Abstract
Alcohol dependence (AD) affects individuals from all racial/ethnic groups and previous research suggests that there is considerable variation in AD among ethnic minority groups in the United States. Although the reasons for these differences are likely due, in part, to contributions of complex sociocultural factors, limited research has attempted to examine whether similar genetic variation plays a role within both ancestral groups. Using a pooled sample of individuals of African and European ancestry (AA/EA) obtained through data shared within the Database for Genotypes and Phenotypes (dbGAP), we estimated the extent of additive genetic similarity for AD between AA and EAs using common single nucleotide polymorphisms (SNPs) that overlapped across the two populations. AD was represented as a factor score using Diagnostic and Statistical Manual (DSM-IV) dependence criteria and genetic data was imputed using the 1000 Genomes Reference Panel. Analyses revealed a significant SNP-based heritability of 22% (SE=5%) in EAs and 27% (SE=13%) in AAs. Further, a significant genetic correlation of 0.77 (SE=0.46) suggests that the allelic architecture influencing the AD factor for EAs and AAs is largely similar across the two populations. Follow-up analysis indicated that investigating the genetic underpinnings of alcohol dependence in different ethnic groups may serve to highlight etiological influences that are otherwise missed.
Introduction
Alcohol dependence (AD) is a global problem that affects individuals from all racial/ethnic groups and all levels of socioeconomic standing. Previous studies suggest that there is considerable variation in patterns of drinking and alcohol use disorders among U.S. ethnic minority groups (Caetano et al., 1998). For example, among college-aged students (i.e., ages 18–24), higher rates of alcohol use and disorders are observed amongst individuals of European and Native American ancestry compared to citizens of African or Asian ancestry. While there is evidence to suggest the risk for AD may be greater, in part, for some individuals as a function of their economic standing and familial background, there have been a limited number of studies that have attempted to examine whether genetic variation might also play a role in observed individual differences among African Americans. A review of the published literature using a combination of search terms (see appendix) in PubMed (in October 2016) revealed 14 of genomewide association studies that have examined genetic variants related to alcohol consumption and/or dependence with some combination of analyses in ancestrally mixed samples or ancestry-specific (i.e., identified using samples(Bierut et al., 2010; Gelernter et al., 2014; Johnson et al., 2011; Panhuysen et al., 2010; Ulloa et al., 2014; Xu et al., 2015; Yang et al., 2014a; Zuo et al., 2012a; Zuo et al., 2014; Zuo et al., 2013a; Zuo et al., 2015; Zuo et al., 2012b; Zuo et al., 2013b). Of these, none have empirically compared/contrasted the genome-wide effects in subjects of African ancestry (AA) and European ancestry (EA), at least as best defined by the International HAPMAP (2003) or 1000 Genomes Projects.
Whole genome association studies across mixed populations are limited by small sample sizes across groups and the methodological limitations that arise from differences in linkage disequilibrium across ancestral groups, as well as variation in minor allele frequencies (MAFs) between groups, all of which affects statistical power. At the same time, these differences highlight the strengths of conducting candidate gene and genome-wide types of association studies within each of these ancestrally defined groups, to the extent that sufficient statistical power is achieved. This has largely been seen in studies of candidate biological systems, some of which have shown that increased power can be gained by studying other ethnic groups where certain alleles are more commonly observed in comparison to subjects of European ancestry. For example, the most reproduced effect on AD in GWAS center around variation in and around the chromosome 4 ADH cluster. Genes that play a role in the alcohol metabolizing system and its associated genes on chromosomes 4 (ADH1B, ADH1C, ALDH2, and ADH4) have been observed in individuals of Korean, Chinese, African, and European ancestry (Frank et al., 2012; Park et al., 2013; Quillen et al., 2014). Among the studies including AAs, the recent Gelernter et al. (2014) genome-wide study of a pooled sample of 16,087 individuals of European and African ancestry was the only one to explore convergence of genomewide significant findings across the sub populations. Notably, ADH1B was identified along with several other novel loci. The Gelernter study supports the hypothesis that genetic variation across similar regions index common biological systems that are susceptible to long-term exposure to alcohol. The report by Gelernter et al., (2014) was the first to provide some indication of shared genetic effects around a nominal GWAS finding. The current study expands upon the Gelernter et al. (2014) paper by using pooled samples of EAs and AAs (respectively) to estimate the extent of genomewide additive genetic similarity for AD between AA and EAs. The data for this project are the result of sharing agreements imposed by the National Institutes of Health and principal investigators that support collaborative work by submitting their data to the Database for Genotypes and Phenotypes (dbGAP). This is, to our knowledge, the first study of its kind to estimate these genomewide effects using molecular data.
Methods
Sample
All study data was accessed as part of the National Human Genome Research Institute’s Gene Environment Association Study Initiative [Database for Genotypes and Phenotypes (dbGaP)]. For all analyses, data from four dbGaP datasets were pooled, including: The Study of Addiction: Genetics and Environment (SAGE; study accession phs000092.v1.p1), the Alcohol Dependence GWAS in European- and African Americans (Yale Study; study accession phs000425.v1.p1), the Australian twin-family study of alcohol use disorder (OZ-ALC; study accession phs000181.v1.p1), the Genome-Wide Association Study of Heroin Dependence (Heroin GWAS study; study accession phs000277.v1.p1). Table 1 describes the set of samples.
Table 1.
Description of samples aggregated to identifying genetic factor related to alcohol dependence
| Study | N | Description |
|---|---|---|
| Study of Addiction: Genetics and Environment (SAGE) | 4,316 | A multi-ethnic sample of unrelated individuals from three large, complementary data sets designed to study drug addiction: the Collaborative Study on the Genetics of Alcoholism (COGA), the Family Study of Cocaine Dependence (FSCD) and the Collaborative Genetic Study of Nicotine Dependence (COGEND). |
| Alcohol Dependence GWAS in European- and African Americans (Yale Study) | 2,909 | A case-control study focusing on AAs and EAs who meet DSM-IV criteria for AD. The sample was collected over the course of ongoing projects that focused on oversampling of alcohol dependent AAs and also included measures on cocaine and opioid dependence. The sample was originally collected to identify sibling pairs suitable for linkage analysis. |
| Australian twin-family study of alcohol use disorder (OZ-ALC) | 6,701 | A family study deriving from two general population volunteer cohorts of twins in Australia totaling over 11,000 families. Two cohorts of twins born between 1940–1961 (cohort 1) or 1964–1971 (cohort 2) were assessed using a shared protocol to discover genes related to alcohol use. Data from these studies were compiled into a case-control family-based GWAS that focused on alcohol use and dependence |
| Genome-Wide Association Study of Heroin Dependence (Heroin GWAS) | 6,410 | A collaboration of investigators from the United States and Australia to identify genes associated with heroin dependence using a case-control study. Data on participants from the Heroin Study who were assessed for dependence on alcohol consisted of the following from ongoing genetic studies of substance dependence conducted by investigators at Yale and collaborating institutions:
|
Assessments
Each study collected DSM-IV symptoms (coded as present or absent) for AD using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA; SAGE study), the adapted SSAGA-OZ (OZ-ALC study), or the Semi Structured Assessment for Drug Dependence and Alcoholism (Yale Study, Heroin GWAS) (Bucholz et al., 1994; Hesselbrock et al., 1999; Pierucci-Lagha et al., 2005). All responses were limited to individuals who had previously been exposed to alcohol (and possibly other drugs).
Genotyping, Quality Control, and Genetic Imputation
GWAS data were obtained through the National Center for Biotechnology Information’s (NCBI) Database for Genotypes and Phenotypes (dbGAP), where more detailed protocols are available. For each sample set, quality control (QC) and imputation of autosomal SNPs were conducted separately by study and are explained below. Genotyping in SAGE was conducted using the Illumina Human 1M BeadChip. Genotyping in the Yale Study was conducted on the Illumina HumanOmni1-Quad v1.0 microarray. Genotyping for the OZ-ALC study was conducted on Illumina HumanCNV370-Duov1 BeadChip. Finally, participants from the Heroin GWAS were genotyped on three separate platforms: Illumina Human610 Quad v1, Illumina Human660W Quad v1, and HumanCNV370 Quad v3.0.
Genomic data across all samples were imputed (within sample [by ethnic group]) up to Phase III of the 1000 Genomes Project (1KG) in order to maximize similar genetic coverage across samples. Data management was conducted using SNP & Variation Suite v8.x (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com)(SNP & Variation Suite (Version 8.4.4)), PLINK version 1.9 (Purcell et al., 2007), and R version 3.1.1. Genetic imputation was conducted using Minimac (version 3) via the Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html#!pages/home). Ancestry determination of sample data and imputation of genotypes utilized data from Phase III of the 1,000 Genomes Project (1KGP) reference sample (Auton et al., 2015).
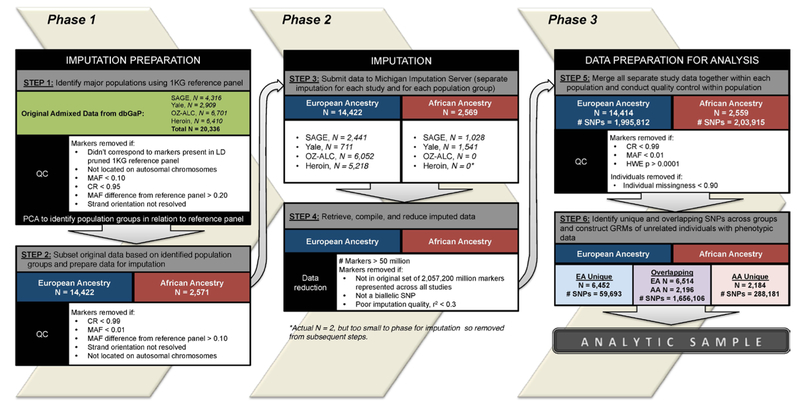
A series of steps across three phases was conducted to prepare data for imputation, impute data, and prepare data for analyses. A flow chart of this procedure is presented in Figure 1.
Figure 1.
Flow chart of data preparation and imputation.
Phase 1: Imputation preparation.
Step 1: Identify major ancestral populations within sample data using the 1KGP reference sample.
Subject ancestry was determined using the Phase 3 reference panel from the 1KGP, which is comprised of 2,504 individuals across 26 populations and contains genotyping data for 84.4 million markers. The major ancestral groups captured in this data are East Asian, South Asian, African, European, and Americas. For ancestry determination, we restricted the number of markers in the 1KGP to include only the union of markers present in each of our sample data sets (2,240,710). We then removed any markers with a minor allele frequency (MAF) less than 5% and a call rate (CR) less than 99%. Finally, we used a subset of the resulting set of 1KGP data based on linkage disequilibrium (LD, r2 < 0.5), resulting in a final set of 423,738 markers to be used for ancestry determination.
Quality control (QC) was conducted in each of the study samples separately prior to being combined with the reference panel. Each study sample set was subset to include autosomal SNPs with MAF greater than 10% and a CR of 95%. Using allele information compiled from the marker map of the reference panel data, we compared the allele frequencies (across all populations) and strand orientation of our data to the reference panel. Markers that had minor allele frequency differences of greater than 20% when compared to the reference panel were removed. Markers whose stand orientation could not be resolved (e.g. flipped) were also removed.
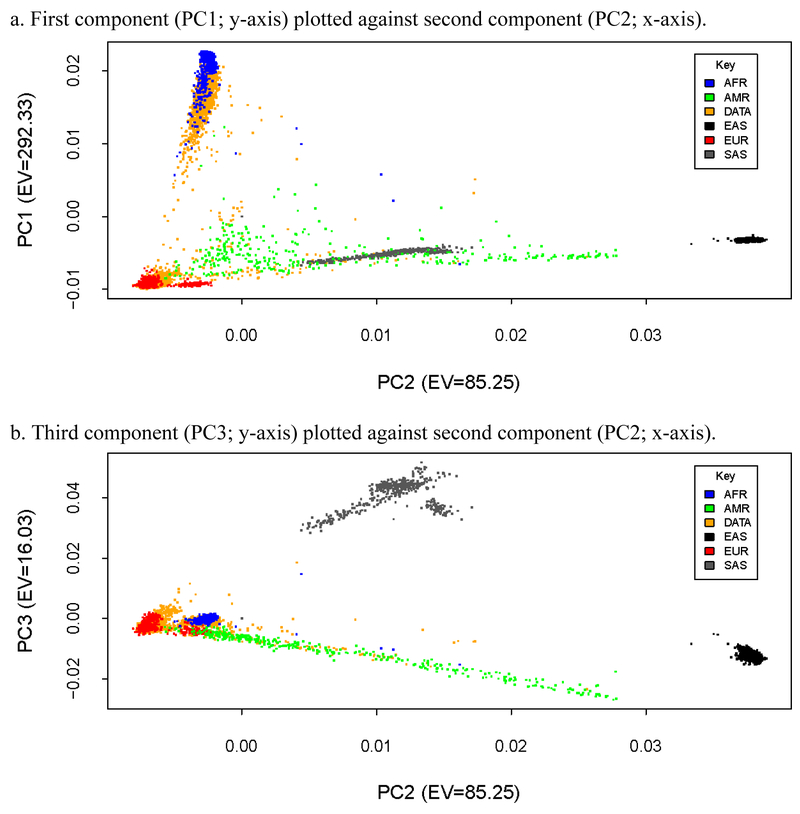
After QC of the sample data and preparation of the reference panel data were complete, the study samples were combined with the reference data (separately) to determine ancestry within each study. Principal components analysis (PCA) was conducted within each study to examine population stratification. Plots of genetic components were examined visually and statistically to determine ancestral groups. First, scatterplots comparing the first, second, and third components, which largely distinguish between African, East Asian, South Asian, and European groups, were plotted to determine ancestry of the sample data compared to the reference data. For example, Figure 2 presents scatterplots of principal components of the SAGE data with the 1KG reference data. In panel a, the first principal component is plotted against the second principal component. The first component in each of the data sets separated African and European ancestral groups, which represented the larges two subgroups in each of samples examined in this study.Subsequently, we calculated the mean and standard deviations of the first principal component in the reference panel and retained individuals in the sample data whose eigenvector value fell within two standard deviations (i.e., 98% of the 1KG ansestral distributions) of the African and European reference panel component means. As such, the current study clusters individuals into two groups, African Ancestry (AA) or European Ancestry (EA), based on their proximity to established ancestral groups within the 1KG reference panel data. To reduce further population stratification, we conducted multidimensional outlier detection within the identified AAEA groups in the sample data using a multiplier value of 1.5. This procedure computed a distance score based on the median centroid vector calcutated using the first three principal components. Any individuals determined to be outliers from the AA and EA samples were removed from the sample data. The resulting set of AA and EA individuals in the sample data were selected for imputation.
Figure 2.
Scatterplots of genetic principal components of the SAGE sample data plotted with 1000 Genomes Reference Panel ancestral groups data.
Note: EV = eigenvalue; 1000 Genomes Referance Panel ancestral groups included: AFR = African; AMR = Americas; DATA = SAGE sample data; EAS = East Asian; EUR = European; SAS = South Asian.
Step 2: Subset original data based on identifed population groups and prepare data for imputation..
After the AA and EA individuals were identified in Step 1, the original sample data were subset into the two respective groups identified by PCA to be imputed separately. QC was conducted in each group and markers with CR < 95% or MAF < 1% were removed. Using allele information compiled from the marker map of the reference panel data, we compared the allele frequencies (specific to expected allele frequencies based on the 1KG African or European populations) and strand orientation of our data to the reference panel. Markers that had minor allele frequency differences of greater than 10% when compared to the reference panel population were removed. Markers whose stand orientation could not be resolved (e.g. flipped) were also removed. Individuals who had greater than 95% missingness were also removed. The final set of data for AA/EA individuals within each study was separated into autosomal chromosome files for submission to the Michigan Imputation Server.
Phase 2: Imputation of genotypes in identified EAs and AAs (separately).
Step 3: Submit data for imputation.
Ancestral groups within each study were imputed separately on the Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html) using Minimac3 with the 1KG Phase 3 reference panel and SHAPEIT phasing.
Step 4: Retrieve, compile, and reduce imputed data.
After imputation was completed, each file totalled over 50 million markers. Files were subset based on the union of the aforementioned 2,057,200 million markers present across all the studies and markers that did not represent biallelic SNPs were removed. Finally. markers with an imputation quality score (r2) < 0.3 were removed.
Phase 3: Data preaparation for analysis.
Step 5: Merge all study data together within each population and conduct QC.
Following imputation, all study data for EAs across each study were merged. Likewise, data for AAs across each study were merged. QC was conducted within each ancestry group separately to select individuals with missingness < 10% and markers with CR > 99%, MAF > 1%, and that passed HWE test (p < 0.0001) (see Supplementary Table S1 for summary of the number of markers across EAs/AAs that drop out at each step).
Step 6: Identify unique and overlapping SNPs across ancestral groups and construct genetic-relatedness matrices of unrelated individuals.
Following QC, we identified a common set of SNPs across both populations (1,656,106 in EAs and AAs) and a set of SNPs that survived QCs in in one group but not the other (N=288,181 unique to AAs; N=59,693 unique to EAs) . Each set of SNPs were then used to contruct genetic relationship matrices (GRMs). The GRMs were computed using the Genomewide Complex Trait Analysis (GCTA) software tool [version 1.25.3] to maximally select one of any pair of individuals who were more related than second cousins (Yang et al., 2011) . Subsetting the data for unrelated individuals was done to control for cryptic relatedness, which could artificially inflate SNP heritability estimates (see below). The ancestry specific GRMs used in univariate genetic analyses were comprised of 2,257 unrelated AA individuals and 8,722 unrelated EA individuals. Joint analyses for each ancestry group using the GRM constructed from overlapping markers as well as the GRM constructed from sample-specific markers provided the total amount of variation in AD attributable to genetic variants. In addition, bivariate genetic models described below used a combined GRM of 11,314 individuals who had overlapping SNPs to provide an estimate of genetic correlation between populations.
Derivation of Phenotypes and Sample Characteristics
Data for the seven DSM-IV AD symptoms were pooled to determine and confirm the factor structure of the AD latent variable in EAs and AAs. Data for participants in each study was subset to include only those participants who were unrelated and were genetically determined to be EA or AA; consequently, data for 6,514 genetically determined EAs and 2,196 genetically determined AAs who had phenotypic data were used for factor analysis. The final sample of EA individuals were 53.22% male and ranged in age from 16 to 82 (mean age = 40.16, standard deviation = 10.42). Of these individuals, 34.35% came from the SAGE study, 8.26% came from the Yale study, 15.13% came from the OZ-ALC study, and 42.26% came from the Heroin GWAS study The final sample of AA individuals were 51.09% male and ranged in age from 16 to 79 (mean age = 40.48, standard deviation = 8.84). Of these AA individuals, 37.93% came from the SAGE study and 62.07% came from the Yale study. The OZ-ALC and Heroin GWAS studies did not contain enough individuals of African ancestry to impute genetic data, and thus did not contribute to the final sample of AAs used in the current study.
The factor structure of AD symptoms within each ancestral group was determined by randomly splitting each subpopulation in half to create exploratory and confirmatory subsets. Exploratory and confirmatory factor analyses (EFA/CFA) were conducted in Mplus [version 7] (Muthén and Muthén, 1998–2015) using weighted least-squares mean variance estimation. Missing data was handled in Mplus with full information maximum likelihood estimation. The exploratory subsets consisted of 1,098 AA participants and 3,255 EA participants. The confirmatory subsets consisted of 1,098 AA participants and 3,260 EA participants. Scree plots, consistency with previous empirical research, and examination of fit indices [e.g. root mean square error of approximation (RMSEA), comparable fit index (CFI) and Tucker–Lewis index (TLI)] were used to determine the factor structure for each 1KGP defined ancestral group (Hu and Bentler, 1999; Yu, 2002). EFA and CFA models indicated that a single latent factor represented AD symptoms (see Supplementary Figure S1 for scree plot and Supplementary Table 2 for model fit for EFA/CFA across ancestral groups). Measurement invariance of the single latent factor across the two ancestral groups supported configural invariance, which established that the same factor structure, but different error variances and item thresholds, existed for each group.
Based on consensus from EFA/CFA, separate factor scores (mean = 0, standard deviation = 1) from a one-factor solution were extracted for EAs and AAs to be used in genetic analyses. Specifically, these analyses yielded factor scores derived within each 1KGP defined ancestral group that represent latent indicators of alcohol dependence based on the seven DSM-IV symptoms specific to that ancestral group. The factor model, with unstandardized factor loadings, are presented in Figure 2 along with fit indices for each CFA model. After extraction, each factor score was residualized to account for variation due to age, sex, and study of origin. The residualized factor scores were used for all subsequent genetic analyses.
Estimation of variance/covariance explained by the SNPs
Genomic restricted maximum likelihood estimation (GREML) to was used to decompose phenotypic variance in the EA and AA AD factors into additive effects of genotyped and imputed SNPs (Yang et al., 2013). In this approach, genetic similarity captured in each GRM is modeled as a random effect to account for variance in the residualized AD factor score for each ancestry group. Two separate variance components were included in each ancestry-specific linear model: one component comprised of genetic variance due to markers that overlapped between ancestry groups and one that represented genetic variance due to sample-specific markers (i.e., markers that passed QC for one group but not the other). Total SNP-heritability for each model represents the total variation across each component for EAs and AAs separately. In bivariate GREML models, the covariance between two groups can be described by a standard bivariate linear mixed model in which covariance is reflected by the covariance between the genetic and environmental/residual factors influencing each trait. Only genetic variance attributable to overlapping markers was used in the bivariate model. As such, with the current data, the additive genetic correlation (rG-SNP) reflects shared genetic variance tagged by the genotyped SNPs is interpreted as the extent to which the genetic variants influencing AD in EAs and AAs are correlated (ranging in value from −1 to 1)(de Candia et al., 2013; Lee et al., 2012). Consequently, analyses were designed to determine the SNP heritability (h2SNP) within each ancestral group as well as the genetic correlation across EAs and AAs (using the set of overlapping SNPs that survive QC across EAs’ and AAs). Given the lack of direct evolutionary pressures related to alcohol use, we hypothesized that SNP-heritability estimates would be similar across EAs and AAs and that the genetic correlation would be large (>0.60). Given, our observation of non-overlapping SNP sets following sample QC, we also explored the extent to which these SNPs might be an additional source of genetic variation for EAs and AAs.
Results
Prevalence of alcohol dependence items across ancestral groups
Demographics of sample data by ancestral group are presented in Table 3. Prevalence rates and correlations between AD symptoms are presented in Table 4. For AAs, “using longer than intended” (58% endorsed) and “great time spent” (57%) were the highest endorsed items, with all items being endorsed at rates between 33–58%. For EAs, “using longer than intended and “tolerance” (49%) were the highest endorsed items, with all items being endorsed at rates between 25–57%. Phenotypic tetrachoric correlations among all items were generally high (all greater than 0.58).
Table 3.
Demographics of sample data by ancestral group.
| EA | AA | |||||
|---|---|---|---|---|---|---|
| N | % | N | % | |||
| Sex | ||||||
| Male | 3467 | 53.22% | 1122 | 51.09% | ||
| Female | 3048 | 46.78% | 1074 | 48.91% | ||
| Study | ||||||
| SAGE | 2238 | 34.35% | 833 | 37.93% | ||
| YALE | 538 | 8.26% | 1363 | 62.07% | ||
| OZ-ALC | 986 | 15.13% | 0 | 0.00% | ||
| Heroin | 2753 | 42.26% | 0 | 0.00% | ||
| M | SD | M | SD | |||
| Age | 40.16 | 10.42 | 40.48 | 8.84 | ||
Note: EA = European ancestry; AA = African ancestry; N = sample size; M = mean; SD = standard deviation.
Table 4.
Item endorsement and polychoric correlations of DSM-IV Alcohol Dependence for each ancestral group.
| Symptom | % | N | Correlation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| African Ancestry | 1. | 2. | 3. | 4. | 5. | 6. | |||
| 1. Tolerance | 47% | 1033 | |||||||
| 2. Withdrawal | 53% | 1170 | 0.66 | ||||||
| 3. Longer than intended | 58% | 1279 | 0.70 | 0.63 | |||||
| 4. Attempt to quit | 56% | 1238 | 0.76 | 0.68 | 0.69 | ||||
| 5. Time spent | 57% | 1251 | 0.69 | 0.89 | 0.68 | 0.74 | |||
| 6. Acts foregone | 33% | 724 | 0.66 | 0.59 | 0.66 | 0.68 | 0.67 | ||
| 7. Continued use | 46% | 998 | 0.69 | 0.66 | 0.66 | 0.71 | 0.70 | 0.72 | |
| European Ancestry | |||||||||
| 1. Tolerance | 49% | 2880 | |||||||
| 2. Withdrawal | 26% | 1660 | 0.58 | ||||||
| 3. Longer than intended | 57% | 3278 | 0.67 | 0.71 | |||||
| 4. Attempt to quit | 40% | 2608 | 0.65 | 0.75 | 0.74 | ||||
| 5. Time spent | 30% | 1656 | 0.60 | 0.86 | 0.71 | 0.72 | |||
| 6. Acts foregone | 25% | 1421 | 0.62 | 0.82 | 0.73 | 0.75 | 0.86 | ||
| 7. Continued use | 41% | 2292 | 0.65 | 0.78 | 0.78 | 0.75 | 0.76 | 0.85 | |
Phenotypic variance attributable to AD among EAs and AAs
Total SNP-based heritability estimates of the AD factor were similar across EAs and AAs. See Table 5 for a summary of univariate results within EA and AA groups. Partitioning of the total genetic variance for EAs using multiple GRMs in a single linear model revealed a significant SNP heritability estimate of 0.17 (SE = 0.05, p < 0.001) for variation in AD attributable to SNPs that overlapped with AAs and a non-significant estimate of 0.10 (SE = 0.04, p < 0.001) for variation in AD attributable to SNPs that were sample-specific to EAs (e.g. these markers include markers that passed QC for EAs but not for AAs).
Table 5.
Univariate SNP-heritability [h2SNP (SE)] of AD factor for EAs and AAs.
| h2SNP Gene set A | h2SNP Gene set B | Total SNP-Heritability | |
|---|---|---|---|
| European Ancestry | 0.17 (0.05)*** | 0.10 (0.04)** | 0.27 (0.05)a |
| African Ancestry | 0.24 (0.15)* | 0.07 (0.14) | 0.30 (0.15)a |
Note: Table presenting the univariate SNP-heritability of AD factors for EAs and AAs using subsets of SNPs (Gene set A comprises SNPs that survive within-ancestral-group quality control procedures [QC] across both populations; Gene set B includes SNPs that differentially survive QC across ancestral groups; Total SNP-heritability reflects the genomewide effects of Gene sets A & B within each ancestral group). Notations:
p < 0.05,
p < 0.01,
p < 0.001;
significance test not available for total heritability in model, as the likelihood ratio test is conducted only on the specific variance components within the model.
Partitioning of the total genetic variance for AAs revealed a significant SNP heritability estimate of 0.24 (SE = 0.15, p = 0.028) for variation in AD attributable to SNPs that overlapped with EAs and a non-significant estimate of 0.07 (SE = 0.14, p = 0.313) for variation in AD attributable to SNPs that were sample-specific to AAs (e.g. these markers include markers that passed QC for AAs but not for EAs).
Further examination of the additive genetic effects (i.e., due to SNPs that survive QC across EAs and AAs) for AD in EAs and AAs partitioned by chromosomes (see Supplementary Figure S2) indicated that longer chromosomes did not account for significantly more phenotypic variation among overlapping markers than shorter ones (For EAs: R2=0.15, β=6.96×10−11, t(20)=1.87, p=0.076; For AAs: R2=0.01, β=1.98×10−11, t(20)=0.39, p=0.704).
Genetic correlation attributable to overlapping markers across EAs and AAs
Bivariate analyses revealed a significant genetic correlation between EAs and AAs (rG-SNP = 0.77, SE = 0.46, p = 0.030) for SNPs that survived QC across both ancestral groups. Overall, this suggests that there is moderate evidence for convergence across EA and AAs for a subset of genomewide SNPs that contribute to the additive genetic variance of AD.
Discussion
Results from this study are, to our knowledge, the first to directly compare the SNP-based genetic liability for AD across individuals of African and European ancestry. The inclusion of racial and ethnic minority groups in genetic research (when used appropriately and ethically) is essential to progress in understanding the role that genetic and socio-cultural factors play in racial and ethnic health disparities. Large scale GWA studies have primarily concentrated on European populations, with very little representation of individuals of African ancestry (Need and Goldstein, 2009). Yet despite the tendency for genetically informed studies to focus on populations of European ancestry, psychological research has found that compared to their European American counterparts, African Americans initiate drinking at an older age and report overall lower rates and levels of use and higher levels of abstinence (Quality, 2016; Zapolski et al., 2014). Further, African American drinkers report significantly higher rates of social consequences and alcohol dependence symptoms compared to European Americans (Mulia et al., 2009).
Evidence from the current study supported a moderately shared genetic liability for AD across EA and AA groups, yet empirical research has identified social, cultural, health, environmental, historical, economic and numerous other demographic factors that contribute to observed disparities in AD risk and consequences among African Americans (Zapolski et al., 2014). It is also possible that the intersectionality of multiple other risk factors (Mereish and Bradford, 2014), as well as specific individual and environmental influences, may impart an impact on risk for substance use above and beyond the observed genetic effects (McGue et al., 2000). Although the present study does not explore sociocultural factors, it lays the groundwork for beginning to explore these potential sources of variation in the context of genetic variation (i.e., Gene x Environment interaction) that likely have similar patterns of effects in European populations.
SNP-based heritability estimates found in this study are consisted with previous work using the GREML method using various parameterizations of the alcohol dependence phenotype. Our findings indicate that 27% of the phenotypic variation of the alcohol dependence factor in EAs and 30% of the phenotypic variation in AA was attributable to additive genetic effects when examining a set of the same genetic markers across the two populations. These estimates are similar to the 30% SNP-based heritability estimated by Palmer and colleagues (2015) using an alcohol dependence factor in a sample of EAs, within the margin of error. Recent studies that utilized alcohol dependence diagnosis, rather than the factor score, have estimated a heritability of 21% in a Caucasian sample (Vrieze et al., 2013) and 22% in an African American sample (Yang et al., 2014b). Kos and colleagues (2013) recently estimated that 38% and 35% of the variation in AD diagnosis risk is attributable to common SNPs in EAs and AAs, respectively; however, their study did not limit the SNPs used in the estimations of GRMs to be overlapping across populations and thus different markers for each ancestral groups could have contributed to the observed genetic variance in the heritability estimates. For example, the current data suggested that SNPs that differentially survive QC across our groups may contribute an additional 7% genetic variance to the AA AD factor and an additional 10% genetic variance to the EA AD factor. Though the statistical significance of the AA value is precluded by the large standard error, power simulations conducted by Visscher et al. (2014) indicate that with a larger sample, the standard error will decrease rendering the effect significant. As such, the additional sources of genetic variance for EAs or AAs beyond what is captured through overlapping markers may simply represent variance attributable to markers that are more easily detected in one group relative to the other.
The significant genetic correlation found in this study suggests that the allelic architecture influencing the AD factor for EAs and AAs is partly shared across the two populations. In a single population, genetic correlations arise from pleiotropy or co-segregations of causal variants among genes influencing multiple traits. In the current analysis, the significant genetic correlation represents these genetic contributions influencing a single trait (AD) measured across two populations. Consistent with the conclusion reached by de Candia et al. (2013) and given that it is unlikely that different causal variants across ancestral populations would be in LD with the same SNP, the common causal effects tagged by the SNPs that survive QC in both 1KGP-defined ancestral groups likely predate the European-African divergence. It should be noted that we assume, by design, that variation that postdates the European-African divergence would emerge as markers that differentially survive QC across both ancestral groups. Future work should examine the AD factor in other populations to delineate whether these results apply broadly.
One important consideration for the results of this study, and for future studies, is the fact that genetic markers may have different allele frequencies and different effect sizes in relation to a given phenotype across different ancestral populations. Aside from removing rare variants (MAF < 0.01), the current study did not control for differences in allele frequencies across populations during the calculation of genetic correlation. It is possible that genetic influences on AD may be mitigated or exacerbated by differences in allele frequency across populations (e.g., variants that are more common in one population may contribute more to the overall effect), rendering our estimates to be slightly biased. However, one recent study concluded that when variants common in both populations are examined, differences in allele frequencies impart minimal effects on genetic correlation based solely on effect sizes (Brown et al., 2016). The GREML approach used in this study treats SNP effects as statistically random and therefore does not estimate individual effects, but future work could tease out individual SNP-effects to determine which markers contribute the largest effects. Another point of considerations is that the present study focused on common biallelic variants that were present across both ancestral populations, little is known about how rare variation (e.g., copy number variants, multiallelic makers, exome variation) contributes to AD. Kos et al. (2013) show support for modest effects of rare and uncommon loci on the susceptibility for AD that were captured from GWAS signals and then aggregated.
In summary, the demonstration that (1) approximately 60% of the genetic variation for AD that is tagged by measured and retained genome-wide SNPs is shared across EAs and AAs, and (2) additional sources of genetic variation may be captured by studying variants that differentially survive QC in one population but not the other. Overall, these observations underscore the reciprocal value of whole genome alcohol studies of ethnically divergent populations, such as such as Native Americans and Asians that have been well characterized phenotypically.
Supplementary Material
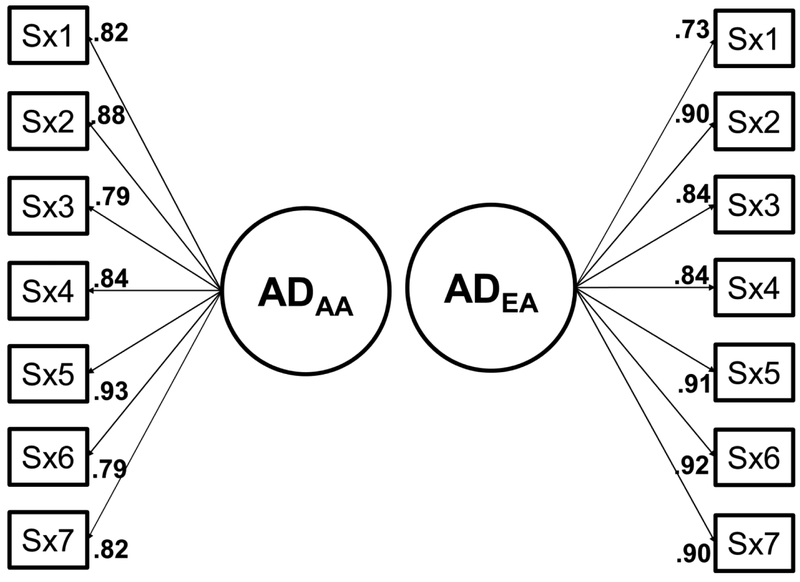
Figure 3.
Confirmatory factor analysis of DSM-IV Alcohol Dependence among individuals of African or European ancestry.
Note: AD = alcohol dependence; AA = African ancestry; EA = European ancestry; Sx1 = Tolerance, Sx2 = Withdrawal, Sx3 = Use longer than intended, Sx4 = Failure to quit, Sx5 = Great time spent using/recovering, Sx6 = Activities foregone, Sx7 = Continued use despite problems.
Table 2.
Model fit for Alcohol Dependence factors among African Ancestry and European Ancestry groups.
| Model Fit Information | ADAA | ADEA |
|---|---|---|
| χ2(14) | 259.800 | 302.649 |
| p-value | <0.001 | <0.001 |
| CFI | 0.986 | 0.995 |
| RMSEA [90% CI] | 0.089 [0.080, 0.099] | 0.056 [0.051, 0.062] |
Note: AD = alcohol dependence; AA = African ancestry; EA = European ancestry; CFI = confirmatory fit index; RMSEA = root mean square error of approximation.
Acknowledgements
The authors gratefully acknowledge Mr. Bryce Christensen and Golden Helix (Bozeman, Montana) for their assistance with data preparation for genomic imputation analyses. Further, we acknowledge the services of the Michigan Imputation Server team for the use of their publicly available resource for genomic imputation. Lastly, we acknowledge the many grant funded research projects (described below) that assembled all of the data used in the current study, as well as the NIH’s database for Genotypes and Phenotypes, which provides a platform for data sharing.
Funding support for the SAGE study was supported by NIH and NHLBI grant # R01HL117004; study enrollment supported by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II.
Funding support for the OZ-ALC GWAS was provided through the Center for Inherited Disease Research (CIDR) and the National Institute on Alcohol Abuse and Alcoholism (NIAAA). CIDR-OZ-ALC GWAS was funded as part of the NIAAA grant 5 R01 AA013320–04. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the CIDROZ-ALC GWAS. Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of OZ-ALC datawas provided by the MARC: Risk Mechanisms in Alcoholism and Comorbidity (MARC; P60 AA011998–11). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
Funding support for the Yale Study was provided through the Center for Inherited Disease Research (CIDR) and the Genetics of Alcohol Dependence in American Populations (CIDR-Gelernter Study). CIDR-Gelernter Study is a genome-wide association studies funded as part of the Genetics of Alcohol Dependence in American Populations. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the Genetics of Alcohol Dependence in American Populations. Assistance with data cleaning was provided by the National Center for Biotechnology Information. Cleared 1/4/10 Gelernter: Whole Genome Association (CIDR-Gelernter Study) Dataset January 21, 2010 version Page 5 of 7 Support for collection of datasets and samples were provided by the Genetics of Alcohol Dependence in American Populations (R01 AA011330). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap through dbGaP accession number [phs000425].
Finally, funding support for the GWAS of Heroin Dependence was provided by R01DA17305.
Declaration of interests
Role of Funding Sources
This project was supported by a K01 grant (K01AA021113 awarded to Dr. Palmer) from the National Institute on Alcohol Abuse and Alcoholism (NIAAA), a T32 (T32MH019927; which supports the postdoctoral training of Dr. Brick), and an R01 (MH100141) from the National Institute on Mental Health (NIMH; awarded to Dr. Keller). The views expressed in this article do not necessarily reflect the position or policy of the Department of Veteran Affairs.
Footnotes
Conflict of Interest
All of the listed authors declare that they have no conflicts of interests.
References
- (2003) The International HapMap Project. Nature 426:789–796. [DOI] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP (2010) A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A 107:5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BC, Ye CJ, Price AL, Zaitlen N, Consortium AGENTD (2016) Transethnic Genetic-Correlation Estimates from Summary Statistics. The American Journal of Human Genetics 99:76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr., Reich T, Schmidt I, Schuckit MA (1994) A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of studies on alcohol 55:149–158. [DOI] [PubMed] [Google Scholar]
- Caetano R, Clark CL, Tam T (1998) Alcohol consumption among racial/ethnic minorities: theory and research. Alcohol Health Res World 22:233–241. [PMC free article] [PubMed] [Google Scholar]
- de Candia TR, Lee SH, Yang J, Browning BL, Gejman PV, Levinson DF, Mowry BJ, Hewitt JK, Goddard ME, O’Donovan MC, Purcell SM, Posthuma D, Visscher PM, Wray NR, Keller MC (2013) Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. Am J Hum Genet 93:463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mossner R, Gaebel W, Dahmen N, Scherbaum N, Schmal C, Steffens M, Lucae S, Ising M, Muller-Myhsok B, Nothen MM, Mann K, Kiefer F, Rietschel M (2012) Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol 17:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA (2014) Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V (1999) A validity study of the SSAGA--a comparison with the SCAN. Addiction 94:1361–1370. [DOI] [PubMed] [Google Scholar]
- Hu LT, Bentler PM (1999) Cutoff criteria for fit indices in covariance structure analysis: conventional criteria versus new alternatives. Structural Equation Modeling 6:1–55. [Google Scholar]
- Johnson C, Drgon T, Walther D, Uhl GR (2011) Genomic regions identified by overlapping clusters of nominally-positive SNPs from genome-wide studies of alcohol and illegal substance dependence. PLoS One 6:e19210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos MZ, Yan J, Dick DM, Agrawal A, Bucholz KK, Rice JP, Johnson EO, Schuckit M, Kuperman S, Kramer J (2013) Common biological networks underlie genetic risk for alcoholism in African‐and European‐American populations. Genes, Brain and Behavior 12:532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR (2012) Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics 28:2540–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Elkins I, Iacono WG (2000) Genetic and environmental influences on adolescent substance use and abuse. Am J Med Genet 96:671–677. [DOI] [PubMed] [Google Scholar]
- Mereish EH, Bradford JB (2014) Intersecting identities and substance use problems: sexual orientation, gender, race, and lifetime substance use problems. J Stud Alcohol Drugs 75:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulia N, Ye Y, Greenfield TK, Zemore SE (2009) Disparities in alcohol-related problems among white, black, and Hispanic Americans. Alcohol Clin Exp Res 33:654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén LK, Muthén BO (1998–2015) Mplus User’s Guide. Seventh Edition ed. Muthén & Muthén: Los Angeles, CA. [Google Scholar]
- Need AC, Goldstein DB (2009) Next generation disparities in human genomics: concerns and remedies. Trends Genet 25:489–494. [DOI] [PubMed] [Google Scholar]
- Palmer RH, McGeary JE, Heath AC, Keller MC, Brick LA, Knopik VS (2015) Shared additive genetic influences on DSM‐IV criteria for alcohol dependence in subjects of European ancestry. Addiction 110:1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuysen CI, Kranzler HR, Yu Y, Weiss RD, Brady K, Poling J, Farrer LA, Gelernter J (2010) Confirmation and generalization of an alcohol-dependence locus on chromosome 10q. Neuropsychopharmacology 35:1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, Kang TC, Nam YW, Kim GB, Shin HD, Choi IG (2013) Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet 132:657–668. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR (2005) Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA). Drug Alcohol Depend 80:303–312. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quality CfBHSa (2016) 2015 National Survey on Drug Use and Health: Detailed Tables. Substance Abuse and Mental Health Services Administration, Rockville, MD. . [Google Scholar]
- Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, Wang XY, Liu TQ, Hao W, Deng HW, Kranzler HR, Gelernter J (2014) ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet 165B:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNP & Variation Suite (Version 8.4.4) SNP & Variation Suite (Version 8.x) [Software]. Bozeman, MT: Golden Helix, Inc; Available from http://www.goldenhelix.com. [Google Scholar]
- Ulloa AE, Chen J, Vergara VM, Calhoun V, Liu J (2014) Association between copy number variation losses and alcohol dependence across African American and European American ethnic groups. Alcohol Clin Exp Res 38:1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hemani G, Vinkhuyzen AA, Chen GB, Lee SH, Wray NR, Goddard ME, Yang J (2014) Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS genetics 10:e1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Miller MB, Hicks BM, Iacono WG (2013) Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: twin biometry, GCTA, and genome-wide scoring. Behav Genet 43:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, Zhao H, Farrer LA, Gelernter J (2015) Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcohol Clin Exp Res 39:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Li C, Kranzler HR, Farrer LA, Zhao H, Gelernter J (2014a) Exploring the genetic architecture of alcohol dependence in African-Americans via analysis of a genomewide set of common variants. Hum Genet 133:617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Li C, Kranzler HR, Farrer LA, Zhao H, Gelernter J (2014b) Exploring the genetic architecture of alcohol dependence in African-Americans via analysis of a genomewide set of common variants. Human genetics 133:617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM (2013) Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Methods Mol Biol 1019:215–236. [DOI] [PubMed] [Google Scholar]
- Yu C (2002) Evaluation of model fit indices for latent variable models with categorical and continuous outcomes. .
- Zapolski TC, Pedersen SL, McCarthy DM, Smith GT (2014) Less drinking, yet more problems: understanding African American drinking and related problems. Psychol Bull 140:188–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X (2012a) Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology 37:557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Wang K, Wang G, Pan X, Zhang X, Zhang H, Luo X (2014) Common PTP4A1-PHF3-EYS variants are specific for alcohol dependence. Am J Addict 23:411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Wang K, Zhang XY, Krystal JH, Li CS, Zhang F, Zhang H, Luo X (2013a) NKAIN1-SERINC2 is a functional, replicable and genome-wide significant risk gene region specific for alcohol dependence in subjects of European descent. Drug Alcohol Depend 129:254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Sayward FG, Cheung KH, Wang K, Krystal JH, Zhao H, Luo X (2015) Gene-based and pathway-based genome-wide association study of alcohol dependence. Shanghai Arch Psychiatry 27:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang F, Zhang H, Zhang XY, Wang F, Li CS, Lu L, Hong J, Krystal J, Deng HW, Luo X (2012b) Genome-wide search for replicable risk gene regions in alcohol and nicotine co-dependence. Am J Med Genet B Neuropsychiatr Genet 159B:437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X (2013b) Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol Clin Exp Res 37:730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.