Abstract
Zika virus is a mosquito-transmitted flavivirus that causes devastating fetal outcomes in the context of maternal infection during pregnancy. An important target for drugs combatting Zika virus pathogenicity is NS2B-NS3 protease, which plays an essential role in hydrolysis and maturation of the flavivirus polyprotein. We identify hydroxychloroquine, a drug that already has approved uses in pregnancy, as a possible inhibitor of NS2B-NS3 protease by using a Food and Drug Administration-approved drug library, molecular docking, and molecular dynamics simulations. Further, to gain insight into its inhibitory potential toward NS2B-NS3 protease, we performed enzyme kinetic studies, which revealed that hydroxychloroquine inhibits protease activity with an inhibition constant (Ki) of 92.34 ± 11.91 μM. Additionally, hydroxychloroquine significantly decreases Zika virus infection in placental cells.
Introduction
Zika virus (ZIKV) belongs to the Flavivirus genus and is a member of the Flaviviridae family. Recent research has revealed that ZIKV is associated with microcephaly in fetuses1,2 and neurological disorders such as Guillain-Barré syndrome in adults.3−5 The rapid spread of this virus, affecting over a million people6 across multiple continents, has spurred researchers to search for effective therapeutic intervention. A broad-spectrum antiviral agent against shared flavivirus proteins would be especially attractive, given the preponderance of related flavivirus infections (such as dengue and West Nile viruses) in areas where ZIKV has been most prevalent.
Zika virus is an enveloped virus like other flaviviruses, encapsulating a single-stranded, positive-sense, RNA genome7 encoding a single polyprotein precursor.8 It is hydrolyzed into three structural proteins (E, prM/M, and C) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5)8−10 by the host and viral proteases.10 Among these viral and host elements, the viral NS2B-NS3 protease is an attractive drug target due to its essential role in the virus life cycle. The crystal structure of NS2B-NS3 protease reveals that NS2B (only the hydrophilic part was taken in the construct for crystallographic studies, approximately residues 49–95 of the full-length NS2B protein constituting ∼130 residues) can be found in two conformations. In the presence of inhibitor/substrate, NS2B forms a β-hairpin and lies near the substrate binding site of NS3 protease, adopting a closed conformation, but in the absence of inhibitor/substrate, it adopts an open conformation.11−14 The NS2B-NS3 protease structure with PDB ID: 5LC013 (hydrophilic part, residues 49–95 of NS2B fused via a Gly4–Ser–Gly4 linker to the N-terminal of the NS3 protease) shows NS2B wrapping around NS3 in such a way that the C-terminal residues of NS2B form a β-hairpin that contributes to the S2 pocket of the NS3 protease.13−15 As reported NS2B protein has a high abundance of disorder promoting residues containing a 37-residue disordered region (62–98).16,17 The NS2B interaction with NS3 protease facilitates NS3-mediated cleavage of polyprotein thus, it acts as an important cofactor for the activity of NS3 protease.18 Generally, disordered proteins lead to functionality only upon interaction with its binding partner such as transactivation domain of cMyb, in which cMyb becomes functional only upon binding its ordered counterpart, KIX.19−22 Together, NS2B and NS3 form the NS2B-NS3 protease complex that hydrolyzes the ZIKV polyprotein into functional proteins used for viral propagation and maturation.11
Viral proteases are considered excellent targets for the identification of potential drug candidates, as protease plays an indispensable role in viral replication.23−25 Recently, the NS2B-NS3 protease has been investigated as a target to identify potential inhibitors.26−29 Repurposing approved drugs can be an efficient method to identify drug compounds, which may be capable of activating or inhibiting new targets.30 This approach has several advantageous features, including reduced development time and expense and improved safety.30 In our study, we screened Food and Drug Administration (FDA)-approved drugs for their ability to target NS2B-NS3 protease. We hypothesize that specific drugs in the library will show potential to specifically target NS2B-NS3 protease of ZIKV, especially given that some drug compounds are already being used to target NS2B-NS3 protease in other flaviviruses.31−33 Here, we use molecular docking and molecular dynamics (MD) simulation studies to identify potential drug candidates from the library based on their predicted ability to target the active site of NS2B-NS3 protease. Our top hits include hydroxychloroquine (HCQ), mitoxantrone, miglustat, nadolol, carteolol, and pindolol. Among these potential candidates, hydroxychloroquine (HCQ) was shown in a recent study to have an inhibitory effect on ZIKV in a mouse model of ZIKV infection during pregnancy.34 Therefore, we pursued further studies with HCQ and showed that HCQ likely targets the active site of NS2B-NS3 protease; thus, it may block its ability to hydrolyze the single polypeptide product of ZIKV into functional proteins required for ZIKV survival and replication. Molecular dynamic simulation reveals a significant gain in stability associated with the binding of HCQ to the protease complex, suggesting a strong binding affinity between HCQ and the active site of NS2B-NS3 protease. Further, inhibition assays support the inhibition of NS2B-NS3 protease by HCQ. Finally, a recent study from our group demonstrated that ZIKV infection activates the cellular recycling pathway, autophagy, and that HCQ treatment blocks this activation, leading to a reduction in ZIKV vertical transmission.34 Also, HCQ shows antiviral potential in the treatment of dengue virus infection35 by activating the innate immune signaling pathways and inducing the production of reactive oxygen species. However, the molecular mechanism of action of HCQ against ZIKV infection is still unclear. Our work, which describes the interaction between HCQ and NS2B-NS3 protease in great detail, may provide a foundation to leverage this compound and other derivatives in treating patients infected with ZIKV.
Results
Virtual Screening of FDA-Approved Drugs
To identify drug candidates with the potential to inhibit NS2B-NS3 protease, we performed structure-based virtual screening employing in silico molecular docking techniques. An X-ray crystal structure of ZIKV NS2B-NS3 protease (PDB ID: 5LC0) was utilized as an input in the docking process. Since the dipeptide boronic acid acts as an inhibitor of the protease complex as reported previously,13 we utilized the same binding site as the active site of NS2B-NS3 protease for docking. After extra precision (XP) docking, the top 25 compounds were selected to go through induced fit docking (IFD) protocol. This docking protocol considers the movement of highly flexible residues of the NS2B-NS3 protease complex upon binding of a drug compound at the active site. The IFD scores of top six compounds, viz., mitoxantrone, hydroxychloroquine (HCQ), miglustat, nadolol, carteolol, and pindolol, with NS2B-NS3 protease are shown in Tables 1 and S1. Mitoxantrone and HCQ are associated with the highest docking scores of −12.785 and −10.725 kcal/mol, respectively (Table 1), indicating that they have high potential to bind to NS2B-NS3 protease and to fit in the active site pocket. Both these compounds, along with miglustat, nadolol, carteolol, and pindolol, were stabilized at the NS2B-NS3 protease active site by interactions with key amino acid residues through H-bonds, pi interactions, and salt bridges (Table S1).
Table 1. Summary of Induced Fit Docking (IFD) Results of Top-Hit FDA-Approved Drugs Representing Docking Score and Interacting Amino Acid Residuesa.
| compound name (ID) | docking score (kcal/mol) | interacting amino acid residues |
|---|---|---|
| mitoxantrone (DB01204) | –12.785 | H-bond: Ser81*, Phe84*, Asp75, Pro131; pi–cation: Tyr161, pi–pi: Tyr161; salt bridge: Asp129, Asp75 |
| hydroxychloroquine (DB01611) | –10.725 | H-bond: Asp83*, Tyr130, Asn152, Gly151; pi–pi: Tyr161, pi–cation: Tyr161; salt bridge: Asp129 |
Asterisk (*) for residues that belong to NS2B cofactor.
IFD revealed that mitoxantrone was associated with the highest docking score. Mitoxantrone has a molecular weight (MW) of 444.5 g/mol, and its interaction with the active site of NS2B-NS3 protease is established by H-bonds (Ser81 and Phe84 of NS2B; Asp75 and Pro131 of NS3 protease), pi–pi interaction (Tyr161 of NS3 protease), a pi–cation interaction (Tyr161 of NS3 protease), salt bridges (Asp129 and Asp175 of NS3 protease), and hydrophobic interactions (Trp50, Val72, Tyr130, Ala132, Tyr150, Val154, and Val155 of NS3 protease) (Figures S1A and S2A).
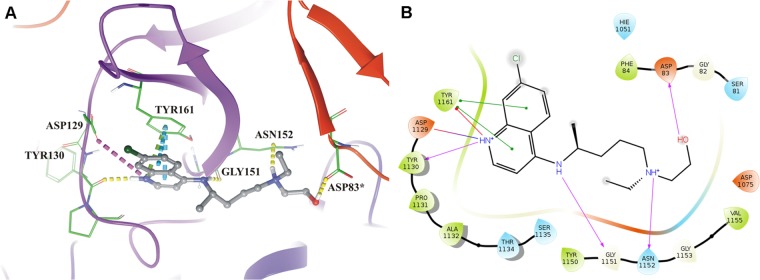
Interactions between HCQ (MW: 335.9 g/mol), the second highest-scoring compound, and protease include H-bonds involving NS2B (Asp83) and NS3 protease (Tyr130, Gly151, and Asn152). HCQ also forms a salt bridge (Asp129 of NS3 protease) and pi interactions (pi–pi and pi–cation interactions with Tyr161 of NS3 protease). Besides, some hydrophobic amino acid residues (Phe84 of NS2B and Pro131, Ala132, Tyr150, and Val155 of NS3 protease) contribute to the interaction between HCQ and the active site of NS2B-NS3 protease (Figure 1). The binding pose of mitoxantrone and HCQ show that the NS3 component of the protease complex has a higher number of interacting amino acid residues than the NS2B cofactor does.
Figure 1.
Molecular interaction of hydroxychloroquine (HCQ) at NS2B-NS3 protease active site. (A) Molecular interaction of HCQ (gray) with NS2B-NS3 protease (three-dimensional view) by H-bonds (yellow dashed lines), pi–pi interactions (cyan dashed lines), pi–cation interactions (green dashed lines), and salt bridges (pink dashed lines). (B) Molecular interaction diagram in two-dimensional illustrates interactions between HCQ and NS2B-NS3 protease by H-bonds (pink arrow), pi–pi interactions (green solid line), pi–cation interactions (red solid line), and salt bridges (blue-red straight line). [A: asterisks (*) indicate residues that belong to NS2B; B: residues 49–87 of NS2B are denoted 49–87, while residues 15–167 of NS3 protease are denoted 1015–1167].
The third-ranking molecule, miglustat (MW: 219.3 g/mol), interacts with NS3 protease using two H-bonds (Tyr130 and Ser135), one pi–pi interaction (Tyr161), and one salt bridge (Ser135). The interaction between miglustat and NS2B-NS3 protease is also established through hydrophobic residues (Leu128, Pro131, Ala132, Tyr150, and Val162) of NS3 protease (Figures S1B and S2B).
The fourth-ranking molecule is nadolol, (MW: 309.4 g/mol), which interacts with NS2B through one H-bond (Asp83) and NS3 protease by five H-bonds (Tyr130, Ser135, Asn152, Gly153, and Tyr161) and two pi interactions (pi–cation at Hie51 and pi–pi at Tyr161). Nadolol is also stabilized at the active site of NS2B-NS3 protease by hydrophobic residues of NS2B and NS3 protease (Pro131, Ala132, Tyr150, Val154, and Val155 of NS3 protease and Phe84 of NS2B) (Figures S1C and S2C).
Carteolol (MW: 292.4 g/mol), the fifth-ranking drug compound, interacts with NS3 protease through four H-bonds (Asp75, Asn152, Tyr161, and Tyr130), two pi interactions (pi–pi at Tyr161 and pi–cation at Hie51), and a salt bridge (Asp75) (Figures S1D and S2D). Pindolol (MW: 248.3 g/mol), the sixth-ranking drug compound, shows interactions with NS3 protease by means of four H-bonds (Tyr130, Asn152, Gly153, and Tyr161) and one pi–pi interaction (Tyr161) (Figures S1E and S2E). The interactions of both carteolol and pindolol with the active site are also established through hydrophobic residues of NS2B (Phe84) and NS3 protease (Pro131, Ala132, Tyr150, and Val155).
Molecular Dynamics Simulation
To better understand the stability of their binding interactions with NS2B-NS3 protease, we subjected these top-scoring six compounds to molecular dynamics simulation in order to monitor the conformational changes and dynamic behavior of protein over the course of a 30 000 ps time period. Changes in the structural integrity of the protease complex were analyzed by calculating the root-mean-square deviation (RMSD) and the root-mean-square fluctuations (RMSFs) over the backbone atoms of NS2B-NS3 protease, which includes a series of carbon, nitrogen, and oxygen. The RMSD plot revealed that the compound HCQ (Figure 2A) and mitoxantrone (Figure S3A) demonstrate greatest stability at approximately 1.5–1.8 Å, indicating that the interaction between HCQ and the active site of the protease is stable throughout the simulation period of 30 000 ps. The other compounds were observed to have stability around RMSD of 1.5–2 Å over the simulation time period (Figure S3). The root-mean-square fluctuations (RMSFs) illustrate the distinction between the NS2B and NS3 components of the ZIKV protease (Figures 2 and S3).
Figure 2.
Molecular dynamics simulation of NS2B-NS3 protease with HCQ. (A) Interaction between HCQ and protease active site shows the greatest stability around 1.5–1.8 Å throughout the simulation period. (B) Protease attains increased stability upon addition of HCQ. (C) HCQ maintains compactness during the simulation period. (D) NS2B-NS3 protease–HCQ complex shows no loss of hydrogen bonds. [5LC0: NS2B-NS3 protease; ps: picosecond; residues 49–87 of NS2B are denoted 49–87, while residues 15–167 of NS3 protease are denoted 1015–1167].
HCQ (Figure 2B) and mitoxantrone (Figure S3B) exhibit the least fluctuation and are associated with the greatest gain in stability upon binding to NS2B-NS3 protease. Similar RMSFs are observed at residues 50–100 of the NS3 protease in the case of all of the compounds, and this may be due to a change in the interacting residues. Further, the radius of gyration (Figure 2C) reveals that bound HCQ maintains compactness during the simulation period. The complex does not lose hydrogen bonds during the simulation period. HCQ (Figure 2D) and mitoxantrone (Figure S3D) show no loss of hydrogen bonds throughout the simulation, and the H-bond interactions were maintained during the simulation period. Overall, the drug–protease complexes show significant stability throughout the simulation, but compared to other compounds, HCQ and mitoxantrone show the greatest stability over the 30 000 ps time scale.
HCQ Inhibits NS2B-NS3 Protease Activity
Among the top-hit compounds, we selected HCQ, an antimalarial agent that is considered an FDA Pregnancy Class C drug, for further investigation. HCQ is also utilized as a chronic suppressive treatment for rheumatological diseases, such as systemic lupus erythematosus (SLE).
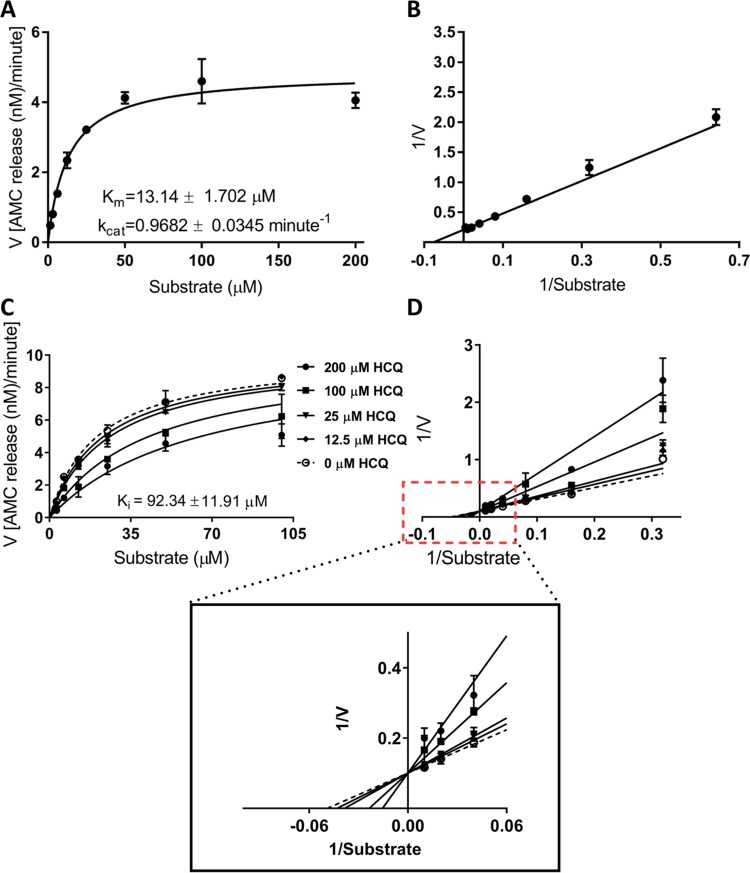
Importantly, women taking HCQ for SLE are recommended to continue their HCQ regimen throughout pregnancy, and several studies illustrate the relative safety of using this drug during pregnancy, for both the mother and the developing fetus.36−38 A recent report on ZIKV transmission in pregnant mice revealed that HCQ successfully inhibits maternal–fetal transmission due to modulation of the autophagy pathway in the placenta.34 HCQ also acts as a potential inhibitor of dengue virus infection by activating the innate immune signaling pathway.35 With these features in mind, HCQ was considered to be a prime candidate for further experiments. Therefore, we pursued further studies with HCQ and showed that HCQ likely targets the active site of NS2B-NS3 protease. The kinetic parameters (Km, kcat, and kcat/Km) of active NS2B-NS3 protease were obtained using the Michaelis–Menten equation, which showed Km, kcat, and kcat/Km to be 13.14 ± 1.702 μM, 0.9682 ± 0.0345 min–1, and 0.0737 ± 0.00989 min–1 μM–1, respectively (Figure 3A, B). To demonstrate the inhibitory potential of the drug against NS2B-NS3 protease, we computed kinetic parameters in the presence of HCQ (Figure 3C, D). Using the substrate–velocity curves, we determined the inhibition constant (Ki) of HCQ to be 92.34 ± 11.91 μM.
Figure 3.
HCQ inhibits NS2B-NS3 protease activity. (A) Substrate–velocity curve and (B) Lineweaver–Burk (LB) plot illustrating the activity of NS2B-NS3 protease (5 nM) with 1.56, 3.125, 6.25, 12.50, 25, 50, 100, and 200 μM substrate benzoyl−norleucine−lysine−lysine−arginine−7-amino-4-methylcoumarin (benzoyl–Nle–KKR–AMC). (C, D) Inhibition of NS2B-NS3 protease activity by HCQ. NS2B-NS3 protease (5 nM) and 3.125, 6.25, 12.50, 25, 50, and 100 μM substrates were used to compute the kinetic curves. LB plot in (D) represents the fitting of the data through competitive inhibition.
HCQ Inhibits ZIKV Burden in Placental Trophoblast Cells
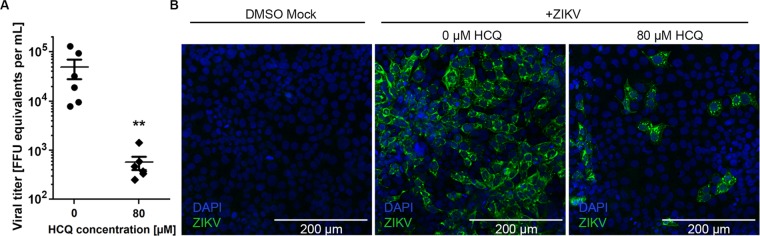
Given that ZIKV is transmitted from mother to the developing fetus in a transplacental manner by damaging and killing placental cells (trophoblasts), we tested the ability of HCQ to block ZIKV activity in a placental trophoblast cell line, JEG3. A dose curve of HCQ (0–80 μM) was performed (data not shown). JEG3 cells were infected for 48 h with ZIKV and treated with 80 μM HCQ, which led to a highly significant reduction in ZIKV titers (Figure 4A). Next, we examined the localization of the inhibitory activity of HCQ. Immunofluorescence analysis of ZIKV-infected trophoblasts reveals that the virus can be found in large foci of cells and that HCQ treatment reduces the number of foci as well as the size of the foci of infected cells (Figure 4B).
Figure 4.
HCQ treatment reduces ZIKV viral burden in placental cells. (A) Titers at 48 h post infection (hpi) of ZIKV-infected JEG3 cells treated with indicated concentrations of HCQ. Symbols represent six biological replicates from two separate experiments. The bars represent mean ± standard error of the mean. *P < 0.01 (Mann–Whitney test). (B) Representative immunofluorescence microscopy for ZIKV-E protein-positive (green) cells following indicated treatments. Nuclei are stained blue.
Discussion
The rapid spread of ZIKV infection, its association with microcephaly and neurological disorders,1−4 and its capacity for human-to-human transmission39,40 have produced an urgent need for an effective drug against ZIKV. In the area of drug discovery, repurposing is an efficient approach to screen drug molecules from an existing drug library for successful inhibition of a new target,30 and in silico drug modeling is a useful and cost-effective strategy to identify drug candidates in a short period. Viral proteases are considered excellent targets for the identification of potential drug candidates, as protease plays an indispensable role in viral replication.23−25 This has been the case in human immunodeficiency virus (HIV) research, which has developed several drugs with the ability to inhibit the HIV protease.41,42 Recently, NS2B-NS3 protease has been investigated as a target to identify potential inhibitors against other flaviviruses.26−28,43−45 In our study, we have used ZIKV NS2B-NS3 protease as a target as it plays a key role in processing a single polyprotein precursor into functional proteins. Therefore, NS2B-NS3 protease is an attractive target for identifying potential drug candidates against ZIKV.46,47
A previous report showed significant inhibition of the ZIKV NS2B-NS3 protease by dipeptide boronic acid;13 hence, the same active site can be utilized to broaden drug discovery research to develop new therapeutic interventions. To explore potential new inhibitors of the ZIKV NS2B-NS3 protease, we employed molecular docking and molecular dynamics simulation studies of FDA-approved drug molecules. Among the top hit, we selected HCQ, an antimalarial and antirheumatic agent that is considered an FDA Pregnancy Class C drug, for further investigation. We identify a possible interaction of HCQ with the NS2B-NS3 protease, where the interactions of certain key amino acid residues of NS2B and NS3 protease are required to stabilize HCQ at the binding site. Molecular docking reveals that HCQ exhibits considerable affinity for the NS2B-NS3 protease active site. Also, refinement in the binding affinity through induced fit docking confirms strong binding of HCQ to NS2B-NS3 protease. The binding and stabilization of HCQ at the active site of NS2B-NS3 protease requires the formation of an H-bond with Asp83 of NS2B and Gly151 of NS3 protease, a pi interaction at Tyr161, and a salt bridge at Asp129 of NS3 protease. A similar mode of interaction has been seen in the NS2B-NS3 protease of Zika virus, where the substrate interacts through Asp129, Gly151, and Tyr161 of NS3 protease and Asp83 of NS2B.48 Also, there are some reports on the NS2B-NS3 protease of Zika virus and West Nile virus, which demonstrated that a dipeptide boronic acid compound (cn-716) acts as a reversible inhibitor of the NS2B-NS3 protease complex.13,49 In the case of Zika virus protease complex, arginine of cn-716 forms a salt bridge with Asp129 residue of the NS3 protease, and this feature is conserved in other flavivirus protease complexes. A similar kind of interaction has also been seen in the crystal structure of West Nile virus NS3 protease complexed with an inhibitor.50,51 This interaction provides a basis for the strong binding hypothesized between HCQ and the active site of NS2B-NS3 protease. Also, our molecular dynamics simulation result shows the stabilization of the HCQ at the active site of NS2B-NS3 protease throughout simulation period. Further, enzyme inhibition kinetics suggest that HCQ significantly inhibits the activity of NS2B-NS3 protease (Ki = 92.34 ± 11.91 μM). Here, we show that HCQ blocks ZIKV protease activity. On the basis of the previous report on chloroquine inhibiting proteolytic processing of prM to M protein,52 we do not rule out the possibility of other targets of HCQ in Zika virus, but these need to be further explored. Previous studies documented the antiviral properties of chloroquine and HCQ. They suggested that chloroquine inhibits the early and late stages of viral replication, but the antiviral effect of the drug is increased when added before infection compared to post infection.53 From these studies, it is clear that at early stages of viral replication, chloroquine acts as an entry inhibitor. In our studies, we treated the trophoblast cells with HCQ 3 h post infection (hpi); therefore, reduced viral burden may have resulted due to inhibition of late-stage viral replication such as the inhibition of viral particle maturation. HCQ may be inhibiting the maturation of polyprotein by blocking NS2B-NS3 protease activity. Since NS2B-NS3 protease plays an important role in the proteolytic processing of ZIKV polyprotein, we can hypothesize that blocking protease activity with HCQ will inhibit polyprotein processing and subsequent viral assembly and maturation. In our recent report on ZIKV transmission in pregnant mice, we revealed that HCQ successfully inhibits maternal–fetal transmission due to modulation of the autophagy pathway in the placenta.34 Our study demonstrates that HCQ can additionally inhibit ZIKV infection of placental trophoblasts possibly due to its binding affinity to the ZIKV NS2B-NS3 protease.
The five other top-ranked compounds, viz., mitoxantrone, miglustat, nadolol, carteolol, and pindolol, form stable interactions with the amino acid residues of NS2B and NS3 protease at the active site. The residues Asp129, Ser135, Gly153, and Tyr161 of NS3 protease and Asp83 of NS2B are involved in establishing the HCQ interaction, demonstrating a similar mode of interaction between substrate/inhibitor as seen in other flavivirus proteases.50,51,54 In summary, various amino acid residues are involved in stabilizing drug molecules at the active site of the NS2B-NS3 protease of Zika virus. Further studies will test the other top compounds for their inhibitory effect against Zika virus.
Methods
System Configuration
Molecular docking and molecular dynamics simulation studies were executed using a high-performance GPU operated with CentOS V6.6 Linux operating platform and with hardware configuration of the HPC GPU Super Micro Intel Xeon E5-620 v3 series with 8-core processor, 64 GB DDR4-2133 ECC RDIMM of RAM, and graphics card of NvidiaQuadroK2000 with 2 GB. The software specifications we used for docking and molecular dynamics simulation are the commercial version of Schrödinger software package, LLC, NY 2012, and the all-atom optimized potentials for liquid simulations (OPLS-AA) force field academic-licensed molecular dynamics package of Desmond4.4.
Structure-Based Virtual Screening
The X-ray crystal structure of the NS2B-NS3 protease (PDB ID: 5LC0) was retrieved from the Protein Data Bank. The crystal structure of NS2B-NS3 protease (dimer form, chains A and B) is crystallized with dipeptide boronic acid (cn-716; ID: 6T8), where each monomer contains a cn-716. The dipeptide boronic acid acts as an inhibitor, embedded at the substrate binding site of the protease. Each monomeric unit comprises a C-terminal fragment of NS2B (hydrophilic residues 49–95), fused via a linker (Gly)4–Ser–(Gly)4 with the N-terminus of NS3.13 The NS2B-NS3 protease monomer was prepared using the Schrödinger suite protein preparation wizard, which has two main functions: preparation and refinement. These features of the preparation wizard perform various tasks, viz., assigning a bond order, adding hydrogen, creating zero-order bonds to metals, creating disulfide bonds, filling in missing side chains, and removing water molecules beyond 5 Å. Further structural optimization was done at pH 7, and water molecules with fewer than three H-bonds were removed. The embedded dipeptide boronic acid compound has been used as a model to investigate catalytic drug binding. This site was chosen as a prerequisite for receptor grid generation to protease using the Grid generation panel of the Schrödinger suite Glide Module. We used boronate inhibitor as a reference to create the size of the grid (X-, Y-, and Z-coordinates as 81.82, 51.89, and 153.80 Å, respectively), where the incoming ligand binds during the docking process. Receptor grid scaling of van der Waals radii of receptor atoms was done with a scaling factor of one unit and a partial charge cutoff of 0.25 to soften the potential of nonpolar parts of the receptor. The FDA-approved drug compounds were prepared using the Schrödinger suite Ligand Prep module before they underwent ligand docking. This drug library, consisting of 1861 compounds, was selected from the DrugBank database.55 The Ligand Prep module is able to generate different structures of compounds based on stereoisomers, ring conformations, and ionization states for each processed compound. Ligand preparation of 1861 drug compounds produced 5323 structures that were processed through the Lipinski filter (retaining structures with molecular weights ≤500 g/mol to make the screening more effective and to retrieve the compounds that satisfy all conditions of filtration) and a reactive filter. Out of 5323 structures, 2558 passed through the Lipinski and reactive filters. To screen a potential candidate for inhibition, FDA-approved drugs were processed using a Glide program. These drug compounds were docked at the active site of NS2B-NS3 protease and filtered using high-throughput virtual screening (HTVS), standard precision (SP), and extra precision (XP) docking. First, we performed high-throughput virtual screening (HTVS), which rapidly identified the top 40% of input compounds based on docking score. These selected molecules then underwent standard precision (SP) docking at the same active site on the protease, again eliminating all but the top 40% of input compounds. The next phase, extra precision (XP) docking, selected only the top 10% of the input compounds.
To understand the binding between protease and drug compounds more accurately, we employed induced fit docking (IFD). IFD accurately predicts binding poses of ligands to their receptors and occurrences of structural changes at the receptor during ligand binding. It involves the Glide module and Prime module to predict accurate binding, considering the flexibility of both the ligand and receptor.56 We used a standard protocol that generates up to 20 poses using automatic docking settings. It utilizes a receptor van der Waals scaling of 0.50 and a ligand van der Waals scaling of 0.05, and it penalizes nonpolar conformations for amide bonds and sample ring conformations with 2.5 kcal/mol energy windows. Further refinement was done for residues within 5 Å of ligand poses. Glide redocking into structures within 30 kcal/mol of the best structure and within the top 20 structures overall was performed with XP precision. Among different poses and conformations, the best one was selected based on docking score.
Molecular Dynamics (MD) Simulation
The NS2B-NS3 protease−ligand complex was subjected to a 30 ns molecular dynamics simulation to further evaluate their stability. The intramolecular conformational changes that occur in the protein structure have been well explained by molecular dynamics simulation studies.57 Molecular dynamics of the NS2B-NS3 protease–FDA drug complexes were simulated using the Desmond module of Schrödinger package.58 The docking score and the interaction analysis were used to select compounds that would be carried forward to the energy minimization and molecular dynamics simulation studies with the OPLS-AA force field.59 An orthorhombic water box was created using the Desmond system builder, and preequilibrated TIP3P water molecules and 0.15 M Na + Cl– were placed in the system as neutralizing compounds.60 A distance of greater than 5 Å was maintained between the box wall and protein, thereby ensuring that the protein could not directly interact with its own periodic image. Energy minimization of the protein complex in the prepared system for MD simulation was carried out to a maximum of 3000 steps using the steepest descent until a gradient threshold (25 kcal/(mol Å)) was reached. The periodic boundary conditions were applied, and the long-range electrostatic interactions were treated with Ewald sums.61 The molecular dynamics simulation was performed at a constant temperature of 300 K and a pressure of 1.01 bar with a time interval of 2 fs.62,63 A cutoff radius of 9 Å was selected for the coulombic short-range interaction cutoff method. At every 4.8 ps, the frames of the trajectory were captured. The root-mean-square deviation (RMSD) and root-mean-square fluctuation (RMSF) of the backbone were obtained through the simulation using the first frames as a reference.61
Gene Construct and Protein Purification
We chose an expression plasmid vector, pET151/D-T, to insert a DNA fragment encoding NS2B-NS3 protease of ZIKV. An Escherichia coli codon-optimized DNA sequence encoding NS2B-NS3 protease (amino acid sequence taken from NS2B-NS3 protease structure 5LC0 from the Protein Data Bank) was inserted in the vector in-frame with 6XHis tag and TEV cleavage site (synthesized by Invitrogen). Restriction enzymes XhoI and NdeI were used to insert the gene of interest into the vector. The NS2B-NS3 protease gene (with tag, i.e., 6Xhis-V5 epitope–TEV cleavage site) is composed of 263 residues, and it has a molecular weight and a theoretical isoelectric point of 27.99 kDa and 5.40, respectively, as determined by the ExPASy ProtParam tool. This recombinant plasmid encodes a polypeptide whose sequence from N- to C-terminus reads: 6Xhis-V5 epitope–TEV cleavage site–NS2B-NS3 protease gene. Transformation of recombinant plasmid was done in BL21(DE3) E. coli cells, and cells positive for the recombinant plasmid were used to express NS2B-NS3 protease (in LB media and 0.5 mM IPTG at 20 °C for 16 h). The cells were harvested by centrifugation (6000 rpm, 10 min) at 4 °C. Afterward, cell lysis was done using B-PER (Thermo Fisher Scientific) and phenylmethylsulfonyl fluoride (1 mM) at 4 °C (30 min incubation). The NS2B-NS3 protease was purified from the soluble fraction through HisPur Cobalt Resin (Thermo Fisher Scientific) using Tris–HCl binding buffer (25 mM Tris–HCl, 300 mM NaCl, 40 mM imidazole, and 10% glycerol, pH 8.5) and elution buffer (25 mM Tris–HCl, 300 mM NaCl, 500 mM imidazole, and 10% glycerol, pH 8.5). Next, the tag of the purified NS2B-NS3 protease was removed by tobacco etch virus protease (AcTEV, Invitrogen) while incubating at 25 °C for 4.5 h. Hydrolysis by AcTEV produced a polypeptide of 236 residues (24.837 kDa), where 230 residues are from the NS2B-NS3 protease reported as 5LC0,13 and the remaining six residues (at the N-terminus of the polypeptide) are Gly–Ile–Asp–Pro–Phe–Thr. The cleaved NS2B-NS3 protease was purified through HisPur Cobalt Resin using 25 mM Tris–HCl, pH 8.5 containing 10% glycerol. The purity of NS2B-NS3 protease (uncleaved and cleaved) was analyzed with sodium dodecyl sulfate polyacrylamide gel electrophoresis (12%).
Activity Assay
Enzyme kinetics of the NS2B-NS3 protease was determined in a reaction buffer containing 10 mM phosphate, 1 mM TCEP, 1 mM CHAPS, and 20% glycerol, pH 7 and 30 °C. In a 96-well black plate, the substrate, benzoyl–norleucine–lysine–lysine–arginine–7-amino-4-methylcoumarin (benzoyl–Nle–KKR–AMC; Sigma-Aldrich: 98.6% purity; stock preparation: in water), was serially diluted in reaction buffer (10 mM phosphate buffer, 1 mM TCEP, 1 mM CHAPS, and 20% glycerol; pH 7) to achieve a concentration range of 1.56–200 μM. Afterward, NS2B-NS3 protease was added into the wells to a 5 nM final concentration. The fluorescence signal of released AMC was monitored with an emission wavelength of 460 nm at 360 nm excitation using a multiplate reader (infiniteM200PRO: TECAN), while gain was adjusted manually to 80. The amount of released AMC at varying substrate concentrations was determined via a standard curve of fluorescence signal produced as a function of free AMC (concentration range: 0.025–12.8 μM; stock: 50 mM in 100% dimethyl sulfoxide). Further, the initial velocity of the reaction was calculated in terms of the amount of AMC produced per minute during 15 min. The data were fitted to the Michaelis–Menten equation, V = Vmax[S]/(Km + [S]), to determine enzyme kinetic parameters (Km, kcat, and Vmax) using GraphPad Prism 7.0 software. Measurement of data points was taken in triplicates, and data were represented as a mean ± standard error with R2 = 0.95.
Inhibition Assay
For inhibition studies, to determine the enzyme kinetic parameters, we used the same reaction buffer and substrate as we used to determine the activity of the NS2B-NS3 protease. HCQ (Sigma-Aldrich) was dissolved in water to prepare 50 mM stock solutions, which were serially diluted in reaction buffer to make a variable range of concentrations (0, 12.5, 25, 100, and 200 μM). Subsequently, NS2B-NS3 protease (5 nM final concentration for 50 μl reaction volume) was mixed with different concentrations of HCQ and incubated for 10 min at 30 °C. Afterward, the reaction was initiated by adding 25 μl of substrate (to final substrate concentrations of 3.125, 6.25, 12.5, 25, 50, and 100 μM). The fluorescence signal was monitored at 460 nm emission and 360 nm excitation wavelength at every 5 min of interval. Further, the Ki was computed using GraphPad Prism 7.0 software at 15 min. Experiments were set up in triplicate, and values were represented as a mean ± standard error with R2 = 0.95.
ZIKV Infection and Measurement of Viral Burden in Vitro
The Brazilian strain of ZIKV (Paraiba 2015) was used in this study.64 Studies with ZIKV were conducted under biosafety level 2 (BSL2). JEG3, a human cytotrophoblast cell line from the ATCC (HTB-36), was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) at 37 °C with 5% CO2. JEG3 cells were infected with ZIKV at a multiplicity of infection of 0.1 for 3 h, washed twice with warm phosphate-buffered saline, and then cultured in fresh media containing 0 or 80 μM hydroxychloroquine (Sigma-Aldrich). At 48 h post infection (hpi), the cells were harvested for the measurement of viral burden or were fixed for immunofluorescence using a ZIKV-specific mAb, ZV-2, as previously described.64 After 48 h, supernatants were harvested for RNA extraction using the Viral RNA Mini kit (Qiagen). ZIKV RNA levels were determined by one-step quantitative reverse transcriptase polymerase chain reaction on an ABI 7500 Fast Instrument using standard cycling conditions. Viral burden from supernatant samples was expressed on a log10 scale as viral RNA equivalents per mL after comparison with a standard curve produced using serial 10-fold dilutions of ZIKV RNA. A published primer set was used to detect ZIKV RNA:65 Fwd, 5′-CCGCTGCCCAACACAAG-3′; Rev, 5′-CCACTAACGTTCTTTTGCAGACAT-3′; Probe, 5′-/56-FAM/AGCCTACCT/ZEN/TGACAAGCAATCAGACACTCAA/3IABkFQ/-3′ (Integrated DNA Technologies).
For immunofluorescence studies, the cells were grown on Millicell EZ slides (Millipore Sigma), fixed with 4% paraformaldehyde, and permeabilized with 0.3% Triton X. After blocking with 1% bovine serum albumin, the cells were incubated overnight with primary antibodies to the E protein of ZIKV,64 followed by fluorescently labeled secondary antibodies (Thermo Fisher Scientific). Images were obtained using a Zeiss LSM880 Confocal Laser Scanning Microscope with Airyscan. Images were processed and analyzed using Adobe Photoshop and NIH Image J software.
GraphPad Prism 5.0 was used for all cell culture-related analyses. The analyses of virologic data were conducted using a Mann–Whitney test. Data with p-values <0.05 were considered to be statistically significant.
Acknowledgments
This work was partially funded by a DST grant, India (YSS/2015/000613) (to R.G.), and a National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development grant R01HD091218 (to I.U.M.). The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01002.
Molecular interaction and molecular dynamics simulation data of top-hit drug compounds from an FDA-approved drug library and the active site of NS2B-NS3 protease; induced fit docking results of the top six drug molecules (HCQ, pindolol, carteolol, nadolol, miglustat, and mitoxantrone) (Table S1); molecular interactions between drug molecules and NS2B-NS3 protease in three- and two-dimensional views, generated from IFD (Figures S1 and S2); and molecular dynamics simulation data from the association of NS2B-NS3 protease with drug molecules (pindolol, carteolol, nadolol, miglustat, and mitoxantrone) (Figure S3) (PDF)
Author Contributions
R.G., I.U.M., N.G., and S.K.S.: Conception, design, analysis, and interpretation of data, writing and review of the manuscript, and study supervision. A.K. and M.A. performed computational studies. A.K. performed enzymatic assays. B.L. performed infection and HCQ treatment studies in placental cells. A.K., B.L., M.A., S.K.S., I.U.M., and R.G. wrote the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Mlakar J.; Korva M.; Tul N.; Popović M.; Poljšak-Prijatelj M.; Mraz J.; Kolenc M.; Resman Rus K.; Vesnaver Vipotnik T.; Fabjan Vodušek V.; et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374, 951–958. 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Rasmussen S. A.; Jamieson D. J.; Honein M. A.; Petersen L. R. Zika Virus and Birth Defects--Reviewing the Evidence for Causality. N. Engl. J. Med. 2016, 374, 1981–1987. 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Oehler E.; Watrin L.; Larre P.; Leparc-Goffart I.; Lastère S.; Valour F.; Baudouin L.; Mallet H. P.; Musso D.; Ghawche F. Zika Virus Infection Complicated by Guillain-Barré Syndrome—Case Report, French Polynesia, December 2013. Eurosurveillance 2014, 19, 20720 10.2807/1560-7917.ES2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- Cao-Lormeau V.-M.; Blake A.; Mons S.; Lastère S.; Roche C.; Vanhomwegen J.; Dub T.; Baudouin L.; Teissier A.; Larre P.; et al. Guillain-Barré Syndrome Outbreak Associated with Zika Virus Infection in French Polynesia: A Case-Control Study. Lancet 2016, 387, 1531–1539. 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkki H. CNS Infections: Zika Virus Infection Could Trigger Guillain-Barré Syndrome. Nat. Rev. Neurol. 2016, 12, 187 10.1038/nrneurol.2016.30. [DOI] [PubMed] [Google Scholar]
- Samarasekera U.; Triunfol M. Concern over Zika Virus Grips the World In the Past Week, the World Has Mobilised to Tackle the Latest Threat to Global Health Security. Lancet 2016, 387, 521–524. 10.1016/S0140-6736(16)00257-9. [DOI] [PubMed] [Google Scholar]
- Sirohi D.; Chen Z.; Sun L.; Klose T.; Pierson T. C.; Rossmann M. G.; Kuhn R. J. The 3.8 Å Resolution Cryo-EM Structure of Zika Virus. Science 2016, 352, 467–470. 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G.; Chang G.-J. J. Full-Length Sequencing and Genomic Characterization of Bagaza, Kedougou, and Zika Viruses. Arch. Virol. 2007, 152, 687–696. 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- Shiryaev S. A.; Strongin A. Y. Structural and Functional Parameters of the Flaviviral Protease: A Promising Antiviral Drug Target. Future Virol. 2010, 5, 593–606. 10.2217/fvl.10.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati M.; Alvarez K.; Assenberg R.; Baronti C.; Canard B.; Cook S.; Coutard B.; Decroly E.; de Lamballerie X.; Gould E. A.; et al. Structure and Functionality in Flavivirus NS-Proteins: Perspectives for Drug Design. Antiviral Res. 2010, 87, 125–148. 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel P.; Schiering N.; D’Arcy A.; Renatus M.; Kroemer M.; Lim S. P.; Yin Z.; Keller T. H.; Vasudevan S. G.; Hommel U. Structural Basis for the Activation of Flaviviral NS3 Proteases from Dengue and West Nile Virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Noble C. G.; Seh C. C.; Chao A. T.; Shi P. Y. Ligand-Bound Structures of the Dengue Virus Protease Reveal the Active Conformation. J. Virol. 2012, 86, 438–446. 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J.; Hansen G.; Nitsche C.; Klein C. D.; Zhang L.; Hilgenfeld R. Crystal Structure of Zika Virus NS2B-NS3 Protease in Complex with a Boronate Inhibitor. Science 2016, 353, 503–505. 10.1126/science.aag2419. [DOI] [PubMed] [Google Scholar]
- Chen X.; Yang K.; Wu C.; Chen C.; Hu C.; Buzovetsky O.; Wang Z.; Ji X.; Xiong Y.; Yang H. Mechanisms of Activation and Inhibition of Zika Virus NS2B-NS3 Protease. Cell Res. 2016, 26, 1260–1263. 10.1038/cr.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.; Ren J.; Nocadello S.; Rice A. J.; Ojeda I.; Light S.; Minasov G.; Vargas J.; Nagarathnam D.; Anderson W. F.; et al. Identification of Novel Small Molecule Inhibitors against NS2B/NS3 Serine Protease from Zika Virus. Antiviral Res. 2017, 139, 49–58. 10.1016/j.antiviral.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri R.; Kumar D.; Sharma N.; Uversky V. N. Intrinsically Disordered Side of the Zika Virus Proteome. Front. Cell. Infect. Microbiol. 2016, 6, 144 10.3389/fcimb.2016.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. M.; Uversky V. N.; Giri R. Molecular Recognition Features in Zika Virus Proteome. J. Mol. Biol. 2018, 430, 2372–2388. 10.1016/j.jmb.2017.10.018. [DOI] [PubMed] [Google Scholar]
- Murray C. L.; Jones C. T.; Rice C. M. Architects of Assembly: Roles of Flaviviridae Non-Structural Proteins in Virion Morphogenesis. Nat. Rev. Microbiol. 2008, 6, 699–708. 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni S.; Morrone A.; Giri R.; Brunori M. A Folding-after-Binding Mechanism Describes the Recognition between the Transactivation Domain of c-Myb and the KIX Domain of the CREB-Binding Protein. Biochem. Biophys. Res. Commun. 2012, 428, 205–209. 10.1016/j.bbrc.2012.09.112. [DOI] [PubMed] [Google Scholar]
- Giri R.; Morrone A.; Toto A.; Brunori M.; Gianni S. Structure of the Transition State for the Binding of c-Myb and KIX Highlights an Unexpected Order for a Disordered System. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 14942–14947. 10.1073/pnas.1307337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto A.; Giri R.; Brunori M.; Gianni S. The Mechanism of Binding of the KIX Domain to the Mixed Lineage Leukemia Protein and Its Allosteric Role in the Recognition of c-Myb. Protein Sci. 2014, 23, 962–969. 10.1002/pro.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto A.; Camilloni C.; Giri R.; Brunori M.; Vendruscolo M.; Gianni S. Molecular Recognition by Templated Folding of an Intrinsically Disordered Protein. Sci. Rep. 2016, 6, 21994 10.1038/srep21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patick A. K.; Potts K. E. Protease Inhibitors as Antiviral Agents. Clin. Microbiol. Rev. 1998, 11, 614–627. 10.1128/CMR.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.; Schiffer C.; Lee S.-K.; Swanstrom R.. Viral Protease Inhibitors. In Antiviral Strategies; Springer: Berlin, Heidelberg, 2009; pp 85–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L. Viral Proteases. Chem. Rev. 2002, 102, 4609–4626. 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- Yang C.-C.; Hsieh Y.-C.; Lee S.-J.; Wu S.-H.; Liao C.-L.; Tsao C.-H.; Chao Y.-S.; Chern J.-H.; Wu C.-P.; Yueh A. Novel Dengue Virus-Specific NS2B-NS3 Protease Inhibitor, BP2109, Discovered by a High-Throughput Screening Assay. Antimicrob. Agents Chemother. 2011, 55, 229–238. 10.1128/AAC.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Bock S.; Snitko M.; Berger T.; Weidner T.; Holloway S.; Kanitz M.; Diederich W. E.; Steuber H.; Walter C.; et al. Novel Dengue Virus NS2B-NS3 Protease Inhibitors. Antimicrob. Agents Chemother. 2015, 59, 1100–1109. 10.1128/AAC.03543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W.; Chik K. K.-H.; Yuan S.; Yip C. C.-Y.; Zhu Z.; Tee K.-M.; Tsang J. O.-L.; Chan C. C.-S.; Poon V. K.-M.; Lu G.; et al. Novel Antiviral Activity and Mechanism of Bromocriptine as a Zika Virus NS2B-NS3 Protease Inhibitor. Antiviral Res. 2017, 141, 29–37. 10.1016/j.antiviral.2017.02.002. [DOI] [PubMed] [Google Scholar]
- Lin K.-H.; Ali A.; Rusere L.; Soumana D. I.; Kurt Yilmaz N.; Schiffer C. A. Dengue Virus NS2B-NS3 Protease Inhibitors Exploiting the Prime Side. J. Virol. 2017, 91, JVI-00045 10.1128/JVI.00045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn T. T.; Thor K. B. Drug Repositioning: Identifying and Developing New Uses for Existing Drugs. Nat. Rev. Drug Discovery 2004, 3, 673–683. 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Leung D.; Schroder K.; White H.; Fang N. X.; Stoermer M. J.; Abbenante G.; Martin J. L.; Young P. R.; Fairlie D. P. Activity of Recombinant Dengue 2 Virus NS3 Protease in the Presence of a Truncated NS2B Co-Factor, Small Peptide Substrates, and Inhibitors. J. Biol. Chem. 2001, 276, 45762–45771. 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Brecher M.; Zhang J.; Li H. The Flavivirus Protease as a Target for Drug Discovery. Virol. Sin. 2013, 28, 326–336. 10.1007/s12250-013-3390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Brecher M.; Deng Y. Q.; Zhang J.; Sakamuru S.; Liu B.; Huang R.; Koetzner C. A.; Allen C. A.; Jones S. A.; et al. Existing Drugs as Broad-Spectrum and Potent Inhibitors for Zika Virus by Targeting NS2B/NS3 Interaction. Cell Res. 2017, 27, 1046–1064. 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B.; Parnell L. A.; Diamond M. S.; Mysorekar I. U. Inhibition of Autophagy Limits Vertical Transmission of Zika Virus in Pregnant Mice. J. Exp. Med. 2017, 214, 2303–2313. 10.1084/jem.20170957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F.; Lin Y.-S.; Huang N.-C.; Yu C.-Y.; Tsai W.-L.; Chen J.-J.; Kubota T.; Matsuoka M.; Chen S.-R.; Yang C.-S.; et al. Hydroxychloroquine-Inhibited Dengue Virus Is Associated with Host Defense Machinery. J. Interferon Cytokine Res. 2015, 35, 143–156. 10.1089/jir.2014.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber K.; Hom C.; Peng Chao C.; Shapiro D.; Ash J.; Aberientos C. Systematic Review of Hydroxychloroquine Use in Pregnant Patients with Autoimmune Diseases. Pediatric Rheumatol. 2009, 7, 9 10.1186/1546-0096-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costedoat-Chalumeau N.; Amoura Z.; Duhaut P.; Huong D. L. T.; Sebbough D.; Wechsler B.; Vauthier D.; Denjoy I.; Lupoglazoff J.-M.; Piette J.-C. Safety of Hydroxychloroquine in Pregnant Patients with Connective Tissue Diseases: A Study of One Hundred Thirty-Three Cases Compared with a Control Group. Arthritis Rheum. 2003, 48, 3207–3211. 10.1002/art.11304. [DOI] [PubMed] [Google Scholar]
- Motta M.; Tincani A.; Faden D.; Zinzini E.; Lojacono A.; Marchesi A.; Frassi M.; Biasini C.; Zatti S.; Chirico G. Follow-Up of Infants Exposed to Hydroxychloroquine Given to Mothers during Pregnancy and Lactation. J. Perinatol. 2005, 25, 86–89. 10.1038/sj.jp.7211208. [DOI] [PubMed] [Google Scholar]
- Yin Y.; Xu Y.; Su L.; Zhu X.; Chen M.; Zhu W.; Xia H.; Huang X.; Gong S. Epidemiologic Investigation of a Family Cluster of Imported ZIKV Cases in Guangdong, China: Probable Human-to-Human Transmission. Emerging Microbes Infect. 2016, 5, e100 10.1038/emi.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grischott F.; Puhan M.; Hatz C.; Schlagenhauf P. Non-Vector-Borne Transmission of Zika Virus: A Systematic Review. Travel Med. Infect. Disease 2016, 14, 313–330. 10.1016/j.tmaid.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Roberts N.; Martin J.; Kinchington D.; Broadhurst A.; Craig J.; Duncan I.; Galpin S.; Handa B.; Kay J.; Krohn A.; et al. Rational Design of Peptide-Based HIV Proteinase Inhibitors. Science 1990, 248, 358–361. 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- Bechtold C. M.; Patick A. K.; Alam M.; Greytok J.; Tino J. A.; Chen P.; Gordon E.; Ahmad S.; Barrish J. C.; Zahler R. Antiviral Properties of Aminodiol Inhibitors against Human Immunodeficiency Virus and Protease. Antimicrob. Agents Chemother. 1995, 39, 374–379. 10.1128/AAC.39.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. C.; Hu H. S.; Wu R. H.; Wu S. H.; Lee S. J.; Jiaang W. T.; Chern J. H.; Huang Z. S.; Wu H. N.; Chang C. M.; et al. A Novel Dengue Virus Inhibitor, BP13944, Discovered by High-Throughput Screening with Dengue Virus Replicon Cells Selects for Resistance in the Viral NS2B-NS3 Protease. Antimicrob. Agents Chemother. 2014, 58, 110–119. 10.1128/AAC.01281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Bock S.; Snitko M.; Berger T.; Weidner T.; Holloway S.; Kanitz M.; Diederich W. E.; Steuber H.; Walter C.; et al. Novel Dengue Virus NS2B-NS3 Protease Inhibitors. Antimicrob. Agents Chemother. 2015, 59, 1100–1109. 10.1128/AAC.03543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouretova J.; Hammamy M. Z.; Epp A.; Hardes K.; Kallis S.; Zhang L.; Hilgenfeld R.; Bartenschlager R.; Steinmetzer T. Effects of NS2B-NS3 Protease and Furin Inhibition on West Nile and Dengue Virus Replication. J. Enzyme Inhib. Med. Chem. 2017, 32, 712–721. 10.1080/14756366.2017.1306521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche C.; Holloway S.; Schirmeister T.; Klein C. D. Biochemistry and Medicinal Chemistry of the Dengue Virus Protease. Chem. Rev. 2014, 114, 11348–11381. 10.1021/cr500233q. [DOI] [PubMed] [Google Scholar]
- Ekins S.; Mietchen D.; Coffee M.; Stratton T. P.; Freundlich J. S.; Freitas-Junior L.; Muratov E.; Siqueira-Neto J.; Williams A. J.; Andrade C. Open Drug Discovery for the Zika Virus. F1000Research 2016, 5, 150 10.12688/f1000research.8013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruba N.; Ignacio J.; Martinez R.; Grzywa R.; Wysocka M. Substrate Profiling of Zika Virus NS2B-NS3 Protease. FEBS Lett. 2016, 590, 3459–3468. 10.1002/1873-3468.12443. [DOI] [PubMed] [Google Scholar]
- Nitsche C.; Zhang L.; Weigel L. F.; Schilz J.; Graf D.; Bartenschlager R.; Hilgenfeld R.; Klein C. D. Peptide–Boronic Acid Inhibitors of Flaviviral Proteases: Medicinal Chemistry and Structural Biology. J. Med. Chem. 2017, 60, 511–516. 10.1021/acs.jmedchem.6b01021. [DOI] [PubMed] [Google Scholar]
- Robin G.; Chappell K.; Stoermer M. J.; Hu S.-H.; Young P. R.; Fairlie D. P.; Martin J. L. Structure of West Nile Virus NS3 Protease: Ligand Stabilization of the Catalytic Conformation. J. Mol. Biol. 2009, 385, 1568–1577. 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- Hammamy M. Z.; Haase C.; Hammami M.; Hilgenfeld R.; Steinmetzer T. Development and Characterization of New Peptidomimetic Inhibitors of the West Nile Virus NS2B-NS3 Protease. ChemMedChem 2013, 8, 231–241. 10.1002/cmdc.201200497. [DOI] [PubMed] [Google Scholar]
- Randolph V. B.; Winkler G.; Stollar V. Acidotropic Amines Inhibit Proteolytic Processing of Flavivirus prM Protein. Virology 1990, 174, 450–458. 10.1016/0042-6822(90)90099-D. [DOI] [PubMed] [Google Scholar]
- Delvecchio R.; Higa L.; Pezzuto P.; Valadão A.; Garcez P.; Monteiro F.; Loiola E.; Dias A.; Silva F.; Aliota M.; et al. Chloroquine, an Endocytosis Blocking Agent, Inhibits Zika Virus Infection in Different Cell Models. Viruses 2016, 8, 322 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut W.; Zhang L.; Kasperkiewicz P.; Poreba M.; Hilgenfeld R.; Drąg M. Extended Substrate Specificity and First Potent Irreversible Inhibitor/activity-Based Probe Design for Zika Virus NS2B-NS3 Protease. Antiviral Res. 2017, 139, 88–94. 10.1016/j.antiviral.2016.12.018. [DOI] [PubMed] [Google Scholar]
- Law V.; Knox C.; Djoumbou Y.; Jewison T.; Guo A. C.; Liu Y.; Maciejewski A.; Arndt D.; Wilson M.; Neveu V.; et al. DrugBank 4.0: Shedding New Light on Drug Metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabuurs S. B.; Wagener M.; de Vlieg J. A Flexible Approach to Induced Fit Docking. J. Med. Chem. 2007, 50, 6507–6518. 10.1021/jm070593p. [DOI] [PubMed] [Google Scholar]
- Gupta S.; Suryanarayanan V.; Yadav S.; Singh S. K.; Saxena J. K. Delineating the Role of Ionic Interactions in Structural and Functional Integrity of B. malayi Guanylate Kinase. Int. J. Biol. Macromol. 2017, 98, 357–365. 10.1016/j.ijbiomac.2017.01.098. [DOI] [PubMed] [Google Scholar]
- Gupta S.; Yadav S.; Suryanarayanan V.; Singh S. K.; Saxena J. K. Investigating the Folding Pathway and Substrate Induced Conformational Changes in B. malayi Guanylate Kinase. Int. J. Biol. Macromol. 2017, 94, 621–633. 10.1016/j.ijbiomac.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Lenselink E. B.; Louvel J.; Forti A. F.; van Veldhoven J. P. D.; de Vries H.; Mulder-Krieger T.; McRobb F. M.; Negri A.; Goose J.; Abel R.; et al. Predicting Binding Affinities for GPCR Ligands Using Free-Energy Perturbation. ACS Omega 2016, 1, 293–304. 10.1021/acsomega.6b00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran D.; Sridhar J.; Suryanarayanan V.; Manimaran N. C.; Singh S. K. Molecular Modeling and Structural Analysis of nAChR Variants Uncovers the Mechanism of Resistance to Snake Toxins. J. Biomol. Struct. Dyn. 2017, 35, 1654–1671. 10.1080/07391102.2016.1190791. [DOI] [PubMed] [Google Scholar]
- Foloppe N.; Chen I.-J. Towards Understanding the Unbound State of Drug Compounds: Implications for the Intramolecular Reorganization Energy upon Binding. Bioorg. Med. Chem. 2016, 24, 2159–2189. 10.1016/j.bmc.2016.03.022. [DOI] [PubMed] [Google Scholar]
- Dror R. O.; Dirks R. M.; Grossman J. P.; Xu H.; Shaw D. E. Biomolecular Simulation: A Computational Microscope for Molecular Biology. Annu. Rev. Biophys. 2012, 41, 429–452. 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- Sharma N.; Murali A.; Singh S. K.; Giri R. Epigallocatechin Gallate, an Active Green Tea Compound Inhibits the Zika Virus Entry into Host Cells via Binding the Envelope Protein. Int. J. Biol. Macromol. 2017, 104, 1046–1054. 10.1016/j.ijbiomac.2017.06.105. [DOI] [PubMed] [Google Scholar]
- Miner J. J.; Cao B.; Govero J.; Smith A. M.; Fernandez E.; Cabrera O. H.; Garber C.; Noll M.; Klein R. S.; Noguchi K. K.; et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell 2016, 165, 1081–1091. 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R. S.; Kosoy O. L.; Laven J. J.; Velez J. O.; Lambert A. J.; Johnson A. J.; Stanfield S. M.; Duffy M. R. Genetic and Serologic Properties of Zika Virus Associated with an Epidemic, Yap State, Micronesia, 2007. Emerging Infect. Dis. 2008, 14, 1232–1239. 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.