Abstract
Brain organoids are an exciting new technology with the potential to significantly change how diseases of the brain are understood and treated. These three-dimensional neural tissues are derived from the self-organization of pluripotent stem cells, and they recapitulate the developmental process of the human brain, including progenitor zones and rudimentary cortical layers. Brain organoids have been valuable in investigating different aspects of developmental neurobiology and comparative biology. Several characteristics of organoids also make them attractive as models of brain disorders. Data generated from human organoids are more generalizable to patients because of the match in species background. Personalized organoids also can be generated from patient-derived induced pluripotent stem cells. Furthermore, the three-dimensionality of brain organoids supports cellular, mechanical, and topographical cues that are lacking in planar systems. In this review, we discuss the translational potential of brain organoids, using the examples of Zika virus, autism-spectrum disorder, and glioblastoma multiforme to consider how they could contribute to disease modeling, personalized medicine, and testing of therapeutics. We then discuss areas of improvement in organoid technology that will enhance the translational potential of brain organoids, as well as the possibility of their use as substrates for repairing cerebral circuitry after injury.
Keywords: brain structure, cerebral organoid, disease model, personalized medicine, pluripotent stem cells
Introduction
The brain is an extraordinarily complex organ from which the entirety of human experience emanates. Understanding the intricacies of the human brain is thus one of the most fascinating endeavors in science. Equally compelling is the task of illuminating how failures in brain function manifest as neurological, cognitive, and psychiatric disorders. Two obstacles traditionally have stood in the way of these twin undertakings: the sheer complexity of the brain and difficulties in accessing human neural tissue. As such, the bulk of neuroscience research has resorted to the use of model systems, including cultures of dissociated neurons and animals. While these approaches have defined some of the fundamental principles of neurobiology and neurophysiology, they possess inherent shortcomings. Neuronal cultures lack the architecture of native brain tissue, which limits the investigation of interactions among different cell types. Species differences between humans and other animals dictate that information generated in the latter cannot necessarily be generalized to the former. Brain organoids are a relatively new technology that address, at least in part, these concerns and thus have the potential to transform how the brain and its associated diseases are studied.
In the most basic terms, organoids are simplified versions of organs that are grown in vitro (Lancaster and Knoblich, 2014b). Organoid technology has evolved tremendously from its earliest stages, in which dissociated embryonic chick cells were aggregated to form structures with a great deal of similarity to the organ of origin (Weiss and Taylor, 1960; Moscona, 1961). The current instantiation of organoids involves the generation of three-dimensional (3D) tissues from pluripotent stem cells. Multiple cell types within the organoid are arranged in a manner that substantially resembles the emulated organ. This architecture arises from the principle of self-organization, an essential driver of normal development that encompasses the modes of self-assembly, self-patterning, and self-driven morphogenesis (Sasai, 2013).
The current era of human brain organoids began five years ago with the description of whole-brain organoids (commonly called “mini-brains” in the lay press)(Lancaster et al., 2013) and the generation of rudimentary neocortex with a multilayered structure and appropriate axial polarity (Kadoshima et al., 2013). These advances sparked intense interest and excitement for a variety of reasons. The faithful recapitulation of brain-specific structure in a laboratory-grown, 3D tissue hinted that brain organoids could be used to explore human neurodevelopment in previously unattainable ways. Moreover, as described in the initial study on whole-brain organoids (Lancaster et al., 2013), these entities potentially could serve as more relevant platforms for examining the mechanisms underlying disorders of the human nervous system. Several excellent reviews have summarized the issues and literature relevant to these topics (Lancaster and Knoblich, 2014b; Kelava and Lancaster, 2016; Kretzschmar and Clevers, 2016; Di Lullo and Kriegstein, 2017). In this review, we focus on translational aspects of brain organoids that are on the horizon, especially as they pertain to personalized models of pathogenesis, screening for therapeutics, and substrates for brain repair. We also discuss the major areas that require progress before the potential of cerebral organoids can be fully unlocked in a clinical context.
History and current state of brain organoids
Brain organoids represent the culmination of progress in several areas of biology (Lancaster and Knoblich, 2014b; Kelava and Lancaster, 2016). Building upon the chicken embryo studies described above (Weiss and Taylor, 1960; Moscona, 1961), the field of tissue engineering has refined methods for creating reproducible aggregates of cells (Ungrin et al., 2008; Dingle et al., 2015). Advances in stem cell biology have made available both fetus-derived embryonic stem (ES) cells (Thomson et al., 1998) and induced pluripotent stem (iPS) cells generated from adult somatic cells via cellular reprogramming (Takahashi et al., 2007). Lastly, rational application of knowledge gained from the study of neurodevelopment has yielded efficient protocols for producing neurons, such as the dual SMAD inhibition method (Gaspard et al., 2008; Chambers et al., 2009), that replicate the temporal sequence of neocorticogenesis (Shi et al., 2012; Espuny-Camacho et al., 2013).
These strands converged in the technique known as serum free culture of embryoid body-like aggregates with quick aggregation (SFEBq), which originally was developed as a variant of the embryoid body method for differentiating monolayer cultures of neurons (Watanabe et al., 2005). Maintaining the 3D configuration of SFEBq cultures and adjusting media components to support directed differentiation led to the self-formation of stratified structures such as the retinal cup (Eiraku et al., 2011; Nakano et al., 2012) and cortical epithelium (Eiraku et al., 2008; Kadoshima et al., 2013). Both spatial and temporal aspects of normal neocorticogenesis were maintained in the latter. Cortical tissues developed identifiable zones that represented proper apico-basal polarity, including three progenitor zones (ventricular, subventricular, and intermediate) and three neuronal zones (subplate, cortical plate, and Cajal-Retzius cell). Features of rostral-caudal patterning also were present and could be manipulated using growth factors. Similar to adherent cultures, cortical plate neurons were born in the correct temporal sequence (lower layer before upper layer). A similar approach to SFEBq devoid of patterning growth factors and small molecule inhibitors resulted in brain organoids that modeled the developing brain in its entirety (Lancaster et al., 2013). These whole-brain organoids exhibited structural features reminiscent of the meninges, choroid plexus, and brain tissue. Markers of forebrain, midbrain, and hindbrain were present, and within the cortical domain, subregions consistent with different lobes of the cortex were identified.
Subsequent studies focused on improving the structural fidelity of cortex-specific organoids. A simplified approach for organoid generation that dispensed with the need for extracellular matrix supplementation (i.e., Matrigel) and media agitation yielded organoids with the well-defined segregation of superficial and deep cortical layers (Pasca et al., 2015). A miniaturized spinning bioreactor that decreased organoid-to-organoid variability produced organoids exhibiting a distinct layer of outer radial glial cells, an evolutionarily distinct hallmark of embryonic human cortex, and neurons belonging to all six cortical layers with rudimentary laminar separation (Qian et al., 2016; Qian et al., 2018). Formation of ellipsoid embryoid bodies around microfilament cores composed of poly(lactic-co-glycolic acid) and media supplementation with dissolved Matrigel reproducibly generated elongated neuroepithelium with a radially organized cortical plate. Moreover, expansion of cortical folds in cortical organoids was induced by PTEN deletion, which increased the size of the progenitor pool (Li et al., 2017b). The genetic programs (Camp et al., 2015; Pasca et al., 2015; Qian et al., 2016) and epigenetic (Luo et al., 2016) and epitranscriptomic (Yoon et al., 2017a) signatures of these organoids generally matched those found in human fetal cortex up to the second trimester of development.
Although much of the initial emphasis of cerebral organoid studies centered on the development of glutamatergic cortical neurons, there has been growing interest in other cell types. The majority of cortical interneurons are born in the subcortical ganglionic eminences and populate the cortex via tangential migration (Chu and Anderson, 2015). Few interneurons are found in standard cortical organoids (Qian et al., 2016) because they are generated from protocols that promote dorsal cell fates and thus lack tissue resembling the ventral ganglionic eminences. To rectify this problem, ventralized organoids containing GABAergic neurons have been generated and fused to dorsalized glutamatergic organoids (Bagley et al., 2017; Birey et al., 2017; Xiang et al., 2017). These “assembloids” demonstrate the migration of interneurons into the dorsalized organoid and establishment of circuits with glutamatergic cells, mirroring normal developmental. With respect to glial populations, several studies have described the appearance of astrocytes (Pasca et al., 2015; Qian et al., 2016; Dezonne et al., 2017; Sloan et al., 2017) and possibly oligodendrocytes (Matsui et al., 2018) in cortical organoids at late time points. Organoids modeling the midbrain (Jo et al., 2016; Qian et al., 2016; Monzel et al., 2017), hippocampus (Sakaguchi et al., 2015), pituitary gland (Ozone et al., 2016), hypothalamus (Qian et al., 2016), and cerebellum (Muguruma et al., 2015) also have been reported and are collectively referred to as region-specific brain organoids.
Insights into the neural activity of brain organoids have begun to emerge. Initial studies showed the presence of slow neuronal calcium waves and action potentials induced by current injections (Lancaster et al., 2013; Pasca et al., 2015; Qian et al., 2016). Spontaneous action potentials have been recorded with extracellular probes in eight-month-old but not four-month-old organoids, and the loss of this activity with the addition of neurotransmitter receptor antagonists suggests the presence of synaptic networks (Quadrato et al., 2017). Both glutamatergic and GABAergic spontaneous synaptic transmission can be detected in cortical organoids (Qian et al., 2016). Evoked action potential firing rates and excitatory synaptic inputs increase substantially in assembloids comprised of both glutamatergic and GABAergic neurons (Birey et al., 2017).
Translational applications of brain organoids
Organoids are an appealing model of the brain due to their human origin and because many of the cellular, mechanical, and topographical cues that arise from a 3D microenvironment are maintained (Figure 1). These advantages have been exploited to examine basic questions in developmental neurobiology, such as the genetic (Matsui et al., 2017; Mellios et al., 2018) and epitranscriptomic (Yoon et al., 2017a) control of cortical neurogenesis, and to investigate evolutionary differences between human and non-human primate brains (Mora-Bermudez et al., 2016; Otani et al., 2016; Giandomenico and Lancaster, 2017). There has also been a surge in studies modeling human genetic diseases with organoids derived from patient-specific iPS cells and isogenic cell lines (Lancaster et al., 2013; Bershteyn et al., 2017; Iefremova et al., 2017; Ye et al., 2017; Allende et al., 2018). In addition to providing a unique platform for identifying the explanatory mechanisms underlying cerebral disorders, organoids may have considerable value in the realm of therapeutics as personalized test beds for drug screening and genomic manipulation. We will explore these translational themes using the examples of Zika virus-associated microcephaly, autism-spectrum disorder (ASD) as a representative of neurocognitive and neuropsychiatric disorders, and glioblastoma multiforme (GBM).
Figure 1. Benefits and disadvantages of different model systems for human brain disorders.
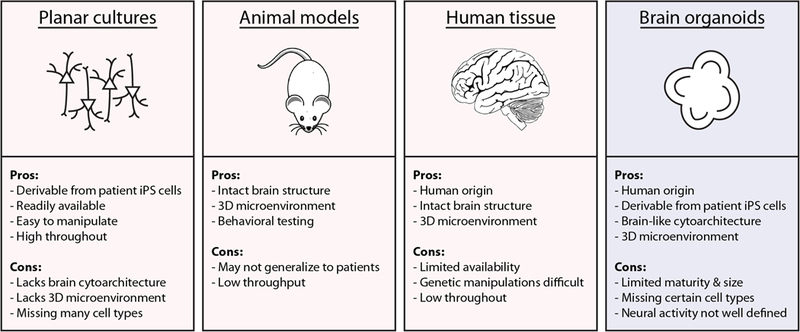
Conventional models of brain disease include monolayer cultures of stems cells, neural progenitors, or neurons; animal models; and human neural tissue. Brain organoids are an intriguing new technology that addresses many of the shortcomings of these other platforms. They may allow disease pathogenesis and therapeutic testing to be explored in previously unachievable ways. 3D: three-dimensional, iPS cells: induced pluripotent stem cells.
Zika virus-associated microcephaly
Zika virus (ZIKV) is a flavivirus first isolated from Ugandan rhesus monkeys in 1947 (Dick et al., 1952). It captured much public attention during an outbreak in Central and South American in 2015 after infections in pregnant women were linked to infants with abnormally small heads, a condition known as microcephaly (Mlakar et al., 2016; Ventura et al., 2016). Within a short period of time, ZIKV infection of the fetal brain was confirmed as the cause of microcephaly and other brain defects, and mechanisms of pathogenesis and potential drug therapies were identified. This rapid progress was facilitated by ZIKV studies in brain organoids, which synergized with other approaches using monolayer cultures of neural progenitor cells, floating neurospheres, and human fetal tissue (Ming et al., 2016; Qian et al., 2017).
The recapitulation of fetal cerebral architecture in brain organoids has been particularly helpful in analyzing the effects of ZIKV infection on brain structure. An early study examining ZIKV pathogenesis in monolayer cultures suggested that human neural progenitors are more vulnerable to ZIKV infection than pluripotent stem cells or differentiated neurons (Tang et al., 2016). Subsequently, multiple studies have demonstrated that ZIKV infection results in smaller organoid size in the context of decreased cortical plate thickness and disruption of laminar organization (Cugola et al., 2016; Garcez et al., 2016; Qian et al., 2016). These findings would not have been possible in monolayer cultures. ZIKV preferentially infects ventricular and outer radial glial cells in cerebral organoids, depleting these progenitor pools through a combination of increased apoptotic cell death and suppressed proliferation (Qian et al., 2016). Brain organoids have been valuable in shedding light on the mechanisms underlying these observations. Upregulation of the innate immune receptor Toll-like Receptor 3 (TLR3) has been identified as a potential mediator of impaired neurogenesis in ZIKV-infected organoids via transcriptomic analyses (Dang et al., 2016). Early switching from horizontal to vertical division planes in dividing progenitors also has been found in ZIKV-infected organoids. This observation is consistent with premature progenitor differentiation as a contributor to progenitor pool depletion and cortical mantle thinning (Gabriel et al., 2017). Expression of the ZIKA-NS2A protein leads to the destabilization of adherens junction complexes and disorganized radial glial scaffolding in both embryonic mouse cortex and human cerebral organoids (Yoon et al., 2017b). This last study is an excellent illustration of the experimental flexibility of organoids, which allows genetic manipulations via techniques such as electroporation and viral transduction to be used to elucidate pathogenic mechanisms.
Thus far, organoids have played a complementary role in the search for viable treatment strategies for ZIKV infection. High-throughput drug screens to identify ZIKV therapies have relied primarily upon monolayer cultures of iPS-derived human neural progenitor cells (Xu et al., 2016; Zhou et al., 2017). These screens and other hypothesis-driven approaches have revealed a number of candidate agents, which have been tested in monolayer cultures, animal models, and brain organoids (Xu et al., 2016; Li et al., 2017a; Sacramento et al., 2017; Watanabe et al., 2017; Zhou et al., 2017). Drug treatment of ZIKV-infected organoids has demonstrated decreased apoptosis (Xu et al., 2016; Watanabe et al., 2017), reduced virus replication (Sacramento et al., 2017; Zhou et al., 2017), and protection from infection (Li et al., 2017a; Watanabe et al., 2017). The use of organoids to assess the efficacy of drugs on preserving the structural integrity of cortex has not yet been performed. This approach may have the most relevance to patients and thus could be especially important in validating drug-testing results from other models.
Cell surface receptors that mediate ZIKV entry into cells highlight another role for organoids in identifying targets for therapeutic intervention. The TAM receptor protein AXL was thought to be a strong candidate for this role in part because of its expression in many tissues that correlate with the path of viral transmission from mother to fetus (Ming et al., 2016). High expression of AXL also is found in radial glial cells, astrocytes, endothelial cells, and microglia in human fetal brain tissue (Nowakowski et al., 2016). However, deletion of AXL using genome-editing techniques did not alter ZIKV entry into or its deleterious effects on human neural progenitors in monolayer cultures or brain organoids (Wells et al., 2016). Other studies have found that AXL mediates ZIKV entry into human glial (Meertens et al., 2017) and endothelial cells (Richard et al., 2017), suggesting an alternative role for this receptor in ZIKV pathogenesis. These findings emphasize the need to consider organoid studies in the context of other disease models.
Autism spectrum disorder
Autism spectrum disorders are a group of neurodevelopmental conditions characterized by deficits in social interactions, communication, and behavioral flexibility (Ilieva et al., 2018). The genetic basis of ASD is quite complex. Many candidate genes, each with multiple pathogenic variants, have been identified, and this molecular diversity is likely responsible for the heterogeneity seen in clinical manifestations (Geschwind, 2011). Explanatory models for ASD include impaired synaptic development (Zoghbi, 2003), abnormal neural connectivity secondary to compromised activity propagation and information processing (Belmonte et al., 2004; Courchesne and Pierce, 2005), and developmental disconnection specifically within the frontal and temporal regions (Geschwind and Levitt, 2007).
It stands to reason that human models are most suitable for studying ASD because of the involvement of the cerebral cortex, which is more highly developed in humans than other species, in ASD pathology (Parikshak et al., 2013). Moreover, the hypothesized role of neural connectivity in ASD pathology is best investigated with networks comprised of discrete groups of neurons to detect problems with neural activity timing and multiple neuron types to faithfully simulate in vivo circuits. Brain organoids fulfill these criteria. It is also expected that organoids, by virtue of their cellular variety, cell-matrix interactions, and re-constitution of in vivo cytoarchitecture and niche-like environments, will better maintain the in vivo genetic and epigenetic landscape than monolayer cultures (Forsberg et al., 2018). Organoids derived from individuals with severe idiopathic ASD demonstrate accelerated cell-cycle dynamics and FOXG1-dependent overproduction of interneurons (Mariani et al., 2015). Patients in this study did not carry any genetic mutations with known associations with ASD, which underscores the potential of organoids to serve as personalized models of disease pathogenesis in highly polygenic disorders (Arlotta, 2018). The opposite approach of examining the contribution of a specific gene to ASD in organoids has also been tried. Isogenic iPS cell lines that are heterozygous for a CHD8 knockout allele have been used to generate cerebral organoids for transcriptomic analysis. DLX6-AS1 and DLX1, which are involved in interneuron differentiation, are among the top differentially expressed genes in CHD8 knockout heterozygote compared to control organoids (Wang et al., 2017). Similar studies using patient-derived organoids and organoids overexpressing a disease gene have been performed in schizophrenia (Ye et al., 2017). These studies are the vanguard of organoid-based investigations of neuropsychiatric and neurocognitive disorders. Additional progress in this direction will need to resolve several questions, including the optimal outcome metrics that are most relevant to clinical symptoms (Ilieva et al., 2018).
Brain tumors
Glioblastoma multiforme (GBM) is the most common primary brain malignancy in adults (Ostrom et al., 2017), and it remains one of the most difficult tumors to treat with a median life expectancy of only 15 months (Stupp et al., 2005; Stupp et al., 2017). A significant part of the challenge in treating this disease is its significant intratumoral heterogeneity (Sottoriva et al., 2013; Parker et al., 2015) and highly infiltrative nature in the brain. Organoid technology may be suitable for modeling both these features with implications for understanding the pathophysiology of GBM and developing patient-specific therapies.
Like many other tumors (Ebben et al., 2010), GBM is thought to arise from mutated stem cells (Sanai et al., 2005). Neurospheres consisting of these putative glioma stem cells have been isolated from patient tumor samples (Ignatova et al., 2002; Galli et al., 2004), and they can differentiate into multiple cell types along neuronal, astroglial, and oligodendroglial lineages (Gunther et al., 2008). However, tumor formation typically occurs only after xenograft transplantation (Gunther et al., 2008; Schulte et al., 2011). Applying the principles of organoid formation to glioma neurospheres and patient tissue in general could result in glioma organoids that reflect the structure, heterogeneity, and perhaps microenvironment of GBM in vivo. In essence, glioma organoids would be 3D human representations of tumors that are free of the constraints of an animal host. An initial report of organoids derived from patient GBM and cerebral metastasis tissues has demonstrated the feasibility of this approach (Hubert et al., 2016). This study showed phenotypic heterogeneity among stem cells in the glioma organoids, as well as hypoxic gradients that modulated mitotic activity in stem cell niches. Glioma organoids also have been generated by introducing the HRasG12V oncogene into whole-brain organoids via CRISPR/Cas9-mediated homologous recombination in the TP53 locus (Ogawa et al., 2018). Transformed cells rapidly proliferate and invade the “healthy” organoid, overtaking the bulk of the organoid after 16 weeks. The molecular profile of these tumor cells is most consistent with the highly aggressive mesenchymal subtype of GBM. Similarly, deletion of DNA repair genes in colonic organoids via CRISPR/Cas9 techniques has been performed to study the accumulation of mutations that drive oncogenesis (Drost et al., 2017). Further investigations along these lines could further elucidate the etiology and evolution of intratumoral heterogeneity, factors influencing the tumor cell-microenvironment interaction, and other pathophysiological mechanisms in GBM.
Glioma organoids also could contribute to the development of personalized therapeutics for GBM patients, which is necessitated by the high degree of genetic variability in this tumor. The effectiveness of different treatment regimens could be trialed in glioma organoids, allowing therapeutic effects on the stem cell versus non-stem cell compartment of the tumor to be evaluated before a course of action is chosen for a patient. This type of organoid biobank approach for treatment screening has been applied to be breast (Sachs et al., 2018), gastrointestinal (Vlachogiannis et al., 2018), and bladder cancers (Lee et al., 2018).
Organoid fusion may be a novel strategy for investigating mechanisms involved in the spread of GBM in the brain. Mimicking the fusion of dorsalized and ventralized organoids to study interneuron migration, glioma organoids could be fused to normal brain organoids to assess for tumor cell infiltration. Along these lines, GBM neurospheres merged with cerebral organoids have demonstrated the ability to infiltrate the organoid (da Silva et al., 2018; Ogawa et al., 2018). This strategy could be used to assess the temporal dynamics and cellular mechanisms of infiltration. In particular, genetic and epigenetic analyses of infiltrative versus non-infiltrative tumor cells in fused organoids could help define molecular signatures of infiltration and suitable targets for therapeutic intervention. Such therapies could then be tested by assaying tumor cell infiltration in fused organoids.
Areas for improvement
The above examples demonstrate the ample advantages of using organoids to model brain diseases. First and foremost, brain organoids are human systems that better match the genetic background and structural characteristics of the human brain than animal models. They are also much more accessible and available in greater quantities than human brain tissue. Lastly, organoids are 3D structures that recapitulate many aspects of the in vivo environment of brain tissue, including its cytoarchitecture, cellular variability, cell-cell and cell-matrix interactions, and various cellular niches. Such features are missing in two-dimensional monolayer cultures. Fully exploiting the benefits of brain organoids as models will require disease-specific fine-tuning. There also exist areas of improvement that cut across all organoid applications to clinical problems, including the logistics of organoid generation, fidelity of brain emulation, and characterization of neural circuit structure and activity.
Logistics of organoid generation
Variability in brain organoid generation imposes constraints on their use for translational purposes. Batch-to-batch variability can be significant, especially with whole-brain organoids (Lancaster and Knoblich, 2014a). Single-cell sequencing experiments show that organoids grown in the same bioreactor exhibit a higher degree of similarity than organoids obtained from different bioreactors and that particular cell populations are only present in certain bioreactor batches (Quadrato et al., 2017). This degree of variability necessitates the inclusion of larger numbers of organoids in studies to mitigate outlier effects. Various techniques have been employed to decrease this cellular variability, including miniaturized spinning bioreactors that may produce a more consistent environment for organoid growth (Qian et al., 2016) and increasing the surface area of embryoid bodies relative to their volume to improve the efficiency of neural induction (Lancaster et al., 2017). An additional source of variability is the number of cortical zones present in each organoid. The factors governing the formation of cortical zones are not yet clear but may include the initial attributes of embryoid bodies (e.g., number of cells, size, and shape), biomechanical variables (Varner et al., 2015), and developmental cues. Simple dissection, as has been done with retinal organoids (Volkner et al., 2016), could be a straightforward way of isolating individual cortical zones. Further application of principles drawn from developmental neurobiology and bioengineering, as well as quality control practices, will greatly improve the reproducibility of organoid generation and efficiency of organoid-based disease models.
Another limitation of cerebral organoids for modeling purposes is the long length of time required for growth and maturation. Similar concerns are relevant to any human stem cell-based platform. Several months are necessary to obtain organoids with molecular signatures similar to second-trimester fetuses (Pasca et al., 2015; Qian et al., 2016). Ostensibly, additional lengths of time would be needed to mature organoids to the third trimester and post-natal stage. While current brain organoids are adequate for modeling neurodevelopmental disorders and fetal infections, there are legitimate concerns about their applicability to cerebral disorders with later onsets, such as schizophrenia, which typically manifests in adolescence (Soliman et al., 2017), and neurodegenerative diseases (Vera and Studer, 2015). Methods for accelerating maturation are being sought and include over-expression of genes implicated in premature aging disorders (Miller et al., 2013; Zhang et al., 2015) and PTEN deletion (personal communication, S. Anderson).
Finally, high-throughput screening methods based on organoids are just beginning to emerge. The previously described miniaturized spinning bioreactor has helped scale up the generation of cerebral organoids (Qian et al., 2016). However, most analyses still require multiple, time-consuming steps, including those associated with immunohistochemistry and microscopy. Newer imaging techniques such as tissue clearing (Chung et al., 2013; Renier et al., 2014) and imaging mass cytometry (Giesen et al., 2014) have the potential to streamline image analysis, but there is a need to develop the methods to analyze multiple samples simultaneously. To this end, radiofrequency identification chips have been embedded in human liver organoids, enabling the identification of disease phenotypes (Kimura et al., 2018). Combining organoids with other tissue matrix arrays (Beachley et al., 2015) or organ-on-a chip technologies (Bhatia and Ingber, 2014) may be alternative solutions to this multiplexing issue.
Augmenting cerebral organoid complexity
Brain organoids in their current iterations already model a great deal of the brain’s complexity, but there are important features that are still missing. Most recent efforts in the field have focused on brain region-specific organoids because it is easier to control their development and structure than whole-brain organoids. This approach is limited in examining interactions between different areas of the brain. Fusion of different brain region-specific organoids provides a means of studying these systems-level interactions. For example, assembloids of cortical and thalamic organoids could be used to model human cortico-thalamic circuitry, which underlie all cortical functions. One downside of this strategy and of cerebral organoids in general is the lack of axon tracts, which serve as the substrate for cerebral connectivity. Novel bioengineering techniques, including axon stretch growth (Pfister et al., 2004) and hydrogel micro-conduits (Cullen et al., 2012; Struzyna et al., 2015), could incorporate axon bundles into organoid models, which would enable the study of neural activity propagation between network nodes (Chen et al., 2017).
Another issue with current brain organoids is the lack of endothelial cells, microglia, and other immune cells, which are valuable for disease modeling. The available protocols for generating brain organoids direct cellular differentiation exclusively along ectodermal pathways. As such, they do not contain any cells that originate from the mesoderm. The blood-brain barrier is imperative for maintaining cerebral homeostasis, and its disruption is implicated in a number of diseases (Zhao et al., 2015b). Interactions between neural stem cells and the cerebral vasculature, the so-called perivascular niche, may be involved in normal progenitor proliferation and differentiation (Shen et al., 2008; Tavazoie et al., 2008; Codega et al., 2014). Microglia are the resident immune cells of the brain, and they have been implicated in both healthy brain function and pathological neuroinflammation (Blank and Prinz, 2013; Colonna and Butovsky, 2017). Infiltrating monocytes and T cells contribute to the complex microenvironment of GBM (Domingues et al., 2016; Gabrusiewicz et al., 2016), and understanding their role in the immunosuppressed status of this tumor is crucial for optimizing immunotherapies such as CAR-T cells (O’Rourke et al., 2017). Co-culturing techniques could be adequate to introduce these missing cell types into cerebral organoids (Pham et al., 2018), especially in the case of immune cells that are naturally infiltrative. Other methods may need to be developed in order to fully integrate vascular networks and immunological compartments into brain organoids.
Increasing the size of cerebral organoids and maintaining their health during prolonged periods of growth are likely pre-requisites for promoting their maturation beyond the 2nd trimester of human fetal development. As discussed above, developing more mature organoids is particularly important for more accurately modeling cerebral diseases affecting later stages of the human life cycle. The major obstacle to achieving this goal has been inadequate nutrient, gas, and waste exchange in in vitro culture, which leads to a necrotic organoid core and suboptimal cell health beyond the superficial layer of the organoid. Media agitation (Lancaster et al., 2013; Qian et al., 2016) and hyperoxic culture conditions (Kadoshima et al., 2013) have maximized the efficiency of diffusion and mass transport. Further organoid growth and development will need the equivalent of a circulatory system. Micro-fluidics and other bioengineering techniques have improved the ability to perfuse 3D engineered tissues (Miller et al., 2012; Hasan et al., 2014). Another approach is to use host animals as “bioreactors” to support the growth of cerebral organoids. It was shown recently that human brain organoids survive and integrate after transplantation into mouse cortex (Mansour et al., 2018). Grafted organoids are extensively vascularized by the host two weeks after transplantation and contain significantly fewer apoptotic cells than age-matched organoids in vitro. Furthermore, progressive differentiation of neurons, gliogenesis, and host microglia infiltration is observed within the transplanted organoid.
Organoid transplantation could be an end unto itself as a strategy for brain repair. The vast majority of the current approaches for rebuilding cerebral circuitry results in grafted neurons that are arranged in a fairly haphazard manner (Guo et al., 2014; Michelsen et al., 2015; Somaa et al., 2017; Tornero et al., 2017). Especially in highly structured areas of the brain such as the cerebral cortex and hippocampus, a reasonable expectation is that grafts with a greater degree of brain-specific architecture would more effectively restore cerebral function (Chen et al., 2016). Brain organoids are one of the few available substrates that recapitulate cerebral architecture and thus could be the starting point for investigating a new approach to brain repair. Many of the improvements discussed in this section may prove beneficial for the usage of brain organoids as repair substrates as well.
Defining neural activity and networks in brain organoids
A better understanding of how neural activity develops in brain organoids will enhance their modeling of neuropsychiatric and neurocognitive disorders (Di Lullo and Kriegstein, 2017). Neural activity arising from and between neural ensembles is the closest proxy to behavior and clinical function available in the laboratory. Thus, it is likely to be one of the best outcome metrics available for organoid-based models.
The current picture of neural activity evolution in brain organoids is incomplete. Spontaneous action potentials measured with extracellular techniques have been observed in eight-month-old but not four-month-old organoids (Quadrato et al., 2017). A more precise timeline for the appearance of these spontaneous action potentials remains to be determined, and it will be interesting to compare this timeline to the evolution of spiking activity in human fetal brain tissue (Moore et al., 2011). Moreover, spontaneous calcium activity and evoked action potentials have been documented in cerebral organoids well before eight months (Pasca et al., 2015; Qian et al., 2016). The relationship between this nascent activity and spontaneous action potentials is not known. Perhaps most importantly, very little is known about the development of local neural circuits and larger neuronal networks in brain organoids. Planar cultures of iPS cell-derived human cortical neurons may provide some guidance in this area. These cultures transition from asynchronous activity to synchronized oscillatory networks to structured asynchronous activity with recurring activity motifs over 100 days (Kirwan et al., 2015). It is clear that the presence of interneurons accelerate the functional maturity of neurons in brain organoids (Birey et al., 2017), but other factors pertinent to this maturation process, including the contribution of astrocytes and other glial cells, have not yet been examined in detail. Making progress on these various topics will require collaborative work among developmental neurobiologists and circuit-/systems-level neuroscientists.
Clinical and ethical considerations
In addition to technical improvements in generating and understanding brain organoids, there are issues associated with the clinical application of these entities that require careful consideration. The long length of time needed to generate patient-specific iPS cells and brain organoids is a major impediment to the practical use of the latter for both brain modeling and circuit repair. Methods for accelerating the growth of brain organoids have been discussed above. An alternative could be the creation of organoid banks for specific patient genetic profiles, immunological phenotypes, and disease types. The complex microenvironment of brain organoids may predispose them to genetic, epigenetic, and phenotypic drift over time, which could impact their ability to effectively model cerebral disorders. Quality-control protocols and thorough validation of organoid-derived data for patient use could help manage such problems. Finally, there are potential risks to using brain organoids for brain repair, which overlap with concerns associated with stem cell-based therapies in general. Even patient-derived iPS cells, and hence brain organoids, could provoke an immune reaction (Zhao et al., 2011; Zhao et al., 2015a). Oncogenic transformation of transplanted progenitor cells remains a worry, and over-exuberant organoid growth that is not neoplastic also could cause neurological problems. Mechanisms to limit organoid growth or induce apoptosis of transplanted cells may be needed to safeguard against these eventualities. Transplanted organoids could trigger unwanted functional outcomes, including seizure activity. Neuromodulatory interventions may be necessary to facilitate the proper integration of organoid neurons and prevent maladaptive plasticity.
The generation of human brain organoids has spurred ethical debates on a range of topics, which prompted two workshops supported by the National Institutes of Health in May 2017 and March 2018 (Farahany et al., 2018). There are three general areas of discussion. The first pertains to the moral status and well being of the organoids themselves. Because of their human origin and brain-like architecture, brain organoids theoretically have the potential to attain more advanced brain states, including sensory perception (e.g., pain) and perhaps consciousness. The likelihood that current brain organoids are conscious is miniscule given their small size and primitive nature. However, as their size and complexity increase, pertinent questions include defining sensitive methods for assessing possible sentience (Lavazza and Massimini, 2018) and determining the need to protect these more advanced organoids in ways similar to laboratory animals. Second, transplantation of human brain organoids into animal hosts will create a spectrum of human-animal chimeras. While chimeras have become more commonplace in biomedical research, brain chimeras may require special consideration. Is it scientifically feasible for brain chimeras to become enhanced or more human-like? If so, are there certain levels of enhancement that would be considered acceptable, and would these enhanced chimeras deserve additional research protections? Lastly, there are issues related to consent and ownership. When subjects donate their cells for the generation of iPS lines, should they be told explicitly that brain organoids will be generated from these lines? Can they request specifically that brain organoids not be generated? As brain organoids become more complex, the question of who controls the fate of these entities (e.g., the researcher, source subject, or perhaps state) also may become more contentious. Further discussions of these issues should include all relevant stakeholders and be based upon appropriately defined scientific frameworks and ethical principles.
Conclusion
Brain organoids have inspired such intense excitement over the past five years because they are the only in vitro platform that recreates the 3D architecture of the human brain and recapitulates the process of human neurodevelopment. Beyond the study of normal brain development, organoids provide a unique opportunity to model human brain disorders and develop therapies for these conditions. The examples of Zika virus, autism-spectrum disorders, and glioblastoma multiforme underscore three translational themes. First, due to their human and 3D nature, brain organoids enable the investigation of aspects of disease pathogenesis that are difficult or impossible to examine using other models. Second, the possibility of generating patient-specific organoids means that personalized models of disease can be created, which may be especially helpful in understanding the pathogenesis of polygenic disorders. Third, brain organoids may provide unique outcome measures that can be used to screen the efficacy of drugs and other therapeutic interventions in a personalized manner. Separate from disease modeling, brain organoids are a potentially transformative substrate for rebuilding cerebral circuitry after injury. All of these translational indications would benefit from continued improvements in the reproducibility, maturity, and biological complexity of organoid technology. While progress is being made on these fronts, it will be important to keep in mind potential ethical quandaries associated with cerebral organoid growth and use. Resolving these biological, engineering, clinical, and ethical issues will help realize the full potential of brain organoids in their application to clinical problems.
Acknowledgements:
The work in the authors’ laboratories is supported by the Department of Veterans Affairs (IK2RX002013 to H.I.C.) and the National Institutes of Health (R37NS047344, U19MH106434, P01NS097206, and R01AG057497 to H.J.S; R01MH105126, R35NS097370, U19AI131130, and R21MH110160 to G-l.M.)
References
- Allende ML, Cook EK, Larman BC, Nugent A, Brady JM, Golebiowski D, Sena-Esteves M, Tifft CJ, Proia RL. 2018. Cerebral organoids derived from Sandhoff disease-induced pluripotent stem cells exhibit impaired neurodifferentiation. J Lipid Res 59:550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P 2018. Organoids required! A new path to understanding human brain development and disease. Nat Methods 15:27–29. [DOI] [PubMed] [Google Scholar]
- Bagley JA, Reumann D, Bian S, Levi-Strauss J, Knoblich JA. 2017. Fused cerebral organoids model interactions between brain regions. Nat Methods 14:743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachley VZ, Wolf MT, Sadtler K, Manda SS, Jacobs H, Blatchley MR, Bader JS, Pandey A, Pardoll D, Elisseeff JH. 2015. Tissue matrix arrays for high-throughput screening and systems analysis of cell function. Nat Methods 12:1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MK, Cook EH Jr., Anderson GM, Rubenstein JL, Greenough WT, Beckel-Mitchener A, Courchesne E, Boulanger LM, Powell SB, Levitt PR, Perry EK, Jiang YH, DeLorey TM, Tierney E. 2004. Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol Psychiatry 9:646–663. [DOI] [PubMed] [Google Scholar]
- Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, Kriegstein AR. 2017. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 20:435–449 e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia SN, Ingber DE. 2014. Microfluidic organs-on-chips. Nat Biotechnol 32:760–772. [DOI] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O’Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Pasca SP. 2017. Assembly of functionally integrated human forebrain spheroids. Nature 545:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank T, Prinz M. 2013. Microglia as modulators of cognition and neuropsychiatric disorders. Glia 61:62–70. [DOI] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Paabo S, Huttner WB, Treutlein B. 2015. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A 112:15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HI, Jgamadze D, Serruya MD, Cullen DK, Wolf JA, Smith DH. 2016. Neural Substrate Expansion for the Restoration of Brain Function. Front Syst Neurosci 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HI, Wolf JA, Smith DH. 2017. Multichannel activity propagation across an engineered axon network. J Neural Eng 14:026016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Anderson SA. 2015. Development of cortical interneurons. Neuropsychopharmacology 40:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. 2013. Structural and molecular interrogation of intact biological systems. Nature 497:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. 2014. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 82:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Butovsky O. 2017. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu Rev Immunol 35:441–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Pierce K. 2005. Why the frontal cortex in autism might be talking only to itself: local over-connectivity but long-distance disconnection. Curr Opin Neurobiol 15:225–230. [DOI] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandao WN, Rossato C, Andrade DG, Faria Dde P, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrao-Braga PC. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen DK, Tang-Schomer MD, Struzyna LA, Patel AR, Johnson VE, Wolf JA, Smith DH. 2012. Microtissue engineered constructs with living axons for targeted nervous system reconstruction. Tissue Eng Part A 18:2280–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva B, Mathew RK, Polson ES, Williams J, Wurdak H. 2018. Spontaneous Glioblastoma Spheroid Infiltration of Early-Stage Cerebral Organoids Models Brain Tumor Invasion. SLAS Discov:2472555218764623. [DOI] [PubMed] [Google Scholar]
- Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, Rana TM. 2016. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell 19:258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezonne RS, Sartore RC, Nascimento JM, Saia-Cereda VM, Romao LF, Alves-Leon SV, de Souza JM, Martins-de-Souza D, Rehen SK, Gomes FC. 2017. Derivation of Functional Human Astrocytes from Cerebral Organoids. Sci Rep 7:45091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E, Kriegstein AR. 2017. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci 18:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. [DOI] [PubMed] [Google Scholar]
- Dingle YT, Boutin ME, Chirila AM, Livi LL, Labriola NR, Jakubek LM, Morgan JR, Darling EM, Kauer JA, Hoffman-Kim D. 2015. Three-Dimensional Neural Spheroid Culture: An In Vitro Model for Cortical Studies. Tissue Eng Part C Methods 21:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues P, Gonzalez-Tablas M, Otero A, Pascual D, Miranda D, Ruiz L, Sousa P, Ciudad J, Goncalves JM, Lopes MC, Orfao A, Tabernero MD. 2016. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun 53:1–15. [DOI] [PubMed] [Google Scholar]
- Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, Nik-Zainal S, Kuiper RP, Cuppen E, Clevers H. 2017. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebben JD, Treisman DM, Zorniak M, Kutty RG, Clark PA, Kuo JS. 2010. The cancer stem cell paradigm: a new understanding of tumor development and treatment. Expert Opin Ther Targets 14:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. 2011. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472:51–56. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3:519–532. [DOI] [PubMed] [Google Scholar]
- Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, Lambert N, Gaspard N, Peron S, Schiffmann SN, Giugliano M, Gaillard A, Vanderhaeghen P. 2013. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron 77:440–456. [DOI] [PubMed] [Google Scholar]
- Farahany NA, Greely HT, Hyman S, Koch C, Grady C, Pasca SP, Sestan N, Arlotta P, Bernat JL, Ting J, Lunshof JE, Iyer EPR, Hyun I, Capestany BH, Church GM, Huang H, Song H. 2018. The ethics of experimenting with human brain tissue. Nature 556:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg SL, Ilieva M, Maria Michel T. 2018. Epigenetics and cerebral organoids: promising directions in autism spectrum disorders. Transl Psychiatry 8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Kronke M, Utermohlen O, Gopalakrishnan J. 2017. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell 20:397–406 e395. [DOI] [PubMed] [Google Scholar]
- Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, Thomas G, Zhou S, Wang Q, Elakkad A, Liebelt BD, Yaghi NK, Ezhilarasan R, Huang N, Weinberg JS, Prabhu SS, Rao G, Sawaya R, Langford LA, Bruner JM, Fuller GN, Bar-Or A, Li W, Colen RR, Curran MA, Bhat KP, Antel JP, Cooper LJ, Sulman EP, Heimberger AB. 2016. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. 2004. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64:7011–7021. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. 2016. Zika virus impairs growth in human neurospheres and brain organoids. Science 352:816–818. [DOI] [PubMed] [Google Scholar]
- Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, Passante L, Schiffmann SN, Gaillard A, Vanderhaeghen P. 2008. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455:351–357. [DOI] [PubMed] [Google Scholar]
- Geschwind DH. 2011. Genetics of autism spectrum disorders. Trends Cogn Sci 15:409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. 2007. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol 17:103–111. [DOI] [PubMed] [Google Scholar]
- Giandomenico SL, Lancaster MA. 2017. Probing human brain evolution and development in organoids. Curr Opin Cell Biol 44:36–43. [DOI] [PubMed] [Google Scholar]
- Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Gunther D, Bodenmiller B. 2014. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11:417–422. [DOI] [PubMed] [Google Scholar]
- Gunther HS, Schmidt NO, Phillips HS, Kemming D, Kharbanda S, Soriano R, Modrusan Z, Meissner H, Westphal M, Lamszus K. 2008. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene 27:2897–2909. [DOI] [PubMed] [Google Scholar]
- Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. 2014. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 14:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A, Paul A, Vrana NE, Zhao X, Memic A, Hwang YS, Dokmeci MR, Khademhosseini A. 2014. Microfluidic techniques for development of 3D vascularized tissue. Biomaterials 35:7308–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, Couce M, McLendon RE, Sloan AE, Rich JN. 2016. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res 76:2465–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iefremova V, Manikakis G, Krefft O, Jabali A, Weynans K, Wilkens R, Marsoner F, Brandl B, Muller FJ, Koch P, Ladewig J. 2017. An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell Rep 19:50–59. [DOI] [PubMed] [Google Scholar]
- Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. 2002. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 39:193–206. [DOI] [PubMed] [Google Scholar]
- Ilieva M, Fex Svenningsen A, Thorsen M, Michel TM. 2018. Psychiatry in a Dish: Stem Cells and Brain Organoids Modeling Autism Spectrum Disorders. Biol Psychiatry 83:558–568. [DOI] [PubMed] [Google Scholar]
- Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Goke J, Tan ZY, Saw TY, Tan CP, Lokman H, Lee Y, Kim D, Ko HS, Kim SO, Park JH, Cho NJ, Hyde TM, Kleinman JE, Shin JH, Weinberger DR, Tan EK, Je HS, Ng HH. 2016. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 19:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. 2013. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A 110:20284–20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I, Lancaster MA. 2016. Dishing out mini-brains: Current progress and future prospects in brain organoid research. Dev Biol 420:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Asuma M, Zhang R, Thompson W, Mayhew CN, Takebe T. 2018. Digitalized human organoid for wireless phenotyping. iScience [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan P, Turner-Bridger B, Peter M, Momoh A, Arambepola D, Robinson HP, Livesey FJ. 2015. Development and function of human cerebral cortex neural networks from pluripotent stem cells in vitro. Development 142:3178–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Clevers H. 2016. Organoids: Modeling Development and the Stem Cell Niche in a Dish. Dev Cell 38:590–600. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA. 2017. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol 35:659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. 2014a. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 9:2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. 2014b. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345:1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazza A, Massimini M. 2018. Cerebral organoids: ethical issues and consciousness assessment. J Med Ethics [DOI] [PubMed] [Google Scholar]
- Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, Bergren SK, Pietzak EJ, Anderson CB, Benson MC, Coleman JA, Taylor BS, Abate-Shen C, McKiernan JM, Al-Ahmadie H, Solit DB, Shen MM. 2018. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 173:515–528 e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Deng YQ, Wang S, Ma F, Aliyari R, Huang XY, Zhang NN, Watanabe M, Dong HL, Liu P, Li XF, Ye Q, Tian M, Hong S, Fan J, Zhao H, Li L, Vishlaghi N, Buth JE, Au C, Liu Y, Lu N, Du P, Qin FX, Zhang B, Gong D, Dai X, Sun R, Novitch BG, Xu Z, Qin CF, Cheng G. 2017a. 25-Hydroxycholesterol Protects Host against Zika Virus Infection and Its Associated Microcephaly in a Mouse Model. Immunity 46:446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, Gehrke L, Knoblich JA, Jaenisch R. 2017b. Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell 20:385–396 e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. 2016. Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Rep 17:3369–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour AA, Goncalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, Gage FH. 2018. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 36:432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko EL, Chawarska K, Pelphrey KA, Howe JR, Vaccarino FM. 2015. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162:375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Nieto-Estevez V, Kyrychenko S, Schneider JW, Hsieh J. 2017. Retinoblastoma protein controls growth, survival and neuronal migration in human cerebral organoids. Development 144:1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui TK, Matsubayashi M, Sakaguchi YM, Hayashi RK, Zheng C, Sugie K, Hasegawa M, Nakagawa T, Mori E. 2018. Six-month cultured cerebral organoids from human ES cells contain matured neural cells. Neurosci Lett 670:75–82. [DOI] [PubMed] [Google Scholar]
- Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, Le Charpentier T, Hafirassou ML, Zamborlini A, Cao-Lormeau VM, Coulpier M, Misse D, Jouvenet N, Tabibiazar R, Gressens P, Schwartz O, Amara A. 2017. Axl Mediates ZIKA Virus Entry in Human Glial Cells and Modulates Innate Immune Responses. Cell Rep 18:324–333. [DOI] [PubMed] [Google Scholar]
- Mellios N, Feldman DA, Sheridan SD, Ip JPK, Kwok S, Amoah SK, Rosen B, Rodriguez BA, Crawford B, Swaminathan R, Chou S, Li Y, Ziats M, Ernst C, Jaenisch R, Haggarty SJ, Sur M. 2018. Human cerebral organoids reveal deficits in neurogenesis and neuronal migration in MeCP2-deficient neural progenitors. Mol Psychiatry 23:791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen KA, Acosta-Verdugo S, Benoit-Marand M, Espuny-Camacho I, Gaspard N, Saha B, Gaillard A, Vanderhaeghen P. 2015. Area-specific reestablishment of damaged circuits in the adult cerebral cortex by cortical neurons derived from mouse embryonic stem cells. Neuron 85:982–997. [DOI] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, Studer L. 2013. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13:691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. 2012. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater 11:768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Tang H, Song H. 2016. Advances in Zika Virus Research: Stem Cell Models, Challenges, and Opportunities. Cell Stem Cell 19:690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T. 2016. Zika Virus Associated with Microcephaly. N Engl J Med 374:951–958. [DOI] [PubMed] [Google Scholar]
- Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, Jarazo J, Walter J, Bruggemann I, Boussaad I, Berger E, Fleming RMT, Bolognin S, Schwamborn JC. 2017. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports 8:1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Zhou WL, Jakovcevski I, Zecevic N, Antic SD. 2011. Spontaneous electrical activity in the human fetal cortex in vitro. J Neurosci 31:2391–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Bermudez F, Badsha F, Kanton S, Camp JG, Vernot B, Kohler K, Voigt B, Okita K, Maricic T, He Z, Lachmann R, Paabo S, Treutlein B, Huttner WB. 2016. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A 1961. Rotation-mediated histogenetic aggregation of dissociated cells. A quantifiable approach to cell interactions in vitro. Exp Cell Res 22:455–475. [DOI] [PubMed] [Google Scholar]
- Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. 2015. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep 10:537–550. [DOI] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. 2012. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10:771–785. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval-Espinosa C, Bershteyn M, Kriegstein AR. 2016. Expression Analysis Highlights AXL as a Candidate Zika Virus Entry Receptor in Neural Stem Cells. Cell Stem Cell 18:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke DM, Nasrallah MP, Desai A, Melenhorst JJ, Mansfield K, Morrissette JJD, Martinez-Lage M, Brem S, Maloney E, Shen A, Isaacs R, Mohan S, Plesa G, Lacey SF, Navenot JM, Zheng Z, Levine BL, Okada H, June CH, Brogdon JL, Maus MV. 2017. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa J, Pao GM, Shokhirev MN, Verma IM. 2018. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep 23:1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. 2017. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol 19:v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T, Marchetto MC, Gage FH, Simons BD, Livesey FJ. 2016. 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell 18:467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozone C, Suga H, Eiraku M, Kadoshima T, Yonemura S, Takata N, Oiso Y, Tsuji T, Sasai Y. 2016. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat Commun 7:10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155:1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker NR, Khong P, Parkinson JF, Howell VM, Wheeler HR. 2015. Molecular heterogeneity in glioblastoma: potential clinical implications. Front Oncol 5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Pasca SP. 2015. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister BJ, Iwata A, Meaney DF, Smith DH. 2004. Extreme stretch growth of integrated axons. J Neurosci 24:7978–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham MT, Pollock KM, Rose MD, Cary WA, Stewart HR, Zhou P, Nolta JA, Waldau B. 2018. Generation of human vascularized brain organoids. Neuroreport 29:588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming GL. 2018. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat Protoc 13:565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Jacob F, Song H, Ming GL. 2017. Using brain organoids to understand Zika virus-induced microcephaly. Development 144:952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL. 2016. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165:1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P. 2017. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. 2014. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159:896–910. [DOI] [PubMed] [Google Scholar]
- Richard AS, Shim BS, Kwon YC, Zhang R, Otsuka Y, Schmitt K, Berri F, Diamond MS, Choe H. 2017. AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Proc Natl Acad Sci U S A 114:2024–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, Korving J, van Boxtel R, Duarte AA, Lelieveld D, van Hoeck A, Ernst RF, Blokzijl F, Nijman IJ, Hoogstraat M, van de Ven M, Egan DA, Zinzalla V, Moll J, Boj SF, Voest EE, Wessels L, van Diest PJ, Rottenberg S, Vries RGJ, Cuppen E, Clevers H. 2018. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 172:373–386 e310. [DOI] [PubMed] [Google Scholar]
- Sacramento CQ, de Melo GR, de Freitas CS, Rocha N, Hoelz LV, Miranda M, Fintelman-Rodrigues N, Marttorelli A, Ferreira AC, Barbosa-Lima G, Abrantes JL, Vieira YR, Bastos MM, de Mello Volotao E, Nunes EP, Tschoeke DA, Leomil L, Loiola EC, Trindade P, Rehen SK, Bozza FA, Bozza PT, Boechat N, Thompson FL, de Filippis AM, Bruning K, Souza TM. 2017. The clinically approved antiviral drug sofosbuvir inhibits Zika virus replication. Sci Rep 7:40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y. 2015. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun 6:8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N, Alvarez-Buylla A, Berger MS. 2005. Neural stem cells and the origin of gliomas. N Engl J Med 353:811–822. [DOI] [PubMed] [Google Scholar]
- Sasai Y 2013. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 12:520–530. [DOI] [PubMed] [Google Scholar]
- Schulte A, Gunther HS, Phillips HS, Kemming D, Martens T, Kharbanda S, Soriano RH, Modrusan Z, Zapf S, Westphal M, Lamszus K. 2011. A distinct subset of glioma cell lines with stem cell-like properties reflects the transcriptional phenotype of glioblastomas and overexpresses CXCR4 as therapeutic target. Glia 59:590–602. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, Temple S. 2008. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell 3:289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. 2012. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci 15:477–486, S471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, Pasca SP. 2017. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 95:779–790 e776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman MA, Aboharb F, Zeltner N, Studer L. 2017. Pluripotent stem cells in neuropsychiatric disorders. Mol Psychiatry 22:1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somaa FA, Wang TY, Niclis JC, Bruggeman KF, Kauhausen JA, Guo H, McDougall S, Williams RJ, Nisbet DR, Thompson LH, Parish CL. 2017. Peptide-Based Scaffolds Support Human Cortical Progenitor Graft Integration to Reduce Atrophy and Promote Functional Repair in a Model of Stroke. Cell Rep 20:1964–1977. [DOI] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 110:4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struzyna LA, Wolf JA, Mietus CJ, Adewole DO, Chen HI, Smith DH, Cullen DK. 2015. Rebuilding Brain Circuitry with Living Micro-Tissue Engineered Neural Networks. Tissue Eng Part A 21:2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for R, Treatment of Cancer Brain T, Radiotherapy G, National Cancer Institute of Canada Clinical Trials G. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. [DOI] [PubMed] [Google Scholar]
- Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, Di Meco F, Lieberman F, Zhu JJ, Stragliotto G, Tran D, Brem S, Hottinger A, Kirson ED, Lavy-Shahaf G, Weinberg U, Kim CY, Paek SH, Nicholas G, Bruna J, Hirte H, Weller M, Palti Y, Hegi ME, Ram Z. 2017. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 318:2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. [DOI] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, Didier RA, Jin P, Song H, Ming GL. 2016. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 18:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. 2008. A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3:279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147. [DOI] [PubMed] [Google Scholar]
- Tornero D, Tsupykov O, Granmo M, Rodriguez C, Gronning-Hansen M, Thelin J, Smozhanik E, Laterza C, Wattananit S, Ge R, Tatarishvili J, Grealish S, Brustle O, Skibo G, Parmar M, Schouenborg J, Lindvall O, Kokaia Z. 2017. Synaptic inputs from stroke-injured brain to grafted human stem cell-derived neurons activated by sensory stimuli. Brain 140:692–706. [DOI] [PubMed] [Google Scholar]
- Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. 2008. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One 3:e1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner VD, Gleghorn JP, Miller E, Radisky DC, Nelson CM. 2015. Mechanically patterning the embryonic airway epithelium. Proc Natl Acad Sci U S A 112:9230–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura CV, Maia M, Bravo-Filho V, Gois AL, Belfort R, Jr. 2016. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 387:228. [DOI] [PubMed] [Google Scholar]
- Vera E, Studer L. 2015. When rejuvenation is a problem: challenges of modeling late-onset neurodegenerative disease. Development 142:3085–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh DM, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S, Watkins D, Fotiadis N, Bali M, Darvish-Damavandi M, Lote H, Eltahir Z, Smyth EC, Begum R, Clarke PA, Hahne JC, Dowsett M, de Bono J, Workman P, Sadanandam A, Fassan M, Sansom OJ, Eccles S, Starling N, Braconi C, Sottoriva A, Robinson SP, Cunningham D, Valeri N. 2018. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359:920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkner M, Zschatzsch M, Rostovskaya M, Overall RW, Busskamp V, Anastassiadis K, Karl MO. 2016. Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Reports 6:525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, Lachman HM. 2017. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol Autism 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. 2005. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci 8:288–296. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, Coppola G, Pearson CA, Yamauchi K, Gong D, Dai X, Damoiseaux R, Aliyari R, Liebscher S, Schenke-Layland K, Caneda C, Huang EJ, Zhang Y, Cheng G, Geschwind DH, Golshani P, Sun R, Novitch BG. 2017. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep 21:517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P, Taylor AC. 1960. Reconstitution of Complete Organs from Single-Cell Suspensions of Chick Embryos in Advanced Stages of Differentiation. Proc Natl Acad Sci U S A 46:1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, Ihry RJ, Kommineni S, Bilican B, Klim JR, Hill EJ, Kane LT, Ye C, Kaykas A, Eggan K. 2016. Genetic Ablation of AXL Does Not Protect Human Neural Progenitor Cells and Cerebral Organoids from Zika Virus Infection. Cell Stem Cell 19:703–708. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, Zhong M, Stanley EG, Elefanty AG, Naegele JR, Lee SH, Weissman SM, Park IH. 2017. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 21:383–398 e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, Jacob F, Nguyen HN, Itkin M, Hanna C, Shinn P, Allen C, Michael SG, Simeonov A, Huang W, Christian KM, Goate A, Brennand KJ, Huang R, Xia M, Ming GL, Zheng W, Song H, Tang H. 2016. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 22:1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, Mao M, Poon RYC, Kim J, Song H, Ming GL, Zhang M. 2017. DISC1 Regulates Neurogenesis via Modulating Kinetochore Attachment of Ndel1/Nde1 during Mitosis. Neuron 96:1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, Kim S, Wang X, Dore LC, Jin P, Regot S, Zhuang X, Canzar S, He C, Ming GL, Song H. 2017a. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 171:877–889 e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, Kim NS, Habela C, Zheng L, Jacob F, Zhang F, Lee EM, Huang WK, Ringeling FR, Vissers C, Li C, Yuan L, Kang K, Kim S, Yeo J, Cheng Y, Liu S, Wen Z, Qin CF, Wu Q, Christian KM, Tang H, Jin P, Xu Z, Qian J, Zhu H, Song H, Ming GL. 2017b. Zika-Virus-Encoded NS2A Disrupts Mammalian Cortical Neurogenesis by Degrading Adherens Junction Proteins. Cell Stem Cell 21:349–358 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, Yuan T, Yang J, Li Y, Shi L, Guan D, Pan H, Duan S, Ding Z, Li M, Yi F, Bai R, Wang Y, Chen C, Yang F, Li X, Wang Z, Aizawa E, Goebl A, Soligalla RD, Reddy P, Esteban CR, Tang F, Liu GH, Belmonte JC. 2015. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348:1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang ZN, Rong Z, Xu Y. 2011. Immunogenicity of induced pluripotent stem cells. Nature 474:212–215. [DOI] [PubMed] [Google Scholar]
- Zhao T, Zhang ZN, Westenskow PD, Todorova D, Hu Z, Lin T, Rong Z, Kim J, He J, Wang M, Clegg DO, Yang YG, Zhang K, Friedlander M, Xu Y. 2015a. Humanized Mice Reveal Differential Immunogenicity of Cells Derived from Autologous Induced Pluripotent Stem Cells. Cell Stem Cell 17:353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Nelson AR, Betsholtz C, Zlokovic BV. 2015b. Establishment and Dysfunction of the Blood-Brain Barrier. Cell 163:1064–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, Evans T, Studer L, Chen S. 2017. High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain. Cell Stem Cell 21:274–283 e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY. 2003. Postnatal neurodevelopmental disorders: meeting at the synapse? Science 302:826–830. [DOI] [PubMed] [Google Scholar]


