and P30 (Figure 7 f, j). Enucleation of HB9-GFP animals did not affect the morphology of these fibers, demonstrating their non-retinal origin (data not shown). We crossed the Sema6A mutant allele to the HB9-GFP line to generate Sema6A+/−; HB9-GFP controls and Sema6A−/−; HB9-GFP mutants. When examined at P4 and at P30, Sema6A−/− animals had fewer GFP+ cells in the MTNv, whereas the MTNd showed a less severe phenotype (Figure 7f-m). In addition, the GFP+ processes that emanated from the MTNv were nearly absent in Sema6A mutants (Figure 7h, l), suggesting that the MTNv neurons that project to the NOT via the dual tracts have deficits in their ability to form projections to their targets in the NOT. Thus, retrograde and genetic labeling experiments suggest that Sema6A is required for the MTNv to NOT axonal projection.
MTN-innervating RGCs that lack Sema6A fail to arborize properly and subsequently undergo apoptosis (Sun et al., 2015a). There are, however, a small number of RGCs that remain and innervate the most ventral and dorsal aspects of the MTN of Sema6A−/− animals (Figure 7e). We examined whether the remaining MTN neurons in Sema6A−/− animals have any interaction with these persistent RGC terminals. When CTB was injected into the eyes of Sema6A+/−; HB9-GFP animals we observed co-localization of presynaptic CTB+ terminals with postsynaptic GFP+ MTNv and MTNd neurons (Figure 7k). In the Sema6A−/−; HB9-GFP animals, however, the remaining CTB+ axon terminals in the MTN were only present in immediate proximity to the remaining HB9-GFP+ MTN neurons and not in any other part of the MTN (Figure 7m). This observation suggests that RGC innervation may promote survival of MTN-resident neurons, and without this RGC innervation, MTN resident neurons undergo apoptosis
4. Discussion
In this study we have used genetic, viral, physiological, and nerve injury paradigms to characterize the ON-DS RGCs that project to the MTN and preferentially respond to ventral motion. We identified two new mouse lines that will facilitate future studies of the AOS, one that selectively labels a subset of ON-DS RGCs that project to the MTN, and a second that labels MTN-resident neurons that project to the NOT. Unlike other DS-RGCs so far examined, this subtype of ON-DS RGC survives after nerve crush. We demonstrated that Sema6A, in addition to its previously established role in directing RGCs to their MTN target cells, is required to form projections between MTN and NOT, demonstrating that this molecule plays multiple roles in wiring visual system circuits.
4.1. New tools to manipulate development and function of AOS circuits
Pcdh9-Cre+ RGCs exhibit highly similar morphologies, electrophysiological properties, and projection patterns compared to the RGCs labeled in the ventral retina of the previously characterized SPIG1-GFP line (Yonehara et al., 2008; 2009). Gene expression profiling experiments of the RGC populations in these lines demonstrate that there is a high degree of similarity at the transcriptomic level as well (T.A-K., A. Kolodkin, unpublished observations), strongly suggesting that these two lines label an overlapping population of RGCs. Pcdh9-Cre+ RGCs, however, display more diversity in their dendritic stratification patterns. It is unclear whether this diversity reflects simple morphological heterogeneity or whether there might be subclasses of Pcdh9-Cre+ RGCs that have distinct molecular or physiological features. The Pcdh9-Cre mouse line permits genetic access to this subtype of ON-DS RGC, specifically allowing dissection and manipulation of the circuits underlying retinal direction selectivity. Since Pcdh9-Cre+ RGCs project to the MTN, the Pcdh9-Cre line also enables the manipulation of central projections and examination of their roles in image stabilization (Sabbah et al., 2017b). We also characterized the Pdzk1ip1-Cre line that labels MTN-resident neurons that project to the NOT and serve an as-yet- undetermined function within AOS circuitry. These neurons may represent a type of inhibitory projection neuron that receives inputs from ON-DS RGCs and, in the rabbit, displays direction selectivity (Giolli, Blanks, & Lui, 2006). Future studies will involve targeted delivery of genetic tools that allow for control of neuronal subtype activity. We will thus be able to determine the precise roles of these cell types in the control of reflexive eye movements in response to retinal image slip.
4.2. Pcdh9-Cre+ RGCs represent a new RGC subtype that survives after axotomy
Damage to the optic nerve resulting in RGC death occurs in a number of traumatic and degenerative disease states and can lead to permanent blindness (Chang & Goldberg, 2012). Therefore, identifying strategies to keep RGCs alive in the retina after injury is a critical step in developing regeneration- promoting therapies (Welsbie et al., 2013). Previous work demonstrated that different RGC subtypes have different abilities to survive and re-extend axons after injury: α RGCs and M1-ipRGCs survive after optic nerve crush, ON-OFF DS RGCs largely die after nerve crush (Duan et al., 2015). Most of the regeneration-promoting strategies identified to date promote axonal growth of α RGCs. We unexpectedly found that Pcdh9-Cre+ RGCs preferentially survive after optic nerve crush. Future studies will focus on identifying molecular survival strategies used by Pcdh9-Cre+ RGCs as well as potential mechanisms to promote axonal regeneration back to the MTN. Since the function of AOS circuits can be evaluated quantitatively by assessing the optokinetic reflex using video occulography (Faulstich, Onori, & Lac, 2004), examination of functional regeneration of AOS RGCs will be possible.
4.3. The role of Sema6A in wiring visual circuits
Previous work showed that reverse signaling between the transmembrane protein Sema6A, which is expressed on RGCs and acts as a receptor, and PlexA2/A4 transmembrane proteins, expressed in the MTN and acting as ligands, is required to direct RGCs to their correct MTN target cells during development (Sun et al., 2015b). In this present study, we found that Sema6A is additionally required for the MTN to NOT axonal projection. One possible explanation is that innervation by Sema6A-expressing RGCs is required to maintain MTN target neurons. In support of this model,HB9-GFP+ MTN neurons are only found in immediate proximity to the remaining points of RGC innervation in Sema6A mutants. Because HB9-GFP only labels a fraction of MTNv neurons, and because no other markers are known to label the MTN, we cannot say at this point to what extent MTN neuron survival is affected by the loss of Sema6A. An alternative explanation for our observations is that Sema6A, independent of its role in RGCs, plays other roles in the MTN and NOT that regulate either guidance of MTNv neuron axons to the NOT or the maintenance of such projections during development. Is Sema6A is required to maintain these AOS connections, or is it only required developmental to build AOS circuitry? Analysis of conditional Sema6A mutants will enable us to address these questions. Future studies will determine whether other molecular pathways that drive RGC targeting to brain nuclei are re-used for additional aspects of visual system connectivity, and at understanding how loss of specific visual system connections affects visual processing.
Acknowledgements
We thank the viral vector core facilities of the University of North Carolina and HHMI Janelia Research Campus for providing reagents, the Mutant Mouse Resource and Research Center for providing mouse lines, the Johns Hopkins NINDS Center for Neuroscience Research (5P30-NS050274) Murine Mutagenesis Core and Multiphoton Imaging Core facilities, and the Johns Hopkins Integrated Imaging Center for technical assistance, and members of the Kolodkin laboratory for helpful suggestions. This work was supported by NIH R01-EY027713 (A.L.K.). A.L.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors characterize mouse lines labeling retinal ganglion cells and brain neurons in the Accessory Optic System. Pcdh9-Cre+ RGCs prefer ventral motion, target the medial terminal nucleus (MTN) and survive after optic nerve crush. Postsynaptic MTN targets of these RGCs require Sema6A to form connections with other AOS nuclei.
References
- Barlow HB, & Levick WR (1965). The mechanism of directionally selective units in rabbit’s retina. The Journal of Physiology, 178(3), 477–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghuis BG, Marvin JS, Looger LL, & Demb JB (2013). Two-photon imaging of nonlinear glutamate release dynamics at bipolar cell synapses in the mouse retina. Journal of Neuroscience, 33(27), 10972–10985. 10.1523/JNEUROSCI.1241-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EE, & Goldberg JL (2012). Glaucoma 2.0: neuroprotection, neuroregeneration, neuroenhancement. Ophthalmology, 119(5), 979–986. 10.1016/j.ophtha.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J, Wu Z, Choi CHJ, Nguyen L, Tegegne S, Ackerman SE, et al. (2018). Three-Dimensional Adipose Tissue Imaging Reveals Regional Variation in Beige Fat Biogenesis and PRDM16-Dependent Sympathetic Neurite Density. Cell Metabolism, 27(1), 226–236.e3. 10.1016/j.cmet.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Dai X, Zhang H, He Y, Qi Y, Chang B, & Pang J-J (2015). The frequency-response electroretinogram distinguishes cone and abnormal rod function in rd12 mice. PLoS ONE, 10(2), e0117570 10.1371/journal.pone.0117570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann JF, & Buhl EH (1987). Retinal ganglion cells projecting to the accessory optic system in the rat.The Journal of Comparative Neurology, 262(1), 141–158. 10.1002/cne.902620111 [DOI] [PubMed] [Google Scholar]
- Dhande OS, Estevez ME, Quattrochi LE, El-Danaf RN, Nguyen PL, Berson DM, & Huberman AD (2013). Genetic dissection of retinal inputs to brainstem nuclei controlling image stabilization. Journal of Neuroscience, 33(45), 17797–17813. 10.1523/JNEUROSCI.2778-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, la Huerta, De I, & Sanes JR (2014). Type II cadherins guide assembly of a direction-selective retinal circuit. Cell, 158(4), 793–807. 10.1016/j.cell.2014.06.047 [DOI] [PubMed] [Google Scholar]
- Duan X, Qiao M, Bei F, Kim I-J, He Z, & Sanes JR (2015). Subtype-Specific Regeneration of Retinal Ganglion Cells following Axotomy: Effects of Osteopontin and mTOR Signaling. Neuron, 85(6), 1244–1256. 10.1016/j.neuron.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JL, Dumitrescu ON, Wong KY, Alam NM, Chen S-K, LeGates T, et al. (2010). Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron, 67(1), 49–60. 10.1016/j.neuron.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulstich BM, Onori KA, & Lac, du S (2004). Comparison of plasticity and development of mouse optokinetic and vestibulo-ocular reflexes suggests differential gain control mechanisms. Vision Research, 44(28), 3419–3427. 10.1016/j.visres.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Giolli RA, Blanks RHI, & Lui F (2006). The accessory optic system: basic organization with an update on connectivity, neurochemistry, and function. Progress in Brain Research, 151, 407–440. 10.1016/S0079-6123(05)51013-6 [DOI] [PubMed] [Google Scholar]
- Giolli RA, Blanks RH, & Torigoe Y (1984). Pretectal and brain stem projections of the medial terminal nucleus of the accessory optic system of the rabbit and rat as studied by anterograde and retrograde neuronal tracing methods. The Journal of Comparative Neurology, 227(2), 228–251. http://doi.Org/10.1002/cne.902270208 [DOI] [PubMed] [Google Scholar]
- Giolli RA, Blanks RH, Torigoe Y, & Williams DD (1985). Projections of medial terminal accessory optic nucleus, ventral tegmental nuclei, and substantia nigra of rabbit and rat as studied by retrograde axonal transport of horseradish peroxidase. The Journal of Comparative Neurology, 232(1), 99–116. 10.1002/cne.902320109 [DOI] [PubMed] [Google Scholar]
- Govardovskii VI, Calvert PD, & Arshavsky VY (2000). Photoreceptor light adaptation. Untangling desensitization and sensitization. The Journal of General Physiology, 116(6), 791–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, & Barres BA (2009). Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron, 62(3), 327–334. 10.1016/j.neuron.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joesch M, & Meister M (2016). A neuronal circuit for colour vision based on rod-cone opponency. Nature, 532(7598), 236–239. 10.1038/nature17158 [DOI] [PubMed] [Google Scholar]
- Johnson KP, Zhao L, & Kerschensteiner D (2018). A Pixel-Encoder Retinal Ganglion Cell with Spatially Offset Excitatory and Inhibitory Receptive Fields. Cell Reports, 22(6), 1462–1472. 10.1016/j.celrep.2018.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, la Huerta, De I, Kim I-J, Zhang Y, Yamagata M, Chu MW, et al. (2011). Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. Journal of Neuroscience, 31(21), 7753–7762. 10.1523/JNEUROSCI.0907-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjan G, Dolan J, Haumaitre C, Schneider-Maunoury S, Fujisawa H, Mitchell KJ, & Chedotal A (2005). The transmembrane semaphorin Sema6A controls cerebellar granule cell migration. Nature Neuroscience, 8(11), 1516–1524. 10.1038/nn1555 [DOI] [PubMed] [Google Scholar]
- Kim I-J, Zhang Y, Yamagata M, Meister M, & Sanes JR (2008). Molecular identification of a retinal cell type that responds to upward motion. Nature, 452(7186), 478–482. 10.1038/nature06739 [DOI] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, & Sanes JR (2015). Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature, 524(7566), 466–470. 10.1038/nature14682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon S, Yin Y, Nguyen J, Irwin N, & Benowitz LI (2000). Lens injury stimulates axon regeneration in the mature rat optic nerve. The Journal of Neuroscience : the Official Journal of the Society for Neuroscience, 20(12), 4615–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yang C, Zhang L, Gao X, Wang X, Liu W, et al. (2016). Promoting axon regeneration in the adult CNS by modulation of the melanopsin/GPCR signaling. Proceedings of the National Academy of Sciences, 113(7), 1937–1942. 10.1073/pnas.1523645113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience, 13(1), 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, et al. (2017). Diverse Central Projection Patterns of Retinal Ganglion Cells. Cell Reports, 18(8), 2058–2072. 10.1016/j.celrep.2017.01.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauss AS, Vlasits A, Borst A, & Feller M (2017). Visual Circuits for Direction Selectivity. Annual Review of Neuroscience, 40, 211–230. 10.1146/annurev-neuro-072116-031335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naarendorp F, Esdaille TM, Banden SM, Andrews-Labenski J, Gross OP, & Pugh EN (2010). Dark light, rod saturation, and the absolute and incremental sensitivity of mouse cone vision. Journal of Neuroscience, 30(37), 12495–12507. 10.1523/JNEUROSCI.2186-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Nicolas FM, Sobrado-Calvo P, Jimenez-Lopez M, Vidal-Sanz M, & Agudo-Barriuso M (2015). Long-Term Effect of Optic Nerve Axotomy on the Retinal Ganglion Cell Layer. Investigative Ophthalmology & Visual Science, 56(10), 6095–6112. 10.1167/iovs.15-17195 [DOI] [PubMed] [Google Scholar]
- Osterhout JA, Stafford BK, Nguyen PL, Yoshihara Y, & Huberman AD (2015). Contactin-4 mediates axon-target specificity and functional development of the accessory optic system. Neuron, 86(4), 985–999. 10.1016/j.neuron.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyster CW, Takahashi E, & Collewijn H (1972). Direction-selective retinal ganglion cells and control of optokinetic nystagmus in the rabbit. Vision Research, 12(2), 183–193. 10.1016/0042-6989(72)90110-1 [DOI] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, et al. (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science (New York, NY), 322(5903), 963–966. 10.1126/science.1161566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, & Tomancak P (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics, 25(11), 1463–1465. 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockhill RL, Euler T, & Masland RH (2000). Spatial order within but not between types of retinal neurons. Proceedings of the National Academy of Sciences of the United States of America, 97(5), 2303–2307. 10.1073/pnas.030413497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW (1991). The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Visual Neuroscience, 6(2), 95–111. [DOI] [PubMed] [Google Scholar]
- Rodriguez AR, de Sevilla MUller LP, & Brecha NC (2014). The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. The Journal of Comparative Neurology, 522(6), 1411–1443. 10.1002/cne.23521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah S, Berg D, Papendorp C, Briggman KL, & Berson DM (2017a). A Cre Mouse Line for Probing Irradiance- and Direction-Encoding Retinal Networks. eNeuro, 4(2). 10.1523/ENEURO.0065-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah S, Gemmer JA, Bhatia-Lin A, Manoff G, Castro G, Siegel JK, et al. (2017b). A retinal code for motion along the gravitational and body axes. Nature, 546(7659), 492–497. 10.1038/nature22818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, & Masland RH (2015). The types of retinal ganglion cells: current status and implications for neuronal classification. Annual Review of Neuroscience, 38, 221–246. 10.1146/annurev-neuro-071714-034120 [DOI] [PubMed] [Google Scholar]
- Simpson JI (1984). The accessory optic system. Annual Review of Neuroscience, 7, 13–41. 10.1146/annurev.ne.07.030184.000305 [DOI] [PubMed] [Google Scholar]
- Simpson JI, Leonard CS, & Soodak RE (1988). The accessory optic system of rabbit. II. Spatial organization of direction selectivity. Journal of Neurophysiology, 60(6), 2055–2072. [DOI] [PubMed] [Google Scholar]
- Sun LO, Brady CM, Cahill H, Al-Khindi T, Sakuta H, Dhande OS, Noda M, Huberman AD, Nathans J, & Kolodkin AL (2015a). Functional assembly of accessory optic system circuitry critical for compensatory eye movements. Neuron, 86(4), 971–984. 10.1016/j.neuron.2015.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LO, Brady CM, Cahill H, Al-Khindi T, Sakuta H, Dhande OS, Noda M, Huberman AD, Nathans J, & Kolodkin AL (2015b). Functional assembly of accessory optic system circuitry critical for compensatory eye movements. Neuron, 86(4), 971–984. 10.1016/j.neuron.2015.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LO, Jiang Z, Rivlin-Etzion M, Hand R, Brady CM, Matsuoka RL, et al. (2013). On and off retinal circuit assembly by divergent molecular mechanisms. Science (New York, NY), 342(6158), 1241974–1241974. 10.1126/science.1241974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Deng Q, Levick WR, & He S (2006). ON direction-selective ganglion cells in the mouse retina. The Journal of Physiology, 576(Pt 1), 197–202. 10.1113/jphysiol.2006.115857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szel A, Rohlich P, Caffe AR, Juliusson B, Aguirre G, & Van Veen T (1992). Unique topographic separation of two spectral classes of cones in the mouse retina. The Journal of Comparative Neurology, 325(3), 327–342. 10.1002/cne.903250302 [DOI] [PubMed] [Google Scholar]
- Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, et al. (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron, 92(2), 372–382. 10.1016/j.neuron.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholm S, Johnson K, Li X, Smith RG, & Awatramani GB (2011). Parallel mechanisms encode direction in the retina. Neuron, 71(4), 683–694. 10.1016/j.neuron.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp S, & De Zeeuw CI (2015). Forward Signaling by Unipolar Brush Cells in the Mouse Cerebellum. Cerebellum (London, England), 14(5), 528–533. 10.1007/s12311-015-0693-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsbie DS, Yang Z, Ge Y, Mitchell KL, Zhou X, Martin SE, et al. (2013). Functional genomic screening identifies dual leucine zipper kinase as a key mediator of retinal ganglion cell death. Proceedings of the National Academy of Sciences, 110(10), 4045–4050. 10.1073/pnas.1211284110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, & Jessell TM (2002). Directed differentiation of embryonic stem cells into motor neurons. Cell, 110(3), 385–397. [DOI] [PubMed] [Google Scholar]
- Yonehara K, Balint K, Noda M, Nagel G, Bamberg E, & Roska B (2010). Spatially asymmetric reorganization of inhibition establishes a motion-sensitive circuit. Nature 10.1038/nature09711 [DOI] [PubMed] [Google Scholar]
- Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, et al. (2009). Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. PLoS ONE, 4(1), e4320 10.1371/journal.pone.0004320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Shintani T, Suzuki R, Sakuta H, Takeuchi Y, Nakamura-Yonehara K, & Noda M (2008). Expression of SPIG1 reveals development of a retinal ganglion cell subtype projecting to the medial terminal nucleus in the mouse. PLoS ONE, 3(2), e1533 10.1371/journal.pone.0001533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Kolodkin AL, Wong RO, & James RE (2017). Establishing Wiring Specificity in Visual System Circuits: From the Retina to the Brain. Annual Review of Neuroscience, 40(1), 395–424. 10.1146/annurev-neuro-072116-031607 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim I-J, Sanes JR, & Meister M (2012). The most numerous ganglion cell type of the mouse retina is a selective feature detector. Proceedings of the National Academy of Sciences, 109(36), E2391–8. 10.1073/pnas.1211547109 [DOI] [PMC free article] [PubMed] [Google Scholar]