Function al diversity in multicellular organisms is achieved through the differentiation of stem cells. During this process, stem cells must retain both the capacity for self-renewal and the ability to differentiate into highly specialized cell types to produce a diverse array of tissues, each with distinct functions and organization. This plasticity is achieved through alterations to the epigenome, heri-table and reversible modifications to DNA and histones that affect chromatin structure and gene transcription without altering the DNA sequence itself. Alterations to the epigenome enable cell type–specific transcriptional control that can change dynamically over the life of a cell. Such flexibility and responsiveness are instrumental in directing gene expression changes throughout cellular differentiation and lineage specification. The acquisition of more specialized functions during differentiation requires not only that the epigenome turn “on” genes involved in lineage commitment, it also necessitates that genes associated with stemness are simultaneously turned “off” (1). On page 177 of this issue, Pace et al. (2) demonstrate that this phenomenon exists in CD8+ T cells, in which epigenetic repression of stemness-associated genes by the histone methyltransferase SUV39H1 is required for T cell effector differentiation. Understanding these mechanisms addresses important questions in immunology and is applicable to cancer immunotherapy.
The CD8+ T lymphocyte compartment of the adaptive immune system has emerged as a model for developmental biology in adult mammalian cells owing to its remarkable degree of functional plasticity (3). CD8+ T cells can rapidly differentiate from a quiescent, long-lived memory state into an effector state characterized by short-lived cytotoxicity toward cancer cells or cells infected with intracellular pathogens (4). Multiple differentiation models have been proposed to account for the observed changes in CD8+ T cell subsets during an immune response. The linear differentiation model places effector T cells (Teff cells) at the end of the differentiation process after the development of multiple intermediary memory T cell subsets (3). Specialized memory T cells, including the relatively rare T memory stem cells (Tscm cells) and the more common central memory T cells (Tcm cells), have characteristics associated with conventional stem cells. This includes enhanced self-renewal, which is essential for maintaining long-term immunological memory, and the ability to reconstitute other CD8+ T cell subsets, which maintains the functional diversity of the CD8+ T cell compartment (5–7). Tscm cells have enhanced stem cell–like capabilities, whereas Tcm cells are poised to rapidly initiate an effector response. With further T cell activation, memory subsets can differentiate into Teff cells followed by terminal differentiation, functional senescence, and ultimately apoptosis (cell death). An alternative model suggests that naïve T cells (Tn cells) differentiate into Teff cells immediately after activation, with “dedifferentiation” into memory cells occurring after pathogen clearance (8). Because the dedifferentiation of lineage-restricted cells rarely occurs in nature outside of cancer formation (9), we and others (7) feel that the linear differentiation model is more consistent with typical patterns of cellular differentiation.
CD8+ T cell subsets can be partitioned on the basis of distinct patterns of gene expression. Multiple subset-specific transcription factors regulate gene expression throughout differentiation (4). Although transcription factors are critical mediators of gene expression programs, their activity is largely dependent on epigenetic modifications, the profiles of which can also be used to distinguish T cell subsets (10). Indeed, activating epigenetic modifications are progressively gained at Teff cell–associated gene loci after T cell activation (10, 11). Recently, characterization of repressive epigenetic modifications during differentiation, as described by Pace et al. and others (11–14), have highlighted the importance of epigenetic silencing for proper Teff cell differentiation. Specifically, epigenetic silencing of stem cell- and T cell memory–associated genes in activated T cells permits efficient Teff cell differentiation and function, such that elimination of this activity results in defective Teff cells (2, 11–14).
Investigations into the repressive chromatin landscape of CD8+ T cells have focused on DNA methylation and trimethylation (me3) of specific lysine residues (K) on the histone H3 (specifically, H3K27me3 and H3K9me3). The epigenetic “writer” proteins responsible for adding these modifications include DNA methyltransferase 3A (DNMT3A), an enzyme responsible for de novo DNA methylation, and the histone methyltransferase enzymes enhancer of zeste homolog 2 (EZH2) and SUV39H1 (10). In mice, conditional ablation of Dnmt3a (12) and Ezh2 (13) in T cells and germline ablation of Suv39h1 (2) result in an altered phenotypic composition of antigen-specific CD8+ T cells after viral infection: Both the proportion and number of responding Teff cells are reduced and the frequency of memory T cells are increased. In vitro experiments using Ezh2-deficient T cells suggest selective apoptosis within the Teff cell population (14), which accounts for the equal numbers of antigen-specific memory cell subsets as well as the impaired functional efficacy of CD8+ T cells after secondary viral challenge observed in Ezh2- and Suv39h1-deficient mice (2, 13). Preserved memory T cell formation is consistent with the linear differentiation model that places memory cell development before differentiation into Teff cells. By contrast, in a model that predicts that memory T cells originate from Teff cells, one would expect numbers of memory T cells to decrease as well as Teff cells.
Transcriptional and epigenetic profiling of Dnmt3a-, Ezh2-, and Suv39h1-deficient Teff cells illustrates a common defect that is responsible for impaired Teff cell differentiation. Genes encoding master regulators of the stem and memory cell state fail to ac quire repressive epigenetic modifications, leading to aberrant gene expression and differentiation (2, 12, 13). Therefore, epigenetic repression of essential stem and memory genes is required for full Teff cell differentiation (see the figure). That Teff cell differentiation is still possible with loss of any one of these epigenetic writers illustrates the functional redundancy in silencing stem and memory genes, stressing the importance of this mechanism. This mirrors the epigenetic silencing of developmental and pluripotency genes during differentiation of human embryonic stem cells (1) and further highlights transcriptional silencing of stem cell–associated genes as a hallmark of cellular differentiation.
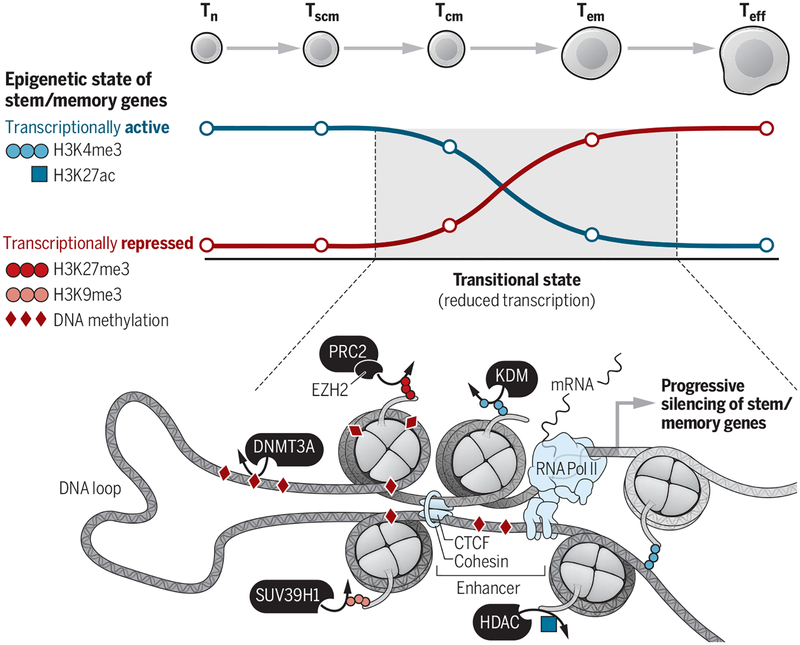
Figure. Shutting down stem and memory genes in CD8+ T cells.
As cells differentiate, stem and memory genes pass through transitional epigenetic states, in which epigenetic modifications associated with transcriptional activation, including H3K4me3 and H3K27ac, are lost via lysine demethylases (KDMs) and histone deacetylases (HDACs). Conversely, repressive modifications such as DNA methylation, H3K27me3, and H3K9me3 are gained because of epigenetic writers, including DNMT3A, EZH2 as part of the Polycomb repressive complex 2 (PRC2), and SUV39H1. Not shown but occurring simultaneously is the acquisition of activating epigenetic modifications at effector-associated genes during T cell differentiation.
Understanding the mechanisms of epigenetic regulation of Teff cell differentiation has considerable implications for multiple fields, including cancer immunotherapy. Less differentiated T cell subsets, such as Tscm and Tcm cells, have enhanced proliferative potential and greater antitumor activity when transferred into both mice and humans compared with the more differentiated T effector memory cell (Tem cell) and Teff cell subsets. This is likely due to their stem cell–like properties (4, 6). Because the majority of cells currently used for T cell–based cancer immunotherapy are Teff cells, the epigenetic silencing of stem and memory genes in these cells poses a considerable therapeutic roadblock. To reacquire therapeutically beneficial stem cell–like properties, Teff cells would need to be epigenetically reprogrammed. This can be experimentally accomplished, albeit inefficiently (15). A greater understanding of the CD8+ T cell epigenome may therefore provide essential clues for how to unlock the potential of highly differentiated, tumor-antigen-specific T cells infiltrating tumors (4). Epi-genetic modifying drugs may reverse the repression of stem and memory genes in differentiated T cells and improve T cell-based immunotherapies.
REFERENCES
- 1.Hawkins RD et al. , Cell Stem Cell 6, 479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pace L et al. , Science 359, 177 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Restifo NP, Gattinoni L, Curr. Opin. Immunol 25, 556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gattinoni L et al. , Nat. Rev. Cancer 12, 671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gattinoni L et al. , Nat. Med 17, 1290 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klebanoff CA et al. , J. Clin. Invest 126, 318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graef P et al. , Immunity 41, 116 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Youngblood B et al. , Nature 10.1038/nature25144 (2017). [DOI] [Google Scholar]
- 9.Friedmann-Morvinski D, Verma IM, EMBO Rep. 15, 244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phan AT et al. , Immunity 46, 714 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crompton JG et al. , Cell Mol. Immunol 13, 502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladle BH et al. , Proc. Natl. Acad. Sci. U.S.A. 113, 10631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray SM et al. , Immunity 46, 596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakaradov B et al. , Nat. Immunol 18, 422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Yamanaka S, Cell 126, 663 (2006). [DOI] [PubMed] [Google Scholar]