Abstract
Background
Neisseria gonorrhoeae resistance to azithromycin has become a significant public health concern globally, and high-level azithromycin-resistant (HL-AzmR) isolates have emerged frequently. However, high-level azithromycin resistance is considered to be caused by mutated alleles of 23S rRNA gene at position 2059, and identification of HL-AzmR isolates mainly relies on agar dilution method or E-test method. This study aimed to assess the accuracy of the molecular assays targeting the mutation A2059G for identifying HL-AzmR isolates and thereby determine the association between the mutation and high-level azithromycin resistance.
Methods
Two researchers independently searched six databases to identify studies published from the launch of each database to October 15, 2017. The fixed effects model was used to estimate the pooled sensitivity rate, specificity rate, positive predictive value (PPV), and negative predictive value (NPV). Summary receiver operating characteristic curves were generated, and the area under the curve (AUC) was determined to estimate the overall performance of the assays. The Deeks’ test was conducted to evaluate potential publication bias.
Results
Ten relevant studies were included in the meta-analysis to assess the synthetic accuracy of the molecular assays. The molecular assays had the synthetic sensitivity rate of 97.8% and the synthetic specificity rate of 99.1%. And the aggregated PPV and NPV were 96.4% and 99.5%, respectively. AUC was 0.99, suggesting a close relation existing between the mutation A2059G and high-level azithromycin resistance. This indicated that the molecular assays targeting the mutation A2059G have relatively high overall accuracy for identifying HL-AzmR N. gonor-rhoeae isolates. Publication bias was statistically significant.
Conclusion
The mutation A2059G is the critical factor causing high-level azithromycin resistance. Hence, molecular methods are recommended to be put into clinical practice by commercialization, which will assist clinicians to prescribe more precisely.
Keywords: Neisseria gonorrhoeae, 23S rRNA, A2059G, azithromycin resistance, systematic review
Introduction
Neisseria gonorrhoeae is a common sexually transmitted pathogen causing male urethritis and female endocervicitis. It also facilitates the transmission of HIV, bringing immense morbidity and socioeconomic consequences.1
Without an effective vaccine against N. gonorrhoeae, antibiotics are the only approach to its treatment. Azithromycin, combined with cephalosporins, is currently recommended as the first-line medicine to treat gonococcal infection in American, Canadian, Australian, European, and WHO guidelines for the treatment of sexually transmitted diseases.2–6 However, there has been a growing number of reports on N. gonorrhoeae isolates with high-level azithromycin resistance in vitro, whose azithromycin minimum inhibitory concentrations (MICs) were commonly defined as ≥256 mg/L.7 Since the first high-level azithromycin-resistant (HL-AzmR) N. gonorrhoeae isolate was identified in Argentina in 2001,8 such isolates have also emerged in UK,9 Europe,10 USA,11 Canada,12 Australia,13 and China.14 High-level azithromycin resistance has become a severe threat to the first-line antimicrobial against gonococcal infection; in Nanjing, China, HL-AzmR N. gonorrhoeae isolates were estimated to account for 10.4% of all the isolates resistant to azithromycin in 2016.15 Their MICs were determined according to the conventional agar dilution method or E-test method; the former is complicated, whereas the latter is easy but has a high cost.
The ribosomal modification in N. gonorrhoeae isolates represents a significant mechanism of azithromycin resistance, which involves mutations in the peptidyl-transferase loop in domain V of 23S rRNA. Mutants with high-level azithromycin resistance commonly have substitutions in three or four alleles at position 2059 (Escherichia coli numbering) of the 23S rRNA gene, with adenine ribonucleotide substituted for guanine ribonucleotide, corresponding to the nucleotide position 2143.16,17 The mutation A2059G is considered to have association with high-level azithromycin resistance because of the above-mentioned phenomenon, which enables researchers to use rapid molecular assays such as PCR technique18,19 or whole-genome sequencing (WGS) technique10,11 coupled with direct sequencing to identify high-level azithromycin resistance. Compared with the agar-dilution method or the E-test method, these molecular assays have the advantages of simplicity in operation, precision in results, and low cost.
To date, though the single point mutation A2059G occurs in three or four alleles of HL-AzmR N. gonorrhoeae isolates,20 the relation between the mutation A2059G and high-level azithromycin resistance has not been explicitly validated.7,11 We systematically appraised the accuracy of the molecular assays targeting the mutation A2059G for identifying HL-AzmR N. gonorrhoeae isolates and determined the degree of the association between the mutation A2059G and high-level azithromycin resistance.
Methods
This study was performed according to the PRISMA guidelines (Table S1).21
Literature search and study selection
The process of literature search comprised four stages (identification, screening, eligibility assessment, and inclusion). Two researchers independently searched six databases (PubMed, Embase, Web of Science, Sinomed, China National Knowledge Infrastructure, and Wanfang Database) to identify relevant studies published from the launch of each database to October 15, 2017. Search terms included “Neisseria gonorrhoeae” in Medical Subject Headings or “Neisseria gonorr*” or “gonococcus” and their combination with “azithromycin”, and with “23S rRNA” or “2059” or “2143” in Title/Abstract (Table S2). Appropriate adjustments to search terms were made so that they could adapt to varied databases. References cited in the retrieved articles were also searched. All references were then uploaded into Endnote Software.
Titles and abstracts of all searched studies were screened first, and the full text of each relevant study was scanned after-ward. Eligible researches were identified and included in the current study, each meeting the following inclusion criteria: 1) a research was published in English or Chinese; 2) had specific breakpoint MICs to detect high-level azithromycin resistance; 3) indicated the numbers of HL-AzmR and non-HL-AzmR N. gonorrhoeae isolates and results of molecular assays targeting the position 2059 of the 23S rRNA gene.
Data extraction and quality assessment
Using a standardized form, data were extracted from each included article and compiled under the following categories: 1) publication year and first author; 2) location where the isolates were collected; 3) isolates collection period; 4) the breakpoint MICs to determine HL-AzmR N. gonorrhoeae isolates; 5) technique used for detecting mutation A2059G; 6) numbers of HL-AzmR N. gonorrhoeae isolates with mutant or without mutant at position 2059; and 7) numbers of non-HL-AzmR N. gonorrhoeae isolates with mutant or without mutant at position 2059. Some included studies did not report these numbers in the results sections directly, but showed relevant data in their supplemental tables and/or discussion sections; accordingly, we derived values from these data for each of these studies.
The methodological quality of each study was assessed using the validated Quality Assessment of Diagnostic Accuracy Studies (QUADAS) tool.22 The courses of literature search, study selection, data extraction, and quality assessment of included studies were completed by two researchers independently. Disagreements were settled by consensus.
Statistical analyses
We defined the phenotype of N. gonorrhoeae isolates as the gold standard. Moreover, numbers of HL-AzmR N. gonorrhoeae isolates with mutant or without mutant and non-HL-AzmR isolates with mutant or without mutant were defined as true positive (TP), false negative (FN), false positive (FP), and true negative (TN), respectively, for the systematic analysis of diagnostic tests (Table 1). The sensitivity rate (TP/(TP+FN)×100%) and specificity rate (TN/(TN+ FP)×100%) and their corresponding 95% CIs were calculated for each study. The sensitivity rate indicated the percentage of HL-AzmR isolates with the mutation A2059G, and the specificity rate meant proportion of non-HL-AzmR isolates which were not mutant at position 2059. Statistical analysis was performed using meta-analysis of Diagnostic and Screening Tests (Meta-DiSc,23 version 1.4, developed by the Unit of Clinical Biostatistics team of the Ramón y Cajal Hospital in Madrid). A fixed effects model was used to perform a group analysis. The pooled positive predictive value (PPV, TP/(TP+FP)×100%) and negative predictive value (NPV, TN/(TN+ FN)×100%) were calculated (PPV and NPV ranging from 0 to 1; higher values mean more effects). The pooled positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic OR (DOR) were calculated (PLR >35, NLR <0.1, and DOR >1, indicating the method with high diagnostic accuracy.24,25 The summary receiver operating characteristic (sROC) curve was plotted, based on which area under the sROC curve (AUC, ranging from 0 to 1; higher values mean more effects) was calculated to assess the overall accuracy of the molecular assays targeting the mutation A2059G for identifying HL-AzmR N. gonorrhoeae isolates.26 Between-study heterogeneity was evaluated by performing the Q test (P<0.05 indicating statistical significance) and calculating I2 values (range, 0%–100%, with higher values meaning greater heterogeneity). The Deeks’ funnel plot asymmetry test was generated using STATA 14.2 (Stata Corp., College Station, TX, USA) to detect potential publication bias (P<0.10 indicating statistical significance).27
Table 1.
Summary of different variables for the meta-analysis of diagnostic test
| Azithromycin susceptibility | With mutants at position 2059 | Without mutants at position 2059 |
|---|---|---|
| HL-AzmR isolates | True positive | False negative |
| Non-HL-AzmR isolates | False positive | True negative |
Abbreviation: HL-AzmR, high-level azithromycin resistant.
Results
Study selection
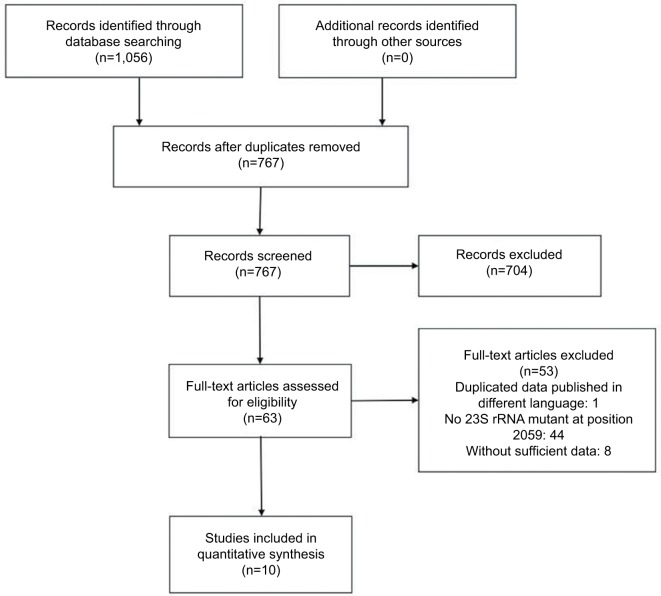
A total of 1,056 potentially relevant abstracts were identified, of which 289 were duplicates and thus removed. The remaining 767 abstracts were read over; 704 of them were subsequently excluded, which neither indicated azithromycin MICs nor indicated nucleotide mutants. Consequently, 63 full-text articles were assessed for eligibility. Ten10,12,15,19,20,28–32 of them were included in the meta-analysis, with seven10,12,20,29–32 in English and three15,19,28 in China (Figure 1).
Figure 1.
Process of selecting published studies for the meta-analysis according to PRISMA guidelines.
Quality assessment
Among the 63 eligible studies, 45 were eliminated for the reason of duplicate or without information of mutation A2059G. And two9,13 and six11,18,33–36 studies only showed data related to HL-AzmR N. gonorrhoeae isolates and non-HL-AzmR isolates, respectively, and they were excluded (Figure 1). The high-level azithromycin-resistance breakpoint was set at MICs ≥256 mg/mL in nine studies, but even higher (MICs ≥512 mg/mL) in one study. In the current research, the high-level azithromycin-resistance breakpoint was defined as MICs ≥256 mg/mL. In terms of methodological quality, the included ten studies had the mean score of 9.4 (range, 7–11) according to the criteria of QUADAS (Table 2).
Table 2.
Overview of ten included studies in the meta-analysis
| Study number | Year, first author | Location | Isolate collection period | Breakpoint MICs | Technique | QUADAS | Diagnostic test results of molecular assays
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| TP | FN | FP | TN | |||||||
|
| ||||||||||
| 1 | 2017, Jiang29 | Hefei, China | 01/2014–11/2015 | ≥256 | PCR | 10 | 13 | 0 | 0 | 28 |
| 2 | 2016, Jacobsson10 | 17 countries, Europe | 2009–2014 | ≥256 | WGS | 10 | 4 | 0 | 0 | 71 |
| 3 | 2016, Demczuk12 | Canada | 1997–2014 | ≥256 | WGS | 9 | 5 | 0 | 0 | 241 |
| 4 | 2015, Xue30 | Hangzhou, China | 2011, 2012 | ≥256 | PCR | 9 | 21 | 0 | 0 | 4 |
| 5 | 2010, Galarza32 | Argentina | – | ≥512 | PCR | 7 | 1 | 0 | 0 | 2 |
| 6 | 2010, Chisholm20 | England | 2004 | ≥256 | WGS | 10 | 19 | 0 | 3 | 35 |
| 7 | 2015, Demczuk31 | Canada | 1989–2013 | ≥256 | WGS | 8 | 1 | 0 | 1 | 3 |
| 8 | 2017, Lan19 | Anhui, China | 2014–2015 | ≥256 | PCR | 9 | 13 | 0 | 0 | 23 |
| 9 | 2017, Zhang28 | Shenzhen, China | 2011–2015 | ≥256 | PCR | 11 | 18 | 3 | 1 | 86 |
| 10 | 2016, Wan15 | Nanjing, China | 2013–2014 | ≥256 | PCR | 11 | 40 | 0 | 0 | 84 |
Note: “–” means information unavailable.
Abbreviations: FN, false negative; FP, false positive; HL-AzmR, high-level azithromycin resistant; MICs, minimum inhibitory concentrations; QUADAS, Quality Assessment of Diagnostic Accuracy Studies; TN, true negative; TP, true positive; WGS, whole-genome sequencing.
Meta-analysis
Assessment of sensitivity and specificity rates in the molecular assays for identifying HL-AzmR N. gonorrhoeae isolates
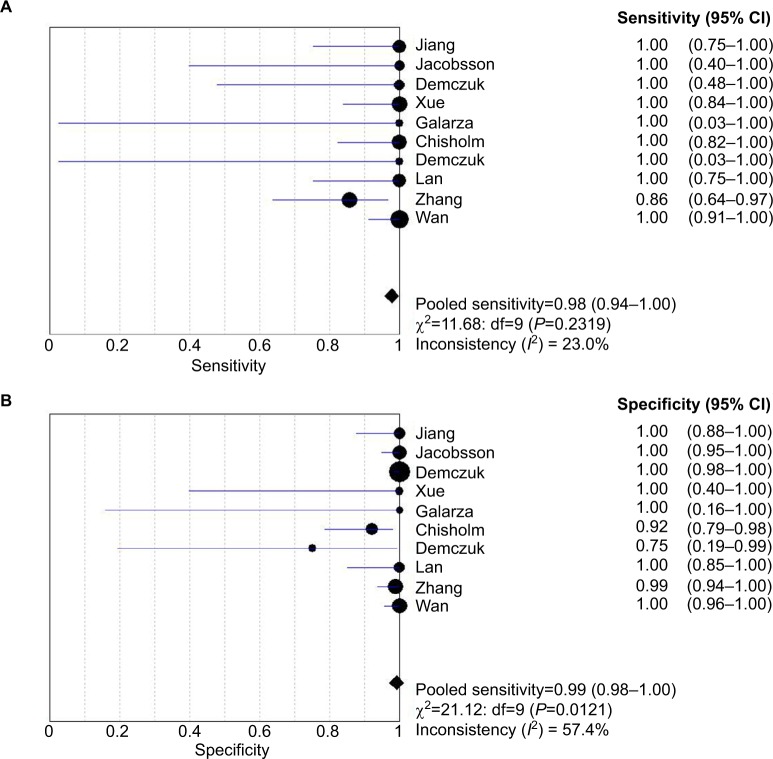
Ten studies (138 isolates) were shown the data for determining the sensitivity rate of detection of HL-AzmR N. gonorrhoeae isolates based on the mutation A2059G; the sensitivity rates in these studies ranged from 85.7% to 100.0%. By performing a meta-analysis with the fixed effects model, the pooled sensitivity rate of the molecular assays was determined to be 97.8% (95% CI, 93.8%–99.5%), and the evidence for between-study heterogeneity (I2=23.0%, P=0.232) was not significant (Figure 2A). On the other hand, the molecular assays conducted in these studies (582 isolates) had specificity rates ranging from 75.0% to 100.0%. The pooled specificity rate of the molecular assays was 99.1% (95% CI, 98.0%–99.7%), and the heterogeneity between included studies (I2=57.4%, P=0.012) cannot be excluded (Figure 2B).
Figure 2.
Analysis of sensitivity and specificity rates from included studies. Forest plot of sensitivity of the molecular assays in (A) and specificity in (B).
Notes: Point estimates of sensitivity and specificity from each study are shown as solid square. Error bars indicate 95% CI. Diamond is the estimated rates of pooled studies.
Diagnostic accuracy of the molecular assays for identifying HL-AzmR N. gonorrhoeae isolates
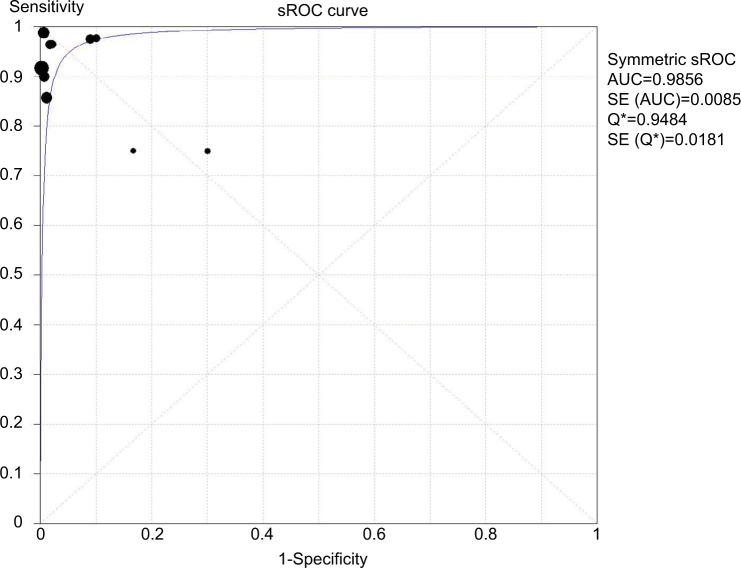
In total, ten studies were included to estimate the pooled diagnostic accuracy of the molecular assays for detecting HL-AzmR N. gonorrhoeae isolates based on the identification of the mutation A2059G. Pooled PPV and NPV were 96.4% (95% CI, 91.9%–98.8%) and 99.5% (95% CI, 98.5%–99.9%). Pooled PLR was 29.6 (95% CI, 16.0–54.6), whereas pooled NLR was 0.06 (95% CI, 0.03–0.12). DOR ranged from 95.0 to 1297.4 (mean, 351.1). An sROC curve was plotted to display sensitivity against “1-specificity” from an individual study. The AUC derived from the sROC curve was 0.99 (Figure 3), suggesting that the molecular assays have a high overall accuracy.
Figure 3.
sROC curve of ten studies with both sensitivity and specificity rates.
Notes: The size of each solid square represents the sample size of individual study. The regression sROC curve summarizes the overall diagnostic accuracy.
Abbreviations: AUC, area under the curve; SE, standard error; sROC, summary receiver operating characteristic.
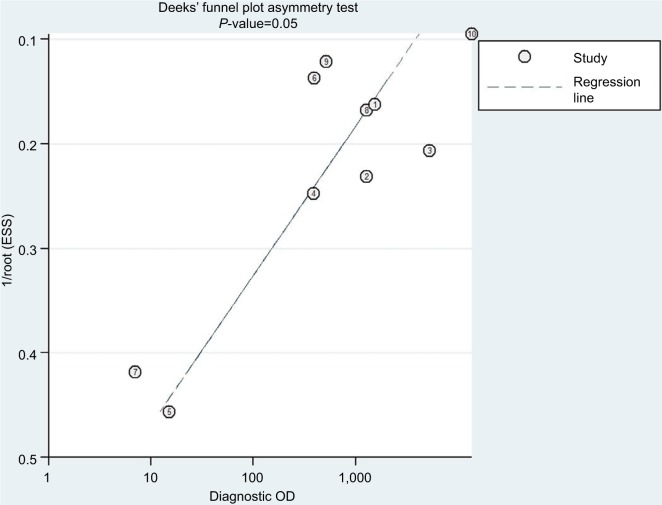
Publication bias
The Deeks’ funnel plot asymmetry test was conducted to estimate potential publication bias of included studies (Figure 4); as a result, the Deeks’ test yielded a P-value of 0.05, meaning that the asymmetries were statistically significant, which indicated the likelihood of publication bias.
Figure 4.
Deeks’ funnel plot asymmetry test indicating the risk of publication bias (P=0.05) of ten included studies.
Note: Circles and numbers represent each study included in this meta-analysis.
Abbreviation: ESS, effective sample size.
Discussion
To our knowledge, the present study is the first systematic review of published papers on the exact association between the mutation A2059G in the 23S rRNA gene and high-level azithromycin resistance. To date, high-level azithromycin resistance is still identified by using the agar-dilution method or the E-test method, and these two methods have notable weaknesses. The agar dilution method has been applied for decades as the golden standard for determining antimicrobial susceptibility of clinical isolates. Nonetheless, it is cumbersome in operation; and its results can be affected by a number of factors: agar medium composition, pH, and incubation parameters such as CO2 level. Therefore, though the MICs estimated by different laboratories are comparable, their values may vary by one or more twofold dilution due to slight technical differences, hence affecting the clinical interpretation.37 In terms of the E-test method, it is costly, requiring the use of experimental materials that are very expensive because of manufacturer’s patent protection, and these materials were not available in some areas.38 On the other hand, the molecular assays (PCR and WGS) detecting the mutation A2059G could be an alternative method for identifying isolates with high-level azithromycin resistance; however, they have not been developed into a commercial diagnostic kit for clinical use nowadays. In this review, we systematically appraised the accuracy of the molecular assays for identification of high-level azithromycin resistance to verify the association between the mutation A2059G and high-level azithromycin resistance.
The present study showed that these molecular assays had the pooled sensitivity rate of 98%, which agreed with the fact that many isolates with high-level resistance to macrolides have the mutation at position 2059 of the 23S rRNA,7,39 and the pooled specificity rate of 99% indicated that among the HL-AzmR isolates, almost none of them had mutation A2059G. These molecular assays also had the pooled PPV of 96% and the pooled specificity rate of 99%, both of which were close to 100%, indicating that almost all isolates with mutation A2059G in 23S rRNA are high-level resistant to azithromycin and nearly all isolates without mutated alleles at position 2059 have no high-level azithromycin resistance. These four rates are in accordance with the previous research that high-level resistance to azithromycin occurred as a result of a single point mutation in the peptidyl-transferase region of domain V of the 23S rRNA.20 Regarding the overall accuracy of the molecular assays, the synthetic PLR and NLR were 29.6 and 0.06, respectively; DOR, the ratio of PLR and NLR, stood at 351, suggesting that these molecular assays can precisely detect high-level azithromycin resistance. We also combined sensitivity and specificity rates to create the sROC curve; as a result, AUC of the curve was 0.99, very close to 1, pointing to the exact association between the mutation A2059G and high-level resistance to azithromycin, as well as the feasibility of using these assays for identification of HL-AzmR N. gonorrhoeae isolates clinically.
The present study has the following strengths. The association between the mutation A2059G and high-level azithromycin resistance was assessed by analyzing the MICs and the statistical data resulting from the molecular methods PCR and WGS. In methodology, this study is a first systematic analysis of the mechanisms underlying the antimicrobial resistance of N. gonorrhoeae isolates. Our findings not only confirm that the mutation A2059G is the unique factor, which can directly result in high-level resistance to azithromycin, but also provide a prospective future for using the molecular methods to detect high-level azithromycin resistance in N. gonorrhoeae isolates clinically.
On the other hand, there are also a few limitations to our study. Primarily, eight studies9,11,13,18,33–36 did not have sufficient data on results of molecular assays from isolates either with or without high-level azithromycin resistance, so these studies were not taken into account for this systematic analysis. Furthermore, the P-value for publication bias was 0.05, less than the breakpoint value (0.10), which was caused by the data extracted from the included studies mostly containing high sensitivity or specificity rates. Lastly, the statistical data from the studies using PCR or WGS were pooled for the meta-analysis; thus, the resultant synthetic accuracy of the molecular assays did not reflect the diagnostic accuracy of each of the two techniques applied alone.
Conclusion
Rapid molecular assays for detecting specific gene mutations in clinical isolates are enabling targeted antimicrobial therapy that could give patients precise therapy and curb the emergence of antimicrobial resistance.40 With high diagnostic accuracy, the molecular assays targeting the mutation at position 2059 of 23S rRNA have excellent prospects, which can be developed into diagnostic kits for the quick identification of HL-AzmR isolates clinically and can be the essential foundation of the molecular assays for detecting azithromycin resistance. However, the mutation A2059G does not occur in most of the identified N. gonorrhoeae isolates with low- to moderate-level resistance to azithromycin (1 mg/mL<MICs<256 mg/mL),8 which, therefore, cannot be detected by the molecular assays targeting this mutation. Moreover, the new point mutation at position 2611 of 23S rRNA and mutations in the efflux pump gene have been found in N. gonorrhoeae isolates with low- to moderate-level azithromycin resistance,7,10,34 which can be utilized to identify all levels of azithromycin-resistant isolates in the future.
Supplementary materials
Table S1.
PRISMA 2009 checklist of the paper
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| Title | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both. | |
| Abstract | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number. | |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known. | |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS). | |
| Methods | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (eg, Web address), and, if available, provide registration information including registration number. | |
| Eligibility criteria | 6 | Specify study characteristics (eg, PICOS, length of follow-up) and report characteristics (eg, years considered, language, publication status) used as criteria for eligibility, giving rationale. | |
| Information sources | 7 | Describe all information sources (eg, databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched. | |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated. | |
| Study selection | 9 | State the process for selecting studies (ie, screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis). | |
| Data collection process | 10 | Describe method of data extraction from reports (eg, piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators. | |
| Data items | 11 | List and define all variables for which data were sought (eg, PICOS, funding sources) and any assumptions and simplifications made. | |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis. | |
| Summary measures | 13 | State the principal summary measures (eg, risk ratio, difference in means). | |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (eg, I2) for each meta-analysis. | |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (eg, publication bias, selective reporting within studies). | |
| Additional analyses | 16 | Describe methods of additional analyses (eg, sensitivity or subgroup analyses, meta-regression), if done, indicating which were prespecified. | |
| Results | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram. | |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (eg, study size, PICOS, follow-up period) and provide the citations. | |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome-level assessment (see item 12). | |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study (a) simple summary data for each intervention group and (b) effect estimates and CIs, ideally with a forest plot. | |
| Synthesis of results | 21 | Present results of each meta-analysis done, including CIs and measures of consistency. | |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see item 15). | |
| Additional analysis | 23 | Give results of additional analyses, if done (eg, sensitivity or subgroup analyses, meta-regression [see Item 16]). | |
| Discussion | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (eg, healthcare providers, users, policymakers). | |
| Limitations | 25 | Discuss limitations at study and outcome level (eg, risk of bias), and at review level (eg, incomplete retrieval of identified research, reporting bias). | |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research. | |
| Funding | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (eg, supply of data), role of funders for the systematic review. | |
Note: From Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. PLoS Med 6(7): e1000097.1
Table S2.
PubMed search strategy and result (October 15, 2017)
| Search | Query | Item number |
|---|---|---|
|
| ||
| #1 | ((“Neisseria gonorrhoeae”[Mesh]) OR Neisseria gonorr*) OR gonococcus | 12634 |
| #2 | (azithromycin[MeSH Terms]) OR (azithro*) OR azithromycin | 7630 |
| #3 | (rRNA[Title/Abstract]) OR (23S[Title/Abstract]) | 67822 |
| #4 | (#1 AND #2) AND #3 | 28 |
Reference
- 1.Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
The authors acknowledge the grants from the Chinese Academy Medical Sciences Initiative for Innovative Medicine (2016-I2M-3–021) and the Sanming Project of Medicine in Shenzhen (SZSM201611077).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cohen MS. Classical sexually transmitted diseases drive the spread of HIV-1: back to the future. J Infect Dis. 2012;206(1):1–2. doi: 10.1093/infdis/jis303. [DOI] [PubMed] [Google Scholar]
- 2.Kimberly AW, Gail AB. Sexually transmitted diseases treatment guidelines, 2015. Curr Opin Pediatr. 2006;15(4):391–397. doi: 10.1097/00008480-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 3.PHAC/EWG Canadian guidelines on sexually transmitted infections. 2013. [Accessed Dec 9, 2018]. Available from: https://www.canada.ca/en/public-health/services/infectious-diseases/sexual-health-sexually-transmitted-infections/canadian-guidelines/sexually-transmitted-infections/canadian-guidelines-sexually-transmitted-infections-34.html.
- 4.Australasian Sexual Health Alliance Australian STI Management Guidelines. 2014. [Accessed July 24, 2018]. webpage on the Internet. Available from: http://www.sti.guidelines.org.au/sexually-transmissible-infections/gonorrhoea.
- 5.Bignell C, Unemo M, Radcliffe K, et al. 2012 European guideline on the diagnosis and treatment of gonorrhoea in adults. Int J STD AIDS. 2013;24(2):85–92. doi: 10.1177/0956462412472837. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization . WHO guidelines for the treatment of Neisseria gonorrhoeae. Geneva, Switzerland: World Health Organization; 2016. [Accessed July 24, 2018]. Available from: https://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0093492/ [Google Scholar]
- 7.Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. 2014;27(3):587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galarza PG, Alcalá B, Salcedo C, et al. Emergence of high level azithromycin-resistant Neisseria gonorrhoeae strain isolated in Argentina. Sex Transm Dis. 2009;36(12):787–788. doi: 10.1097/OLQ.0b013e3181b61bb1. [DOI] [PubMed] [Google Scholar]
- 9.Chisholm SA, Wilson J, Alexander S, et al. An outbreak of high-level azithromycin resistant Neisseria gonorrhoeae in England. Sex Transm Infect. 2016;92(5):365–367. doi: 10.1136/sextrans-2015-052312. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsson S, Golparian D, Cole M, et al. WGS analysis and molecular resistance mechanisms of azithromycin-resistant (MIC >2 mg/L) Neisseria gonorrhoeae isolates in Europe from 2009 to 2014. J Antimicrob Chemother. 2016;71(11):3109–3116. doi: 10.1093/jac/dkw279. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SR, Grad Y, Abrams AJ, Pettus K, Trees DL. Use of whole-genome sequencing data to analyze 23S rRNA-mediated azithromycin resistance. Int J Antimicrob Agents. 2017;49(2):252–254. doi: 10.1016/j.ijantimicag.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demczuk W, Martin I, Peterson S, et al. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol. 2016;54(5):1304–1313. doi: 10.1128/JCM.03195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens K, Zaia A, Tawil S, et al. Neisseria gonorrhoeae isolates with high-level resistance to azithromycin in Australia. J Antimicrob Chemother. 2015;70(4):1267–1268. doi: 10.1093/jac/dku490. [DOI] [PubMed] [Google Scholar]
- 14.Yuan LF, Yin YP, Dai XQ, et al. Resistance to azithromycin of Neisseria gonorrhoeae isolates from 2 cities in China. Sex Transm Dis. 2011;38(8):764. doi: 10.1097/OLQ.0b013e318219cdb5. [DOI] [PubMed] [Google Scholar]
- 15.Wan C, Li Y, Le WJ, et al. Increasing resistance to azithromycin in Neisseria gonorrhoeae in Eastern Chinese cities: resistance mechanisms and genetic diversity among isolates from Nanjing. Antimicrob Agents Chemother. 2018;62(5) doi: 10.1128/AAC.02499-17. pii:e02499-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisblum B. Erythromycin resistance by ribosome modification. Anti-microb Agents Chemother. 1995;39(3):577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45(1):1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trembizki E, Buckley C, Donovan B, et al. Direct real-time PCR-based detection of Neisseria gonorrhoeae 23S rRNA mutations associated with azithromycin resistance. J Antimicrob Chemother. 2015;70(12):3244. doi: 10.1093/jac/dkv274. [DOI] [PubMed] [Google Scholar]
- 19.Lan Q, Jiang FX. Analysis of drug resistance mechanism and molecular epidemiological characteristics of azithromycin resistant strains. Acta Universitatis Medicinalis Anhui. 2017;52(3):360–364. [Google Scholar]
- 20.Chisholm SA, Dave J, Ison CA. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother. 2010;54(9):3812–3816. doi: 10.1128/AAC.00309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mark V. Preferred reporting items for systematic reviews and meta-analyses. Revista Española De Nutrición Humana Y Dietética. 2009;18(3):e123. [Google Scholar]
- 22.Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3(1):25–25. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodacre S, Sutton AJ, Sampson FC. Meta-analysis: the value of clinical assessment in the diagnosis of deep venous thrombosis. Ann Intern Med. 2005;143(2):129–139. doi: 10.7326/0003-4819-143-2-200507190-00012. [DOI] [PubMed] [Google Scholar]
- 24.Vamvakas EC. Meta-analyses of studies of the diagnostic accuracy of laboratory tests: a review of the concepts and methods. Arch Pathol Lab Med. 1998;122(8):675–686. [PubMed] [Google Scholar]
- 25.Rennie D. Improving reports of studies of diagnostic tests: the STARD initiative. JAMA. 2003;289(1):89–90. doi: 10.1001/jama.289.1.89. [DOI] [PubMed] [Google Scholar]
- 26.Walter SD. Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat Med. 2002;21(9):1237–1256. doi: 10.1002/sim.1099. [DOI] [PubMed] [Google Scholar]
- 27.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Zhang LJ, Wang F, Peng Y, Mo JL. Antibiotic resistant mechanism and epidemiological characteristics of azithromycin-resistant Neisseria gonorrhoeae strains in Shenzhen. Chinese J Microbiol Immunol. 2017;37(3):219–224. [Google Scholar]
- 29.Jiang FX, Lan Q, Le WJ, Su XH. Antimicrobial susceptibility of Neisseria gonorrhoeae isolates from Hefei (2014-2015): genetic characteristics of antimicrobial resistance. BMC Infect Dis. 2017;17(1):366. doi: 10.1186/s12879-017-2472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue J, Ni C, Zhou H, Zhang C, van der Veen S. Occurrence of high-level azithromycin-resistant Neisseria gonorrhoeae isolates in China. J Antimicrob Chemother. 2015;70(12):3404–3405. doi: 10.1093/jac/dkv266. [DOI] [PubMed] [Google Scholar]
- 31.Demczuk W, Lynch T, Martin I, et al. Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J Clin Microbiol. 2015;53(1):191–200. doi: 10.1128/JCM.02589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galarza PG, Abad R, Canigia LF, et al. New mutation in 23S rRNA gene associated with high level of azithromycin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2010;54(4):1652–1653. doi: 10.1128/AAC.01506-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen VG, Seah C, Martin I, Melano RG. Azithromycin resistance is coevolving with reduced susceptibility to cephalosporins in Neisseria gonorrhoeae in Ontario, Canada. Antimicrob Agents Chemother. 2014;58(5):2528–2534. doi: 10.1128/AAC.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belkacem A, Jacquier H, Goubard A, et al. Molecular epidemiology and mechanisms of resistance of azithromycin-resistant Neisseria gonorrhoeae isolated in France during 2013-14. J Antimicrob Chemother. 2016;71(9):2471–2478. doi: 10.1093/jac/dkw182. [DOI] [PubMed] [Google Scholar]
- 35.Endimiani A, Guilarte YN, Tinguely R, et al. Characterization of Neisseria gonorrhoeae isolates detected in Switzerland (1998-2012): emergence of multidrug-resistant clones less susceptible to cephalosporins. BMC Infect Dis. 2014;14(1):106. doi: 10.1186/1471-2334-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takayama Y, Nakayama S, Shimuta K, Morita-Ishihara T, Ohnishi M. Characterization of azithromycin-resistant Neisseria gonorrhoeae isolated in Tokyo in 2005-2011. J Infect Chemother. 2014;20(5):339–341. doi: 10.1016/j.jiac.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Woodford N, Ison CA. The effect of media on antimicrobial susceptibility testing of Neisseria gonorrhoeae. J Antimicrob Chemother. 1988;22(4):463–471. doi: 10.1093/jac/22.4.463. [DOI] [PubMed] [Google Scholar]
- 38.Jönsson A, Jacobsson S, Foerster S, Cole MJ, Unemo M. Performance characteristics of newer MIC gradient strip tests compared with the Etest for antimicrobial susceptibility testing of Neisseria gonorrhoeae. APMIS. 2018;126(10):822–827. doi: 10.1111/apm.12887. [DOI] [PubMed] [Google Scholar]
- 39.Ohneck EA, Zalucki YM, Johnson PJ, et al. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. MBio. 2011;2(5):119. doi: 10.1128/mBio.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grad YH, Harris SR, Kirkcaldy RD, et al. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis. 2016;214(10):1579–1587. doi: 10.1093/infdis/jiw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
PRISMA 2009 checklist of the paper
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| Title | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both. | |
| Abstract | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number. | |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known. | |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS). | |
| Methods | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (eg, Web address), and, if available, provide registration information including registration number. | |
| Eligibility criteria | 6 | Specify study characteristics (eg, PICOS, length of follow-up) and report characteristics (eg, years considered, language, publication status) used as criteria for eligibility, giving rationale. | |
| Information sources | 7 | Describe all information sources (eg, databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched. | |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated. | |
| Study selection | 9 | State the process for selecting studies (ie, screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis). | |
| Data collection process | 10 | Describe method of data extraction from reports (eg, piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators. | |
| Data items | 11 | List and define all variables for which data were sought (eg, PICOS, funding sources) and any assumptions and simplifications made. | |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis. | |
| Summary measures | 13 | State the principal summary measures (eg, risk ratio, difference in means). | |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (eg, I2) for each meta-analysis. | |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (eg, publication bias, selective reporting within studies). | |
| Additional analyses | 16 | Describe methods of additional analyses (eg, sensitivity or subgroup analyses, meta-regression), if done, indicating which were prespecified. | |
| Results | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram. | |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (eg, study size, PICOS, follow-up period) and provide the citations. | |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome-level assessment (see item 12). | |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study (a) simple summary data for each intervention group and (b) effect estimates and CIs, ideally with a forest plot. | |
| Synthesis of results | 21 | Present results of each meta-analysis done, including CIs and measures of consistency. | |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see item 15). | |
| Additional analysis | 23 | Give results of additional analyses, if done (eg, sensitivity or subgroup analyses, meta-regression [see Item 16]). | |
| Discussion | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (eg, healthcare providers, users, policymakers). | |
| Limitations | 25 | Discuss limitations at study and outcome level (eg, risk of bias), and at review level (eg, incomplete retrieval of identified research, reporting bias). | |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research. | |
| Funding | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (eg, supply of data), role of funders for the systematic review. | |
Note: From Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. PLoS Med 6(7): e1000097.1
Table S2.
PubMed search strategy and result (October 15, 2017)
| Search | Query | Item number |
|---|---|---|
|
| ||
| #1 | ((“Neisseria gonorrhoeae”[Mesh]) OR Neisseria gonorr*) OR gonococcus | 12634 |
| #2 | (azithromycin[MeSH Terms]) OR (azithro*) OR azithromycin | 7630 |
| #3 | (rRNA[Title/Abstract]) OR (23S[Title/Abstract]) | 67822 |
| #4 | (#1 AND #2) AND #3 | 28 |