Abstract
Objectives
mcr-1 gene has been widely reported in the world. This study aimed to analyze the prevalence and molecular characteristics of mcr-1 gene in Salmonella typhimurium from Quzhou People’s Hospital.
Materials and methods
A total of 62 S. typhimurium isolates were isolated and preserved in our laboratory from 2007 to 2016. PCR was used to screen plasmid-mediated colistin resistance gene, mcr-1. For mcr-1-positive isolates, susceptibilities to colistin and other antibiotics were assessed using broth microdilution or agar dilution methods. The genetic location of mcr-1 was determined by analysis of pulsed-field gel electrophoresis profiles of S1-digested genomic DNA and subsequent Southern blot hybridization. The multi-locus sequence type and other drug resistance genes found in the mcr-1-positive isolates were analyzed by performing whole genome sequencing. Genetic environment of mcr-1 gene was analyzed by RAST and Easyfig.
Results
A total of three S. typhimurium isolates were identified to be mcr-1 positive, with the prevalence rate of 4.8% (3/62). The minimum inhibitory concentration values of colistin for all these isolates were 8 µg/mL. The three mcr-1-positive isolates carried mcr-1 gene on two different types of plasmids having the sizes of ~54.7–78.2 kb and 310.1 kb, respectively. All the three isolates belonged to ST34 and carried various resistant genes.
Conclusion
Colistin-resistant, mcr-1-positive S. typhimurium isolates belonging to ST34 have been isolated from Quzhou People’s Hospital. Surveillance needs to be strengthened to identify colistin resistance and prevent the spread of drug-resistant bacteria in the hospital.
Keywords: Salmonella typhimurium, colistin, mcr-1
Introduction
Polymyxins, including polymyxin B and E (colistins), are generally regarded as the last-resort therapy for infections caused by multidrug resistant (MDR) Gram-negative bacilli, especially carbapenem-resistant Enterobacteriaceae.1,2 In recent years, polymyxin resistance has gained wide attention in the field of global medical research due to the increasing reports of polymyxin-resistant bacteria. In late 2015, Chinese researchers reported the first plasmid-mediated colistin resistance gene, mcr-1,3 and since then, it has been reported in other continents as well, including Asia, Europe, America, Africa, and Australia,4 as well as in several provinces (Zhejiang, Sichuan, Beijing, Guangdong, etc) in China.5–9 The discovery of mcr-1 gene makes it possible for the horizontal transmission of polymyxin resistance in Enterobacteriaceae, which may in turn lead to a rapid increase in polymyxin resistance. So far, mcr-1 gene has been found only in Enterobacteriaceae, such as Escherichia coli, Klebsiella pneumoniae, Salmonella spp., Enterobacter spp., Kluyvera ascorbata, Raoultella ornithinolytica, Citrobacter braakii, etc.4,6,10,11
Studies focusing on the mcr-1 gene-carrying E. coli and K. pneumonia strains are common, while the studies on prevalence and molecular characteristics of the mcr-1 gene in Salmonella spp. are still lacking. Salmonella typhimurium is an important zoonotic pathogen that usually causes foodborne diseases, and humans, especially infants, are highly susceptible to this infection.12 Furthermore, the occurrence of MDR isolates of S. typhimurium have become common now and have had great impact on the effectiveness of current strategies to prevent and control foodborne diseases.13 This study aims to analyze the prevalence and molecular characteristics of mcr-1 gene in S. typhimurium strains isolated from Quzhou People’s Hospital and provide a basis for the prevention and control of polymyxin resistance in the hospital strains of S. typhimurium.
Materials and methods
Bacterial isolates
A total of 62 S. typhimurium isolates were collected from Quzhou People’s Hospital from 2007 to 2016. All the strains were isolated from patients who visited the hospital during the period. Isolates were identified using the automated Vitek 2 system (BioMérieux, Marcy l’Etoile, France).
Screening for mcr-1 gene
All the isolates were screened for the presence of mcr-1 gene by PCR with primers mcr-1-F (5′-GCTCGGTCAGTC-CGTTTG-3′) and mcr-1-R (5′-GAATGCGGTGCG-GTCTTT-3′), and positive amplicons were subsequently sequenced. All the sequences were further analyzed on the BLAST server (http://blast.ncbi.nlm.nih.gov/).
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) of colistin and other ten antimicrobial agents (including levofloxacin, ciprofloxacin, cefotaxime, cefepime, cefoperazone/sulbactam, imipenem, meropenem, amikacin, tigecycline, and aztreonam) were determined for the mcr-1-positive isolates by the broth microdilution method, while the MICs of fosfomycin and sulfamethoxazole/trimethoprim were evaluated by the agar dilution method according to the guidelines of Clinical and Laboratory Standards Institute (CLSI). The results were interpreted in accordance with the CLSI and European Committee on Antimicrobial Susceptibility Testing (for colistin, tigecycline, and fosfomycin) breakpoints (http://www.eucast. org/clinical_breakpoints).
Southern blot analysis
To determine the plasmid location of the mcr-1 gene, genomic DNA digested with S1 nuclease (TaKaRa, Tokyo, Japan) was electrophoresed on a CHEF-mapper XA pulsed-field gel electrophoresis system (Bio-Rad Laboratories Inc., Hercules, CA, USA) for 22 hours at 14°C with run conditions of 6 V/cm and pulse times from 2.16 seconds to 63.8 seconds. The DNA fragments were transferred to a positively charged nylon membrane (EMD Millipore, Billerica, MA, USA) and then hybridized with a digoxigenin-labeled mcr-1-specific probe. The fragments were then detected using a NBT/BCIP color detection kit (Hoffman-La Roche Ltd., Basel, Switzerland).
Whole genome sequencing (WGS)
The total genomes of the mcr-1-positive isolates were extracted and then sequenced on an Illumina-Hiseq™ 2000 sequencing system (Illumina Inc, San Diego, CA, USA) using a paired-end 2×100 bp protocol. Sequence reads were assembled using CLC Genomics Workbench software package (CLC Bio 8.0, Aarhus, Denmark). The multi-locus sequence type (MLST) and other resistant genes were analyzed on the website of Center of Genomics Epidemiology (http://www.genomicepidemiology.org/). Gene prediction and annotation were performed using RAST (http://rast.nmpdr.org/). Sequence comparison was performed using BLAST (http://blast.ncbi.nlm.nih.gov), and physical maps were generated by using Easyfig.14
Ethics approval
The clinical isolates were part of the routine hospital laboratory procedure. The Ethics Committee of the Quzhou People’s Hospital approved this study as it mainly focused on bacteria, and not the patients.
Results
Epidemiology data and characteristics of mcr-1-positive isolates
A total of three mcr-1-positive isolates of S. typhimurium were identified (16–541, 16–573, and 16–623), with the prevalence rate of 4.8% (3/62). All the three strains were isolated from feces of patients with infectious diarrhea diseases in 2016. Two patients were infants and the other was 15 years old (male =1, female =2). No underlying diseases were found in all the three patients, and all of them were eventually cured (Table 1).
Table 1.
Characteristics of mcr-1-positive isolates
| Isolates | Age | Sex | Living place | Underlying disease | Outcome | Department | Samples | Isolated date |
|---|---|---|---|---|---|---|---|---|
| 16–541 | 15 years | Female | Village | None | Cured | Gastroenterology department | Feces | 8/27/2016 |
| 16–573 | 8 months | Female | City | None | Cured | Pediatric department | Feces | 9/20/2016 |
| 16–623 | 10 months | Male | Village | None | Cured | Pediatric department | Feces | 11/2/2016 |
Antimicrobial susceptibility of mcr-1-positive isolates
All the three mcr-1-positive S. typhimurium strains were resistant to colistin, with the MIC of 8 µg/mL. Moreover, they were all resistant to the third/fourth-generation cephalosporins (cefotaxime and cefepime) and sulfamethoxazole/trimethoprim, while imipenem, meropenem, amikacin, tigecycline, and fosfomycin showed great activity against these mcr-1-positive isolates (Table 2).
Table 2.
MICs of mcr-1-positive Salmonella typhimurium isolates to colistin and other antibiotics (μg/mL)
| Antibiotics | 16–541 | 16–573 | 16–623 |
|---|---|---|---|
|
| |||
| Colistin | 8 (R) | 8 (R) | 8 (R) |
| Levofloxacin | 4 (I) | 0.25 (S) | 4 (I) |
| Ciprofloxacin | 8 (R) | 0.125 (S) | 8 (R) |
| Cefotaxime | >256 (R) | >256 (R) | >256 (R) |
| Cefepime | 128 (R) | 64 (R) | 128 (R) |
| Cefoperazone/sulbactam | 32 (I) | 16 (S) | 32 (I) |
| Imipenem | 0.5 (S) | 0.5 (S) | 1 (S) |
| Meropenem | 0.0625 (S) | 0.0625 (S) | 0.0625 (S) |
| Amikacin | 8 (S) | 4 (S) | 16 (S) |
| Tigecycline | 1 (S) | 0.5 (S) | 1 (S) |
| Fosfomycin | 0.25 (S) | 64 (R) | 0.25 (S) |
| Sulfamethoxazole/trimethoprim | >32 (R) | >32 (R) | >32 (R) |
| Aztreonam | 8 (I) | 4 (S) | 16 (R) |
Abbreviations: MIC, minimum inhibitory concentration; I, intermediate; R, resistant; S, susceptible.
mcr-1 gene locations
S1-nuclease digestion and Southern blot analysis indicated that all three mcr-1 genes were located on plasmids. mcr-1 genes of isolates 16–541 and 16–623 were located on the same-size plasmids of ~310.1 kb, while the one of isolate 16–573 was located onã60 kb plasmid (Figure 1).
Figure 1.
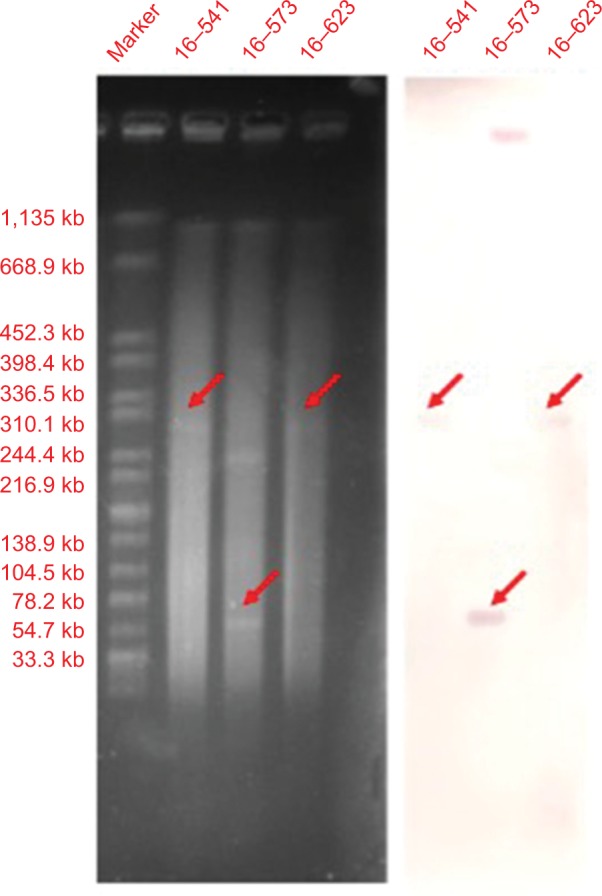
S1-digested plasmid DNA and Southern blot hybridization with mcr-1 gene of Salmonella typhimurium isolates
Notes: The red arrows indicate positive signals via southern blot hybridization with the mcr-1 probes.
WGS analysis
The whole genomes of the three S. typhimurium strains were sequenced and analyzed.
Other mechanisms associated with antimicrobial resistance were analyzed by using ResFinder. In addition to mcr-1, all the three isolates carried a variety of resistance genes to different antibiotics, including aminoglycosides, β-lactams, fluoroquinolones, sulfonamides, tetracyclines, and trimethoprim. In addition, isolate 16–573 carried fosA3 gene, which induced fosfomycin resistance, while isolates 16–541 and 16–623 carried ARR-3 gene, which induced rifampicin resistance (Table 3).
Table 3.
Other resistant genes of mcr-1-positive Salmonella typhimurium isolates
| Isolates/resistant genes | Aminoglycoside | β-Lactam | Fluoroquinolone | Fosfomycin | Rifampicin | Sulfonamide | Tetracyclines | Trimethoprim |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 16–541 | aph(3′)-Ia/Ib, aadA1/A2, aac(6′)Ib-cr, aph(6)- Id, aph(4)-Ia, aac(3)-Iva | blaTEM-1B, blaCTX-M-14, blaOXA-1 | oqxA, oqxB, qnrS2, aac(6′)-Ib-cr | None | ARR-3 | sul1, sul2, sul3 | tet(A), tet(B) | dfrA12 |
| 16–573 | aph(3′)-Ia, aadA1/A2, aph(4)-Ia, aac(3)-Iva | blaCTX-M-14 | oqxA, oqxB | fosA3 | None | sul1, sul2, sul3 | tet(B) | dfrA12 |
| 16–623 | aph(3′)-Ia, aadA1/A2, aac(6′)Ib-cr, aph(4)- Ia, aac(3)-Iva, aph(3′′)-Ib, aph(6)-Id | blaTEM-1B, blaCTX-M-14, blaOXA-1 | oqxA, oqxB, qnrS2, aac(6′)-Ib-cr | None | ARR-3 | sul1, sul2, sul3 | tet(B) | dfrA12 |
Genetic environment of mcr-1 gene
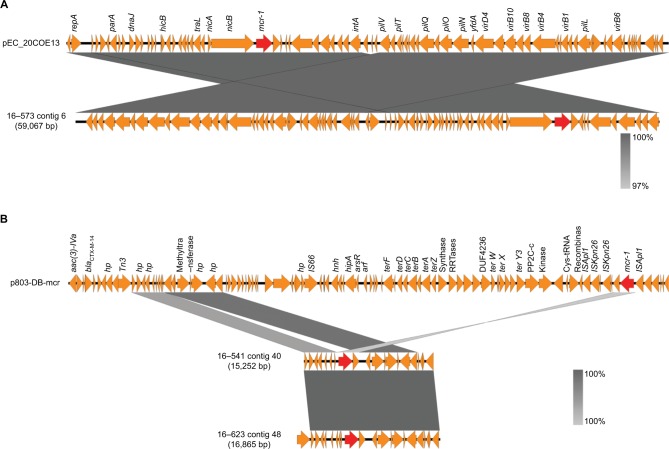
One of the three mcr-1-carrying plasmids (16-573) was greatly similar to pEc20COE13 (accession number: KY012274), which belonged to plasmid type IncI2 (Figure 2A). Although the mcr-1 gene was found to be inserted at different locations of the plasmid, the mcr-1 genes located on the contigs of isolates 16-541 and 16-623 were partly similar to the mcr-1-positive plasmid p803-DB-mcr (accession number: KY012274), which belonged to plasmid type IncHI2 (Figure 2B). No ISApl1 elements were identified at the upstream or downstream of mcr-1 gene.
Figure 2.
Genetic structures of two representative types of plasmids carrying mcr-1 gene.
Notes: (A) Comparison of mcr-1-located contig of isolate 16–573 and pEc_20COE13. (B) Comparison of mcr-1-located contigs of isolates 16–541 and 16–623 and p803-DB-mcr. The red arrows represent the mcr-1 gene.
Discussion
Since the first report of mcr-1 gene, it has been widely reported all over the world, and can be isolated from animals, food, environment, and humans (patients and healthy people). Apart from the common strains like E. coli and K. pneumonia, mcr-1 gene has also been reported from Enterobacter aerogenes and Enterobacter cloaceae.15 Salmonella spp., especially S. typhimurium, is one of the main hosts of mcr-1 gene. Although many countries, such as Britain, Italy, the Netherlands, and Portugal, have reported mcr-1-carrying S. typhimurium isolates, most of them were of animal origin.16–19
This study analyzed the prevalence and molecular characteristics of mcr-1 gene in S. typhimurium isolates from 2007 to 2016 retrospectively, which were preserved in the laboratory of Quzhou People’s Hospital. The results showed that the detection rate of mcr-1 gene was 4.8%, which was slightly higher than that reported previously (0.06%–1.55%).16,20 It is observed that mcr-1-positive S. typhimurium were not detected until 2016. Furthermore, all the mcr-1 genes were located on plasmids. One of the three mcr-1-carrying plasmids belonged to plasmid type IncI2, and the other two belonged to IncHI2, which was the same as the recently reported mcr-1-horbored plasmid of ST34 S. typhimurium isolated from pigs in China.21 All these results suggest that mcr-1 gene may play an important role in polymyxin resistance of clinically isolated S. typhimurium strains and lead to the rapid increase of polymyxin resistance in these bacteria.
S. typhimurium is a common clinical pathogenic serotype of Salmonella spp. In recent years, there has been a rapid increase in the incidence of antimicrobial resistance among the Salmonella spp., which can be attributed to the irrational use of antibiotics in clinics as well as the randomly discharged stools of poultry which can contain the residues of antibiotics.22 According to the previously reported study, mcr-1-positive isolates usually remain susceptible to many other antibiotics.23 In this study, mcr-1-positive strains were resistant to colistin as well as to third/fourth-generation cephalosporins and sulfamethoxazole/trimethoprim. WGS analysis showed that these isolates were all CTX-M-14-type extended-spectrum β-lactamase producing strains and carried sulfonamide and trimethoprim resistance genes, which were in accordance to the resistant phenotype. The MIC of fosfomycin of isolate 16–573 was 64 µg/mL, which was far higher than the MIC of other two isolates (0.25 µg/mL). It was due to the co-expression of fosA3 in isolate 16–573. In addition, although the isolate also carried quinolone-resistance genes oqxA and oqxB, it was susceptible to both levofloxacin and ciprofloxacin, which might be due to the non-expression of the two genes.
In China, the most common ST of S. typhimurium, especially MDR S. typhimurium, is ST34,13 which is also prevalent in Europe.24 In this study, all the three S. typhimurium isolates belonged to ST34. At the same time, most of the S. typhimurium strains carrying mcr-1 gene were isolated from animals and belonged to ST34,21,25 which suggested that ST34 was closely related to antimicrobial resistance and the spread of this clone would pose a great threat to the prevention and control of clinical S. typhimurium infections.
Conclusion
In conclusion, colistin-resistant mcr-1-positive S. typhimurium strains have emerged in Quzhou People’s Hospital and belong to the most common sequence type ST34, which is associated with MDR S. typhimurium. It is necessary to strengthen the surveillance of polymyxin resistance rate in our hospital to prevent the spread of resistant bacteria.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (81802043) and the Medical and Health Technology Project of Zhejiang Province (2018RC011).
Footnotes
Disclosure
None of the authors have any personal or financial involvement with the organizations that have financial interest in the content of this study. The authors report no conflicts of interest in this work.
References
- 1.Nation RL, Li J. Colistin in the 21st century. Curr Opin Infect Dis. 2009;22(6):535–543. doi: 10.1097/QCO.0b013e328332e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim LM, Ly N, Anderson D, et al. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy. 2010;30(12):1279–1291. doi: 10.1592/phco.30.12.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang X, Yu L, Chen X, et al. High prevalence of colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in China. Front Microbiol. 2017;8:562. doi: 10.3389/fmicb.2017.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo J, Yao X, Lv L, et al. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob Agents Chemother. 2017;61(10):e01139–17. doi: 10.1128/AAC.01139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Tian G-B, Zhang R, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1 -positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17(4):390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhao F, Zong Z. Kluyvera ascorbata strain from hospital sewage carrying the mcr-1 colistin resistance gene. Antimicrob Agents Chemother. 2016;60(12):7498–7501. doi: 10.1128/AAC.01165-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H-W, Zhang T, Ma J-H, et al. Occurrence of plasmid- and chromosome-carried mcr-1 in waterborne enterobacteriaceae in China. Antimicrob Agents Chemother. 2017;61(8):e00017–17. doi: 10.1128/AAC.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu BT, Song FJ, Zou M. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob Agents Chemother. 2017;61(2):e01444–16. doi: 10.1128/AAC.01444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sennati S, Di Pilato V, Riccobono E, et al. Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the Chaco region of Bolivia. J Antimicrob Chemother. 2017;72(7):2127–2129. doi: 10.1093/jac/dkx078. [DOI] [PubMed] [Google Scholar]
- 12.Seong M, Lee DG. Silver nanoparticles against Salmonella enterica serotype typhimurium: role of inner membrane dysfunction. Curr Microbiol. 2017;74(6):661–670. doi: 10.1007/s00284-017-1235-9. [DOI] [PubMed] [Google Scholar]
- 13.Wong MH, Yan M, Chan EW, Liu LZ, Kan B, Chen S. Expansion of Salmonella enterica serovar typhimurium st34 clone carrying multiple resistance determinants in China. Antimicrob Agents Chemother. 2013;57(9):4599–4601. doi: 10.1128/AAC.01174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng KJ, Doi Y, Patil S, Huang X, Tian GB. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother. 2016;60(6):3862–3863. doi: 10.1128/AAC.00345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campos J, Cristino L, Peixe L, Antunes P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 2016;21(26) doi: 10.2807/1560-7917.ES.2016.21.26.30270. [DOI] [PubMed] [Google Scholar]
- 17.Anjum MF, Duggett NA, Abuoun M, et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother. 2016;71(8):2306–2313. doi: 10.1093/jac/dkw149. [DOI] [PubMed] [Google Scholar]
- 18.Carnevali C, Morganti M, Scaltriti E, Bolzoni L, Pongolini S, Casadei G. Occurrence of mcr-1 in colistin-resistant salmonella enterica isolates recovered from humans and animals in Italy, 2012 to 2015. Antimicrob Agents Chemother. 2016;60(12):7532–7534. doi: 10.1128/AAC.01803-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veldman K, van Essen-Zandbergen A, Rapallini M, et al. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J Antimicrob Chemother. 2016;71(8):2340–2342. doi: 10.1093/jac/dkw181. [DOI] [PubMed] [Google Scholar]
- 20.Doumith M, Godbole G, Ashton P, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71(8):2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 21.Yi L, Wang J, Gao Y, et al. mcr-1-Harboring Salmonella enterica serovar typhimurium sequence type 34 in pigs, China. Emerg Infect Dis. 2017;23(2):291–295. doi: 10.3201/eid2302.161543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhaomin C, Kai L, Caiying B. Characteristics of molecular typing and drug-resistance for Salmonella typhimurium. Int J Lab Med. 2016;12:1601–1603. [Google Scholar]
- 23.Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 24.Antunes P, Mourão J, Pestana N, Peixe L. Leakage of emerging clinically relevant multidrug-resistant Salmonella clones from pig farms. J Antimicrob Chemother. 2011;66(9):2028–2032. doi: 10.1093/jac/dkr228. [DOI] [PubMed] [Google Scholar]
- 25.Li XP, Fang LX, Song JQ, et al. Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci Rep. 2016;6:38511. doi: 10.1038/srep38511. [DOI] [PMC free article] [PubMed] [Google Scholar]