Abstract
The study of the fecal metabolome is an important area of research to better understand the human gut microbiome and its impact on human health and diseases. However, there is a lack of work in examining the impact of storage and processing conditions on the metabolite levels of fecal water. Furthermore, there is no universal protocol used for the storage of fecal samples and preparation of fecal water. The objective of the current study was to examine the impact of different storage conditions on fecal samples prior to metabolite extraction. Fecal samples obtained from nine healthy individuals were processed under different conditions: (1) fresh samples prepared immediately after collection, (2) fecal samples stored at 4 °C for 24 h prior to processing, and (3) fecal samples stored at −80 °C for 24 h prior to processing. All samples were analyzed using NMR spectroscopy, multivariate statistical analysis, and repeated measures ANOVA. Samples which were frozen at −80 °C prior to extraction of the metabolites exhibited an increase in the number of metabolites including branched-chain amino acids, aromatic amino acids, and tricarboxylic acid cycle intermediates. Storage of fecal samples at 4 °C ensured higher fidelity to freshly processed samples leading to the recommendation that fecal samples should not be frozen prior to extraction of fecal water. Furthermore, the work highlights the need to standardize sample storage of fecal samples to allow for the accurate study of the fecal metabolome.
Introduction
Several recent papers have highlighted the importance of the gut microbiome and its potential role in the human body in functions such as immune response, physiology, and metabolism.1−6 Imbalance in the gut microbiota, also known as dysbiosis, has been linked to various diseases including inflammatory bowel disease, atopy, arthritis, certain cancers, and obesity.7−11 In recent years, the analysis of metabolites in fecal matter has become an important aspect of the study of functional aspects of the gut microbiome and its relationship with human health. Metabolomics has emerged as an important approach for the measurement of metabolites. Using metabolomics, one can identify and quantify many small molecules or metabolites in biological samples such as urine, plasma, or feces using analytical techniques such as nuclear magnetic resonance (NMR) and mass spectrometry coupled to chromatography.12,13 Examination of these metabolites allows detailed metabolic phenotyping and examination of altered pathways under certain conditions.14,15 To this end, application of metabolomics to fecal samples has enhanced our understanding of certain conditions and provided evidence for the link between diet and gut microbiota activity.16−18
However, despite the increasing application of metabolomics to fecal samples, there is a lack of studies in examining the stability of fecal samples. Metabolites and microbial DNA can be degraded in a fecal sample through oxidation, enzymatic degradation, and hydrolysis which can occur during fecal collection and storage before the sample is prepared for analysis.19,20 Owing to this degradation, it is essential to have an optimized method for fecal collection and storage in order to reduce degradation of metabolites and DNA which would allow for more accurate and reproducible results in the area of gut microbiome investigation.20 There are few studies available which investigate the different collection and storage methods and their impact on metabolite and bacteria levels.
Gratton et al. demonstrated that storing a fecal sample over time at different temperatures before processing for analysis has a large impact on the metabolic profiles of human feces. An increase in branched-chain amino acids (BCAAs) and aromatic amino acids was reported after a fecal sample underwent a freeze–thaw cycle before extracting fecal water.21 This information has supported a previous paper by Saric et al. which has also demonstrated an increase in BCAAs after freeze–thaw cycles of the fecal sample prior to fecal water extraction.22 Furthermore, a number of studies have illustrated the effects of different sample-processing procedures on the metabolite levels. An overview of the impact of steps such as homogenization, filtration, centrifugation, and solvent extraction has been previously presented.23−25
The impact of storage conditions on bacterial community levels has been examined. Roesch et al. demonstrated a 10% change in bacterial community levels in a fecal sample when stored at −80 °C at different time points post receiving the sample.26 However, other studies have reported no significant difference in results because of differing storage conditions on bacterial community levels in fecal samples, and27,28 others have recommended a collection protocol to minimize the impact.29 With respect to the fecal metabolome, there is no such consensus, and further studies are warranted.
In summary, analyzing the gut microbiome has become an integral part of many human studies, and assessment of the fecal water metabolome can yield valuable information. However, more work is needed to examine the impact of storage and processing procedures on the metabolite levels in fecal water. The objective of this study was to examine the impact of different storage conditions prior to metabolite extraction on the fecal water metabolome. The work highlights the need for standardized procedures.
Results
A total of nine healthy participants were included in this study including four males and five females. Two of the individuals supplied samples on two separate occasions resulting in a total of 11 sample sets. The mean age was 34 years and participants had a mean body mass index (BMI) of 24.1 ± 2.78 kg/m2. A summary of the baseline characteristics of the participants is presented in Table 1.
Table 1. Demographic Data of Participants Included in the Studya.
| characteristics | male (n = 4) | female (n = 5) |
|---|---|---|
| age (years) | 34 ± 9 | 35 ± 14 |
| weight (kg) | 77.55 ± 7.91 | 66.00 ± 12.24 |
| height (m) | 1.76 ± 0.08 | 1.68 ± 0.08 |
| BMI (kg/m2) | 25.1 ± 2.6 | 23.2 ± 2.9 |
| waist to hip ratio | 0.87 ± 0.05 | 0.82 ± 0.09 |
All values shown are mean ± SD.
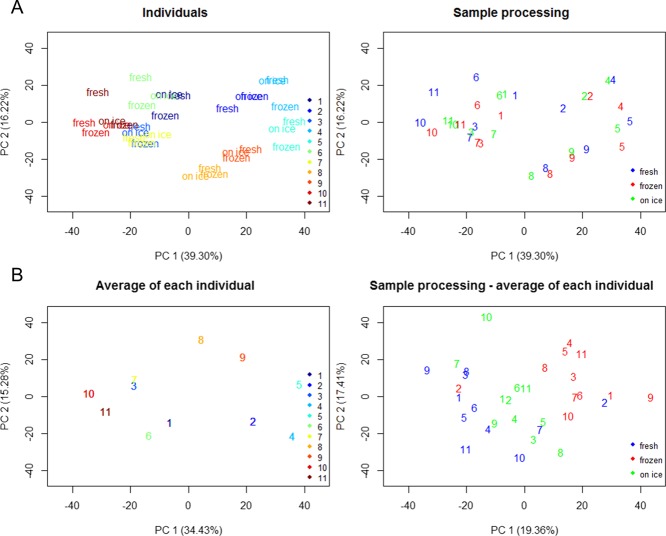
Examination of the principal component analysis (PCA) revealed that the interindividual variation was the dominant source of variation on the dataset. The samples of the individuals were grouped together in the PCA scores plot (Figure 1A). To explore the impact of storage, we employed a row-wise centering of the data and this resulted in separation of the samples according to the storage type (Figure 1 B).
Figure 1.
PCA score plots of samples (A) colored by individual sample sets (top left) and the different sample procedures (top right) used in the study. Average of each individual sample set (B) was performed, plotted (bottom left), and subtracted to each sample (bottom right). Individual sample sets were labeled with numbers (1–11), samples sets 8 and 9 are from the same individual, and samples sets 10 and 11 are from the same individual.
To examine the impact of the storage further, a univariate analysis approach was employed. A total of 14 compounds from the fecal water analysis were significantly different across the three storage conditions. Significant metabolites from repeated measures ANOVA corrected by the false discovery rate (FDR) are presented in Table 2. Interestingly, the metabolites were predominantly increased following freezing at −80 °C prior to preparation of fecal water (Figure 2). The sample stored on ice for 24 h had the highest fidelity to the freshly prepared samples. Closer examination of the significantly different metabolites revealed that the BCAAs, aromatic amino acids, Krebs cycle intermediates, and monosaccharides were found at higher levels in fecal water prepared from frozen samples. However, the short chain fatty acid (SCFA), butyrate, was lower in fecal water from frozen/defrosted, and on ice samples. Figure 3 shows the typical spectra of fecal water analyzed from fresh, frozen, and on ice samples.
Table 2. Differential Fecal Metabolites between the Different Conditions of Sample Storagea.
| metabolite | P-value | FDR | description |
|---|---|---|---|
| aspartate | <0.001 | 0.008 | Krebs cycle: oxaloacetate transamination |
| butyrate | <0.001 | 0.017 | short chain fatty acid |
| fructose | <0.001 | 0.008 | monosaccharide |
| fumarate | <0.001 | 0.004 | dicarboxylic acid related to the Krebs cycle |
| glucose | <0.001 | 0.008 | monosaccharide |
| glutamate | <0.001 | 0.008 | proteinogenic amino acid related to cellular metabolism. |
| isobutyrate | 0.001 | 0.017 | BCAA-derivative |
| isoleucine | <0.001 | 0.004 | BCAA |
| leucine | <0.001 | 0.004 | BCAA |
| nicotinate | <0.001 | 0.004 | energy metabolism in the living cell and DNA repair |
| phenylalanine | <0.001 | 0.009 | aromatic amino acid |
| threonine | <0.001 | 0.008 | alpha amino acid |
| tyrosine | <0.001 | 0.008 | aromatic amino acid |
| valine | <0.001 | 0.004 | BCAA |
BCAA, branched-chain amino acid. P-values of repeated measures ANOVA were corrected for multiple comparisons using the Benjamini–Hochberg (FDR) procedure.
Figure 2.
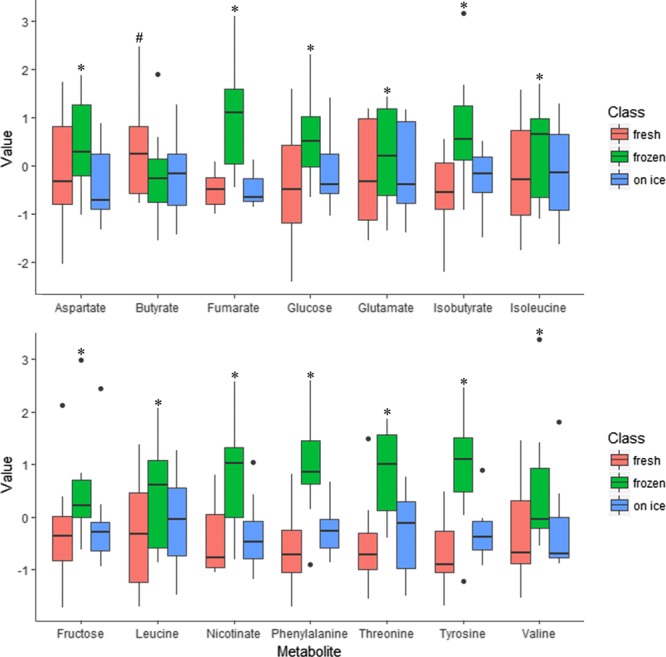
Box plots of significant metabolites from the resulting analysis of fecal water from fresh, frozen/defrosted, and on ice samples. * denotes significant differences between frozen and fresh samples and frozen and on ice samples (Bonferroni post hoc). # denotes significant difference between fresh and frozen sample and fresh and on ice samples (Bonferroni post hoc). All p-values were FDR adjusted.
Figure 3.
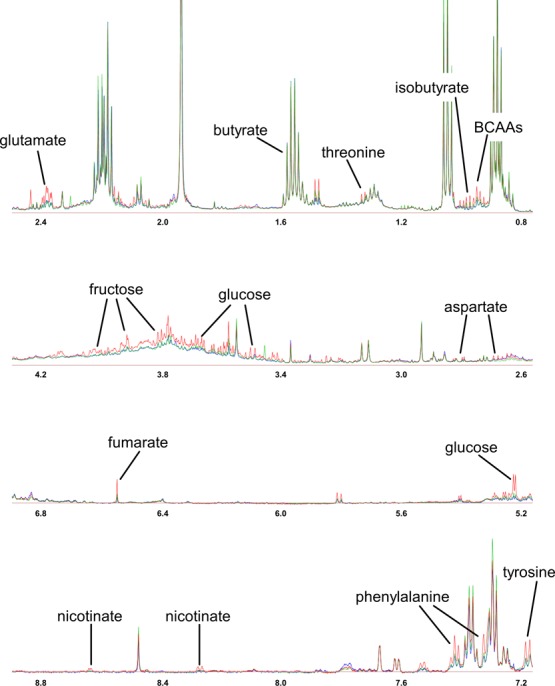
600 MHz 1H NMR spectra of fecal water colored by sample storage: analysis of fecal water from fresh (blue), frozen (red), and on ice (green) samples. Assignations of significant metabolites (FDR < 0.05) are presented. BCAAs: valine, leucine, and isoleucine.
Discussion
The present findings point to differences in levels of several metabolites following different sample storage conditions prior to metabolite extraction. Overall, BCAAs and derivatives, aromatic amino acids, SCFAs, Krebs cycle intermediates, and monosaccharides were sensitive to the storage procedure. Our study highlights the urgent need to standardize processing parameters for the preparation of fecal water in order to obtain meaningful data.
This current lack of standardization is aggravated by the fact that fecal analysis entails a higher sensitivity to differences in study design than most other biofluid analyses.25 Previous work has demonstrated that insufficient sample collection can introduce bias because of the heterogeneity of fecal samples. Typically, despite fecal samples being highly heterogeneous, the fecal sampling method is to take a small portion and extract the metabolites.25 Gratton and colleagues observed that fecal water extracted from 15 g of feces contained different concentrations of certain amino acids and carboxylic acids compared with fecal water extracted from a lower amount of 350 mg of fecal samples.21 Furthermore, proper sample storage is essential to avoid additional bias post sampling. Both microbial and enzymatic activities affect the fecal metabolome. Hence, both storage temperature of the fecal samples and time before preparation of fecal water can have an impact on the metabolome and on the microbial composition.21,30 Based on the findings of the present study, sample storage prior to the extraction of the metabolites is critical for the final levels of fecal metabolites. With the exception of butyrate, in which the highest levels were found in fresh fecal samples, metabolite levels increased in samples prepared from frozen fecal matter.
Similar results were observed in studies by Gratton et al. and Saric et al.21,22 In these previous studies, the relative concentrations of BCAAs and aromatic amino acids were elevated in the fecal samples following a freezing cycle.21,22 In our study, the BCAAs and aromatic amino acids phenylalanine and tyrosine were increased in samples prepared from frozen fecal matter. The impact of freezing was not limited to BCAAs and aromatic amino acids, and indeed significant changes were observed for other metabolites such as fumarate, isobutyrate, nicotinate, and aspartate. Butyrate levels were decreased in samples prepared from fecal samples kept on ice and frozen samples. However, there is no simple interpretation for the unique decrease of this SCFA. The same pattern consisting of an increase of BCAAs and a decrease of SCFAs was observed in lyophilized feces compared with wet feces.22 In this study, the authors suggested that the decreased relative concentrations of SCFAs after the lyophilization process were connected to the instability and variability of these compounds over time.22 This would support the decrease observed in the present study.
Our findings support the call for care in interpretation of metabolites following freeze–thaw cycles of faeces and support the concept that these alterations are not because of improved recovery of fecal metabolites.25 Previous work suggested that these alterations in the fecal metabolome may be related to the lysis of cells causing the release of microbial intracellular contents.21,25 When a fecal sample is frozen, ice crystals form within the cells and these crystals damage the cell walls. Once the sample is thawed, the damaged cell walls allow the release of intracellular contents.31,32 This hypothesis is supported by the fact that several metabolites such as glutamate, BCAAs, Krebs cycle intermediates, and glucose increased only following freezing of the fecal sample. Taken together, it is evident that the procedure used for sample preparation can have an impact on the metabolic profile and care is needed when interpreting results from the fecal water metabolome. Interestingly, our results highlight that avoidance of freezing fecal samples by storage on ice for 24 h increased the fidelity to the freshly processed samples. For many large studies, preparation of fecal water from fresh samples will not be possible, and our results support the potential of storage on ice for 24 h until extraction of fecal water is possible.
The present study has a number of strengths and limitations worth mentioning. The same fecal sample was used for each of the three storage conditions allowing us to disentangle the effects from the larger interindividual variation. Furthermore, the study design enabled comparison to a fresh sample processed immediately following collection. A limitation of the current study is the small sample size; however, it is worth noting that the sample size is larger than previous work in the area.21
Conclusions
Analysis of the fecal water metabolome has the potential to enhance our understanding of the importance of the gut microbiome in a number of health and disease conditions.33,34 However, the lack of standardization in the processing of fecal samples may lead to misinterpretation of the data. The present findings support the need for a standardized and robust sample preparation for fecal water. Importantly, freezing of solid fecal matter prior to fecal water extraction should be avoided.
Materials and Methods
Participants and Sample Collection
Nine participants donated a total of eleven fecal samples. Participants were healthy males or females aged between 18 and 60 years of age with a BMI range of >18.5 and <29 kg/m2. Exclusion criteria included smokers, anyone taking medication or nutritional supplements, pregnant or lactating, or diagnosed with a medical disease. The study was approved by the Human Research Ethics Committee at University College Dublin (LS_16_91_Gibbons_Brennan) and written informed consent was obtained from each participant prior to participation in the study.
Each participant received a sample collection pack consisting of a plastic container, two zip-lock freezer bags, gloves, and two blue plastic bags. Subjects were asked to save the full bowel movement, record the time when the sample was produced, not contaminate the sample with urine or toilet paper, and store the sample in a cool environment prior to attending the visit center. Participants were asked to collect the sample on the day of the study visit to ensure that it was as fresh as possible. The samples were immediately placed on ice using different conditions as described below.
Sample Processing
Each sample was weighed for total weight before being processed. Subsamples from each fecal sample were taken. From these subsamples, the following three conditions were examined. The first sample was processed immediately to obtain a fresh fecal water sample. The second sample was kept for 24 h on ice, and the third sample was placed in the −80 °C freezer for 24 h. The following day, the frozen sample was thawed and the fecal water extraction was performed.
Each subsample was processed in the same way to obtain a fecal water sample regardless of storage conditions. In short, 5 g of the sample was weighed into a 50 mL conical tube and 2× w/v of sterile phosphate-buffered saline was added. The sample was homogenized manually using a tissue grinder and centrifuged at 2654×g for 1 h at 4 °C. Following this, the samples were aliquoted and centrifuged a further two times at 16 000×g for 30 min at 4 °C before filtering (a 0.7 μm pore Minisart Filter followed by a 0.2 μm pore Whatmann filter) and freezing at −80 °C until NMR analysis.
NMR Spectroscopy
To prepare the samples for NMR spectroscopy, 60 μL of D2O and 10 μL of sodium trimethylsilyl propionate-[2,2,3,3-2H4] (TSP) (0.05 g/mL) were added to 540 μL of fecal water. NMR spectra were acquired on a 600 MHz Varian NMR spectrometer by using a CPMG pulse sequence at 25 °C. Spectra were acquired with 16 384 data points and 128 scans. Water suppression was achieved during the relaxation delay of 3 s. All 1H NMR spectra were referenced to TSP at 0.0 parts per million (ppm) and processed manually with Chenomx NMR Suite (version 7.5) by using a line broadening of 0.2 Hz, followed by phase correction and baseline correction. Metabolites were identified by Chenomx NMR Suite. Spectra were then bucketed in domains of 0.005 ppm excluding both TSP and water regions. Data were normalized to the sum of the spectral integral.
Metabolite identification was performed using Chenomx NMR Suite 8.3 Profiler (Chenomx Inc., Edmonton, Canada). Further confirmations to the assignment of proton peaks were provided by comparing the chemical shifts with those available in the Human Metabolome Database (http://www.hmdb.ca).
Statistical Analyses
The dataset containing 1780 variables was log transformed prior statistical analysis. PCA on samples was carried out on all samples to obtain an overview of the data. Because we analyzed the same sample per individual using different storage conditions, the interindividual variability may obscure the actual issue of interest in a multivariate approach.35 Therefore, for visualization purposes, the standardized projection of the individual effects was used by a row-wise mean centering36 of samples, subtracting the within-subject variation for each individual.37 Then, a new “multilevel” PCA score plot was performed exhibiting the variation because of the sample class.36 To assess differences between the three conditions of sample storage, repeated measures ANOVA followed by Bonferroni post hoc comparisons were carried out on the dataset. Benjamini–Hochberg FDR was used to correct analysis for multiple comparisons.38 Statistical significance was considered at a P-value of <0.05. All statistical analyses were performed in the open source statistical programming environment R v3.3.1.
Acknowledgments
The authors acknowledge funding from the European Research Council (647783).
The authors declare no competing financial interest.
References
- Clemente J. C.; Ursell L. K.; Parfrey L. W.; Knight R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio V.; Salvatore F. The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 2015, 451, 97–102. 10.1016/j.cca.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Gerritsen J.; Smidt H.; Rijkers G. T.; de Vos W. M. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011, 6, 209–240. 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J.; Li R.; Raes J.; Arumugam M.; Burgdorf K. S.; Manichanh C.; Nielsen T.; Pons N.; Levenez F.; Yamada T.; Mende D. R.; Li J.; Xu J.; Li S.; Li D.; Cao J.; Wang B.; Liang H.; Zheng H.; Xie Y.; Tap J.; Lepage P.; Bertalan M.; Batto J.-M.; Hansen T.; Le Paslier D.; Linneberg A.; Nielsen H. B.; Pelletier E.; Renault P.; Sicheritz-Ponten T.; Turner K.; Zhu H.; Yu C.; Li S.; Jian M.; Zhou Y.; Li Y.; Zhang X.; Li S.; Qin N.; Yang H.; Wang J.; Brunak S.; Doré J.; Guarner F.; Kristiansen K.; Pedersen O.; Parkhill J.; Weissenbach J.; Bork P.; Ehrlich S. D.; Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J.; Gordon J. I. The core gut microbiome, energy balance and obesity. J. Physiol. 2009, 587, 4153–4158. 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. J.; Guinane C. M.; O’Toole P. W.; Cotter P. D. Beneficial modulation of the gut microbiota. FEBS Lett. 2014, 588, 4120–4130. 10.1016/j.febslet.2014.03.035. [DOI] [PubMed] [Google Scholar]
- Bull M. J.; Plummer N. T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [PMC free article] [PubMed] [Google Scholar]
- DeGruttola A. K.; Low D.; Mizoguchi A.; Mizoguchi E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. 10.1097/mib.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navab-Moghadam F.; Sedighi M.; Khamseh M. E.; Alaei-Shahmiri F.; Talebi M.; Razavi S.; Amirmozafari N. The association of type II diabetes with gut microbiota composition. Microb. Pathog. 2017, 110, 630–636. 10.1016/j.micpath.2017.07.034. [DOI] [PubMed] [Google Scholar]
- Shreiner A. B.; Kao J. Y.; Young V. B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. 10.1097/mog.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Yao M.; Lv L.; Ling Z.; Li L. The Human Microbiota in Health and Disease. Engineering 2017, 3, 71–82. 10.1016/j.eng.2017.01.008. [DOI] [Google Scholar]
- Astarita G.; Langridge J. An Emerging Role for Metabolomics in Nutrition Science. J. Nutrigenetics Nutrigenomics 2013, 6, 181–200. 10.1159/000354403. [DOI] [PubMed] [Google Scholar]
- Gebregiworgis T.; Powers R. Application of NMR metabolomics to search for human disease biomarkers. Comb. Chem. High Throughput Screen. 2012, 15, 595–610. 10.2174/138620712802650522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin K. A.; Moore S. C.; Sampson J. N.; Huang W.-Y.; Xiao Q.; Stolzenberg-Solomon R. Z.; Sinha R.; Cross A. J. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am. J. Clin. Nutr. 2014, 100, 208–217. 10.3945/ajcn.113.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitó M.; Konstantinidou V. Nutritional Genomics and the Mediterranean Diet’s Effects on Human Cardiovascular Health. Nutrients 2016, 8, 218. 10.3390/nu8040218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K.; Chang H.-W.; Yan D.; Lee K. M.; Ucmak D.; Wong K.; Abrouk M.; Farahnik B.; Nakamura M.; Zhu T. H.; Bhutani T.; Liao W. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernocchi P.; Del Chierico F.; Putignani L. Gut Microbiota Profiling: Metabolomics Based Approach to Unravel Compounds Affecting Human Health. Front. Microbiol. 2016, 7, 1144. 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. P.; Gratz S. W.; Sheridan P. O.; Flint H. J.; Duncan S. H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013, 69, 52–60. 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Gorzelak M. A.; Gill S. K.; Tasnim N.; Ahmadi-Vand Z.; Jay M.; Gibson D. L. Methods for Improving Human Gut Microbiome Data by Reducing Variability through Sample Processing and Storage of Stool. PloS One 2015, 10, e0134802 10.1371/journal.pone.0134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton J.; Phetcharaburanin J.; Mullish B. H.; Williams H. R. T.; Thursz M.; Nicholson J. K.; Holmes E.; Marchesi J. R.; Li J. V. Optimized Sample Handling Strategy for Metabolic Profiling of Human Feces. Anal. Chem. 2016, 88, 4661–4668. 10.1021/acs.analchem.5b04159. [DOI] [PubMed] [Google Scholar]
- Saric J.; Wang Y.; Li J.; Coen M.; Utzinger J.; Marchesi J. R.; Keiser J.; Veselkov K.; Lindon J. C.; Nicholson J. K.; Holmes E. Species Variation in the Fecal Metabolome Gives Insight into Differential Gastrointestinal Function. J. Proteome Res. 2008, 7, 352–360. 10.1021/pr070340k. [DOI] [PubMed] [Google Scholar]
- Yen S.; Bolte E.; Aucoin M.; Allen-Vercoe E. Metabonomic Evaluation of Fecal Water Preparation Methods: The Effects of Ultracentrifugation. Curr. Metabolomics 2018, 6, 57–63. 10.2174/2213235x05666161226164121. [DOI] [Google Scholar]
- Deda O.; Gika H. G.; Wilson I. D.; Theodoridis G. A. An overview of fecal sample preparation for global metabolic profiling. J. Pharm. Biomed. Anal. 2015, 113, 137–150. 10.1016/j.jpba.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Karu N.; Deng L.; Slae M.; Guo A. C.; Sajed T.; Huynh H.; Wine E.; Wishart D. S. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Anal. Chim. Acta 2018, 1030, 1–24. 10.1016/j.aca.2018.05.031. [DOI] [PubMed] [Google Scholar]
- Roesch L. F. W.; Casella G.; Simell O.; Krischer J.; Wasserfall C. H.; Schatz D.; Atkinson M. A.; Neu J.; Triplett E. W. Influence of fecal sample storage on bacterial community diversity. Open Microbiol. J. 2009, 3, 40–46. 10.2174/1874285800903010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedjo D. I.; Jonkers D. M. A. E.; Savelkoul P. H.; Masclee A. A.; van Best N.; Pierik M. J.; Penders J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PloS One 2015, 10, e0126685 10.1371/journal.pone.0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. L.; Zhou N.; Gordon J. I.; Knight R.; Fierer N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 2010, 307, 80–86. 10.1111/j.1574-6968.2010.01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona S.; Eck A.; Cassellas M.; Gallart M.; Alastrue C.; Dore J.; Azpiroz F.; Roca J.; Guarner F.; Manichanh C. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012, 12, 158. 10.1186/1471-2180-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo J. M.; Leong L. E.; Rogers G. B. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015, 5, 16350. 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhoff Z. D. V. L.; Franco T. T.; Roberto I. C. A study of cell disruption of Candida mogii by glass bead mill for the recovery of xylose reductase. Sep. Purif. Technol. 2008, 63, 706–709. 10.1016/j.seppur.2008.06.019. [DOI] [Google Scholar]
- Supriya D. S.; Lele S. S. Application of evolutionary optimization technique in maximizing recovery of L-asparginase from E. caratovora MTCC 1428. Global J. Biotechnol. Biochem. Res. 2010, 5, 97–108. [Google Scholar]
- Amiot A.; Dona A. C.; Wijeyesekera A.; Tournigand C.; Baumgaertner I.; Lebaleur Y.; Sobhani I.; Holmes E. 1H NMR Spectroscopy of Fecal Extracts Enables Detection of Advanced Colorectal Neoplasia. J. Proteome Res. 2015, 14, 3871–3881. 10.1021/acs.jproteome.5b00277. [DOI] [PubMed] [Google Scholar]
- Barton W.; Penney N. C.; Cronin O.; Garcia-Perez I.; Molloy M. G.; Holmes E.; Shanahan F.; Cotter P. D.; O’Sullivan O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. 10.1136/gutjnl-2016-313627. [DOI] [PubMed] [Google Scholar]
- Westerhuis J. A.; van Velzen E. J. J.; Hoefsloot H. C. J.; Smilde A. K. Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics 2010, 6, 119–128. 10.1007/s11306-009-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A.; Cloarec O.; Holmes E.; Nicholson J. K.; Lindon J. C. Scaling and Normalization Effects in NMR Spectroscopic Metabonomic Data Sets. Anal. Chem. 2006, 78, 2262–2267. 10.1021/ac0519312. [DOI] [PubMed] [Google Scholar]
- Kemsley E. K.; Le Gall G.; Dainty J. R.; Watson A. D.; Harvey L. J.; Tapp H. S.; Colquhoun I. J. Multivariate techniques and their application in nutrition: a metabolomics case study. Br. J. Nutr. 2007, 98, 1. 10.1017/s0007114507685365. [DOI] [PubMed] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Source J. Roy. Stat. Soc. Seri. B Methodol. 1995, 57, 289–300. [Google Scholar]