Abstract
Selfish genetic elements that manipulate gametogenesis to achieve a transmission advantage are known as meiotic drivers. Sex-ratio X-chromosomes (SR) are meiotic drivers that prevent the maturation of Y-bearing sperm in male carriers to result in the production of mainly female progeny. The spread of an SR chromosome can affect host genetic diversity and genome evolution, and can even cause host extinction if it reaches sufficiently high prevalence. Meiotic drivers have evolved independently many times, though only in a few cases is the underlying genetic mechanism known. In this study we use a combination of transcriptomics and population genetics to identify widespread expression differences between the standard (ST) and sex-ratio (SR) X-chromosomes of the fly Drosophila neotestacea. We found the X-chromosome is enriched for differentially expressed transcripts, and that many of these X-linked differentially expressed transcripts had elevated Ka/Ks values between ST and SR, indicative of potential functional differences. We identified a set of candidate transcripts, including a testis-specific, X-linked duplicate of the nuclear transport gene importin-α2 that is overexpressed in SR. We find suggestions of positive selection in the lineage leading to the duplicate and that its molecular evolutionary patterns are consistent with relaxed purifying selection in ST. As these patterns are consistent with involvement in the mechanism of drive in this species, this duplicate is a strong candidate worthy of further functional investigation. Nuclear transport may be a common target for genetic conflict, as the mechanism of the autosomal Segregation Distorter drive system in D. melanogaster involves the same pathway.
Keywords: testes, spermatogenesis, genetic conflict, nuclear transport, meiotic drive
Introduction
Genetic conflict occurs when one portion of the genome promotes its own transmission to the detriment of another portion of genome. Conflict is pervasive and potentially a major evolutionary force (Burt & Trivers, 2006; Lindholm et al., 2016; Rice, 2013). Meiotic drive is a type of genetic conflict where selfish genes manipulate gametogenesis to subvert Mendel’s law of equal segregation and make their way into over 50% of gametes. The phenomenon itself and its potential evolutionary consequences have been known for decades, but recent developments in engineering synthetic drive systems gives understanding the dynamics and mechanisms of natural driving systems a new urgency (Lindholm et al., 2016; Sandler & Novitski, 1957). Sex-ratio (SR) meiotic drive involves selfish elements located on the X-chromosome that reduce the transmission of Y-bearing sperm in males. This form of drive is particularly interesting because it can bias the sex ratio of the offspring towards daughters, potentially leading to population or species extinction (Carvalho & Vaz, 1999; Hamilton, 1967). Sex-ratio drive is also the most common form of chromosomal meiotic drive known, having evolved independently dozens of times in Dipterans (Jaenike, 2001). The sex chromosomes may be particularly prone to conflict (Hurst & Pomiankowski, 1991), though ascertainment bias also likely contributes to the large number of known sex-ratio drive systems.
Though the first meiotic drive systems were discovered in the 1920s and a variety are known today, only a few have been dissected mechanistically (reviewed in Burt & Trivers, 2006). Of the known SR drive systems two have at least part of their genetic and mechanistic basis identified. Both systems are found in Drosophila simulans (Helleu et al., 2016; Tao, Araripe, et al., 2007). In the Paris system, drive has been pinpointed to a protein that binds to the heterochromatin of the Y-chromosome during meiosis to cause nondisjunction events that result in inviable Y-bearing sperm, though there is another locus associated with this system that is still unknown (Helleu et al., 2016). In the independently evolved Winters sex-ratio system, the genetic loci of two SR distorters, one a duplicate copy of the other, have been identified (Lin et al., 2018; Tao, Araripe, et al., 2007; Tao, Masly, Araripe, Ke, & Hartl, 2007). In this system the mechanistic basis of drive itself remains unclear, but autosomal suppression occurs through small RNA interference (Lin et al., 2018; Tao, Araripe, et al., 2007; Tao, Masly, et al., 2007). One of the best-studied male meiotic drive systems is Segregation Distorter (SD) in D. melanogaster (Larracuente & Presgraves, 2012). Though it is an autosomal driver, it operates in much the same way as sex-ratio X-chromosomes in that half of the sperm fail to develop properly. The distorter gene (sd-RanGAP) is a truncated duplicate of RanGAP (Merrill, Bayraktaroglu, Kusano, & Ganetzky, 1999). Wild-type RanGAP stays in the cytoplasm and powers the nuclear transport cycle, but the sd-RanGAP protein mislocalizes to the nucleus, which disrupts nuclear transport to cause spermatogenesis to fail (Ayumi Kusano, Staber, & Ganetzky, 2001; A. Kusano, Staber, & Ganetzky, 2002; Larracuente & Presgraves, 2012).
In spite of significant interest, the main reason so little is known about the mechanism of drive systems is that most meiotic drivers are associated with chromosomal inversions (Burt & Trivers, 2006; Jaenike, 2001). Inversions are thought to accumulate on driving chromosomes because they link together interacting elements that are required for drive (Charlesworth & Hartl, 1978). Unfortunately, the suppression of recombination caused by such inversions also makes classical genetic analysis difficult. The Drosophila drive systems discussed above are not associated with inversions, allowing for their genetic dissection using traditional mapping techniques. The t-haplotype of Mus musculus remains the only male meiotic driver (either autosomal or sex-linked) with a known mechanistic basis that is associated with inversions (Bauer, Veron, Willert, & Herrmann, 2007; Bauer, Willert, Koschorz, & Herrmann, 2005; Herrmann, Koschorz, Wertz, McLaughlin, & Kispert, 1999; Mary F Lyon, 1991; M. F. Lyon, 2003).
Potential mapping strategies that circumvent chromosomal inversions include large scale transcriptomic or genomic analysis comparing sex-ratio and wild-type chromosomes. To date, the only such investigation is a comparison of gene expression between wild-type and sex-ratio males in the stalk-eyed fly Teleopsis dalmanni (Reinhardt et al., 2014). This analysis showed that meiotic drive has had a significant impact on X-linked evolution in this species, as a large number of X-linked genes were found to be differentially expressed between the wild-type and selfish X-chromosomes (Reinhardt et al., 2014). Though driving X-chromosomes are at found at relatively high frequencies in this species, they appear to have low nucleotide variation and extremely limited recombination with ST, though recombination may occur between SR chromosomes (Christianson, Brand, & Wilkinson, 2011; Paczolt, Reinhardt, & Wilkinson, 2017). Loci involved in drive or very closely linked to drive are expected to show patterns similar to a selective sweep (Derome, Baudry, Ogereau, Veuille, & Montchamp-Moreau, 2008; Kingan, Garrigan, & Hartl, 2010). Unfortunately, the very tight linkage and low polymorphism on the driving X-chromosome suggests that all loci across the X-chromosome – even those not involved in drive – are expected to show the same molecular evolutionary pattern as the drive-associated loci in this species, making its genetic dissection difficult.
The sex-ratio X-chromosome (SR) of D. neotestacea is similarly characterized by large inversions and significant genetic differentiation with the standard X-chromosome (ST), but critically there is substantial genetic variation present on SR (Pieper & Dyer, 2016). In a survey of nucleotide variation on SR and ST at 11 arbitrarily chosen loci uninvolved in drive, there were no fixed differences between the two chromosome types, and linkage disequilibrium on SR was consistent with regular recombination between SR chromosomes (Pieper & Dyer, 2016). This species is broadly distributed through temporal and boreal forests in North America, and there is high gene flow across geographic regions (Dyer, 2012; Pieper & Dyer, 2016). SR is also found at long-term high frequencies in some populations and there are no identified segregating suppressors (Dyer, 2012; Dyer, Bray, & Lopez, 2013; James & Jaenike, 1990). Drive is also extremely effective in this species, with SR males producing 98% daughters; the only sons are sterile and presumed to be XO males that result from nondisjunction (James & Jaenike, 1990).
Altogether, these characteristics make the D. neotestacea SR system an excellent choice for using genomic approaches to investigate the mechanistic basis of an inversion rich drive system. In this study, we used high throughput sequencing to compare gene expression in the testes of males carrying SR or ST in the same genetic background. We determined which chromosomes carry differentially expressed transcripts, and identified nucleotide differences between transcripts from ST and SR. We evaluated the potential functional impact of these differences and generated a list of candidates that were rapidly evolving. We used microscopy to compare spermatogenesis in ST and SR males. We then carried out a population genetic study of the candidates using wild ST and SR males and identified molecular evolutionary patterns at five loci consistent with direct involvement with the mechanism of drive. One of these top candidates is a novel X-linked duplicate of the autosomal gene importin-α2, a key part of the nuclear import pathway. We argue this candidate is highly likely to be involved in the meiotic drive mechanism.
Methods
Samples, sequencing, and transcriptome assembly
The ST and SR D. neotestacea males used in this study come from lab stocks maintained by K. Dyer that were originally collected in New York in 1990. The SR stock was initiated with a single wild-caught SR male and is maintained by crossing to an inbred ST lab stock. Every generation, ST/Y males are crossed to SR/SR females to generate SR/Y males, and SR/Y males are also crossed to SR/SR females to produce more SR/SR females. Thus, the only genetic difference between ST and SR males is the X-chromosome.
cDNA libraries were made from sexually mature adult males. Whole testes were dissected from 25 ST or SR males at 4–7 days after eclosion and flash frozen for each library. The carcass (i.e., the body without the testes) was also prepared for RNA extraction. RNA was extracted using a Qiagen RNEasy kit (Qiagen, Germantown, MD, USA). Six testes cDNA libraries (three ST and three SR biological replicates) and four carcass libraries (two ST and two SR) were prepared for high-throughput 75bp single-end sequencing using the Illumina TruSeq kit (Illumina, Inc, San Diego, CA, USA). All libraries were run two independent times on a single Illumina HiSeq lane (San Diego, CA) and demultiplexed at the Cornell Sequencing Center. Base quality was evaluated using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html) and sequencing adaptors and low-quality bases were trimmed from the ends of reads, and reads with less than 99% of bases with a call quality score of at least 20 were discarded.
All filtered sequencing reads were combined and used to de novo assemble the transcriptome using the Trinity pipeline (Haas et al., 2013). For transcripts that had multiple isoforms identified, the isoform with the highest quality blastx hit (i.e. lowest e-value) against UniProtKB (http://www.uniprot.org/) was included in further analyses (Camacho et al., 2009; The UniProt Consortium, 2014). Reads were then split back into their respective libraries and aligned to the transcriptome using Bowtie2 with the default parameters (Langmead & Salzberg, 2012). SR-only and ST-only transcriptome assemblies were created in the same way. For each library, read abundance per transcript was quantified using RSEM (B. Li & Dewey, 2011). After analysis showed no differences in the abundance of the technical replicates, they were combined together.
Differential and tissue specific expression analysis
We used the program RUVg to normalize abundance counts between all testes libraries (Risso, Ngai, Speed, & Dudoit, 2014). First, a differential expression analysis between ST and SR was carried out in DESeq using the unnormalized read counts and the total 63,821 transcripts in the assembly (Anders & Huber, 2012). The lowest 30% of total expressed transcripts were filtered out to increase the power, leaving 25,484 transcripts in the differential expression analysis. These transcripts were ranked according to their false discovery rate (FDR), and then every transcript except the 10,000 most differentially expressed were chosen for the RUVg empirical normalization control. These remaining 15,484 transcripts were used to estimate unwanted variance, since we assume they are not differentially expressed. That estimation of variance was then included in the general linear model of differential expression between ST and SR in DESeq2 and edgeR (Love, Huber, & Anders, 2014; Robinson, McCarthy, & Smyth, 2010). FDR was calculated to correct for multiple testing, and FDR ≤ 0.01 was required for significance (Benjamini & Hochberg, 1995). Transcripts were considered significantly differentially expressed if they had an FDR ≤ 0.01 in both the DESeq2 and edgeR analyses. All analyses were performed in the R programming language in RStudio (https://www.rstudio.com/) (RCoreTeam, 2014).
The same procedure was carried out for the carcass data, but as there were only two biological replicates each for ST and SR the power of this analysis was low, and no transcripts met the criteria for significance. However, the mean expression estimates from DESeq2 were used to determine the tissue specificity of transcripts expressed in the testes; an estimated carcass base mean expression of < 2 counts was used as the criterion for transcript testis-specificity. The total number of testes-specific transcripts was 14,392.
Assignment of transcripts to chromosomes
To identify X-linked transcripts, homology to the D. melanogaster and D. virilis genomes was used. The Release 6 D. melanogaster whole genome assembly was obtained from Flybase (http://flybase.org/)(Hoskins et al., 2015) and a version of the D. virilis genome assembly with scaffolds assigned to Muller elements was obtained from Yasir Ahmed-Braimah (personal communication). NCBI’s tblastx with cut off values of e-value < 1e−20 and length > 50 bp was used to identify transcript homologs in the D. melanogaster and D. virilis genomes. If no D. virilis homolog was available or the transcript mapped to a scaffold, the location of the D. melanogaster homolog was used. Synteny between Muller elements was used to assign genomic location of transcripts in D. neotestacea (Camacho et al., 2009; Schaeffer et al., 2008). The X-chromosome is homologous in all three species and unlinked to other Muller elements in D. neotestacea (Pieper & Dyer, 2016).
GO term enrichment analysis was carried out using the GOseq program in the Trinity package (Haas et al., 2013). Transcript homologs were identified in the SwissProt database using blastx (e-value < 1e−20) (Camacho et al., 2009; The UniProt Consortium, 2014). GO term enrichment compared to the entire transcriptome was determined for 5 sets of transcripts (DE transcripts, DE transcripts with SR-biased expression, DE transcripts with ST-biased expression, testis-specific DE transcripts with ST-biased expression, and testis-specific DE transcripts with SR-biased expression).
Nucleotide differences in transcripts from ST and SR
Transcripts with nucleotide sequence differences between ST and SR were identified using samtools (H. Li et al., 2009). The mpileup function was used to make a vcf file compiling all the variant sites in the testes libraries. Vcftools was used to remove any variant sites (i.e. SNPs) with a minor allele frequency of less than 50% to remove sites with variation within ST or SR libraries (e.g., one ST library has an A, two ST libraries have a G, and all three SR libraries have a G) (Danecek et al., 2011). Further filtering was performed to remove any nucleotide sites with coverage < 100× across all samples and any transcripts with less than 100 sites and with at least 100× coverage. Finally, any sites with heterozygotes called in any library were removed to ensure that the final set of sites contained only one allele in every SR sample and a different allele in every ST sample, representing a conservative estimate of the sequence divergence between the chromosomes. The percent sequence difference between ST and SR was calculated for each transcript by taking the number of different sites over the total length of the transcript and multiplying by 100. Of the 1,349 transcripts that had at least one sequence difference, 1,067 mapped to the X-chromosome using homology, 32 mapped to the autosomes, and 250 had unknown genomic locations because they did not have homology to a known gene in D. melanogaster or D. virilis (Table S1). The only genetic differences between ST and SR should be on the X-chromosome due to the crossing scheme of the stocks, indicating these transcripts can be assigned to the X-chromosome. We further validated this assumption by performing a blastn search of these 250 transcripts against an unpublished assembly of the D. innubila genome (R. Unckless, unpublished), a species from the quinaria group that is more closely related to D. neotestacea than either D. melanogaster or D. virilis (Perlman, Spicer, Shoemaker, & Jaenike, 2003). Though only 34% of the unknown transcripts matched to the D. innubila genome, 87% of those that did matched to the X-chromosome. We therefore reassigned the 250 unknown transcripts and 32 originally autosome-mapped transcripts with sequence differences to the X-chromosome. This increased the total number of transcripts assigned to the X-chromosome from 2,748 to 3,030.
For each of the 1,349 transcripts with a sequence difference between SR and ST, GATK was used to create a duplicate set of transcript sequences that carried the alternate alleles at each variable site (using the original transcriptome assembly as the reference) (Van der Auwera et al., 2013). The matched alternate and reference allele-containing transcripts were then carefully partitioned between ST and SR based on the assignment of variant sites in the vcf file identifying sequence differences. Open reading frames were identified using Transdecoder (Haas et al., 2013), and the coding sequences were extracted from these transcripts using bedtools (Quinlan & Hall, 2010). The validity of the open reading frames was confirmed by eye in a small subset of transcripts using Geneious (Kearse et al., 2012). The rate of protein evolution, or Ka/Ks, was calculated between the SR and ST sequences for the coding region of each transcript using KaKs_Calculator (Zhang et al., 2006).
Sequencing, gene tree, and population genetic analyses of candidates
Ten candidate loci were chosen and sequenced in each of 10 wild-caught ST and SR D. neotestacea males as well as in one or two ST individuals of the closely-related species D. testacea and D. orientacea. These D. neotestacea males were randomly chosen from the range-spanning dataset used in Pieper and Dyer (2016) (See Table S1 of that paper) and had been identified as carrying an ST or SR X-chromosome by the proportion of female offspring they produced (Dyer, 2012). As the results will show, one of these candidates is an X-linked duplicate of the autosomal importin-α2 gene that we named X-importin-α2. X-importin-α2 was sequenced in an additional seven wild-caught ST males from the same dataset, and both duplicate copies were sequenced in 1 or 2 individuals of several closely-related species (D. testacea, D. orientacea, D. putrida, and D. bizonata). PCR primers were designed with Primer3 in Geneious (Table S2)(Kearse et al., 2012; Untergasser et al., 2012), and fragments were amplified using standard PCR protocols and sequenced on an Applied Biosystems (Foster City, CA, USA) 3730xl DNA Analyzer at the Georgia Genomics Facility. Base calls were confirmed using Geneious and sequences were aligned by hand (Kearse et al., 2012).
The coding sequences of importin-α1, importin-α2, importin-α3, and X-importin-α2 sequences from D. neotestacea, D. melanogaster and D. virilis were aligned in Geneious and corrected by hand and then used to build an unrooted, neighbor-joining phylogenetic tree (Kearse et al., 2012). Sequences of importin-α1, 2, and 3 from D. melanogaster and D. virilis were obtained from Flybase (http://flybase.org/), and the D. neotestacea sequences were identified from the transcriptome using blastx (Camacho et al., 2009). We used the codeml function in PAML v4.8a (Yang, 2007) to infer the rate of protein evolution (ω, or dN/dS) for the importin phylogeny. We compared a model with one ω value across the entire phylogeny versus a free-ratio model where ω was estimated independently for each branch. These models were evaluated using a likelihood ratio test, with the P-value inferred using a χ2-distribution.
Analyses of DNA polymorphism and divergence at these candidate loci were carried out in the program DnaSP (Librado & Rozas, 2009). Population genetic data from a set of five X-linked (marf, mof, pdg, rpl, and spk) and seven autosomal protein coding loci (esc, gl, ntid, mago, tpi, sia, and wee) that were arbitrarily chosen with respect to SR were used as a comparison; these data are from (Pieper & Dyer, 2016). These markers are referred to as the “non-candidate” X-linked markers. Hudson-Kreitman-Aguadé (HKA) tests were carried out in the program MLHKA using an MCMC length of 1,000,000 (Wright & Charlesworth, 2004), with significance determined using likelihood ratio tests.
Sperm microscopy
The testes of 1-day old ST and SR males from the same stock as was used for the RNAseq were dissected and stained with DAPI (4,6-diamidino-2-phenylindole) to identify the heads of developing spermatids. Testes were dissected in PBS (phosphate buffered saline) and the developing spermatids were gently removed and spread apart. The slide was dried at 60 °C for 5–10 minutes, fixed in 3:1 methanol and glacial acetic acid for 5 minutes, rinsed 3 times in PBS, and labelled with 0.5 mg/mL DAPI in glycerol. The developing 64-spermatid bundles in post-meiosis were identified using 650× magnification.
Results
SR and ST chromosomes cause expression differences for genes primarily on the X
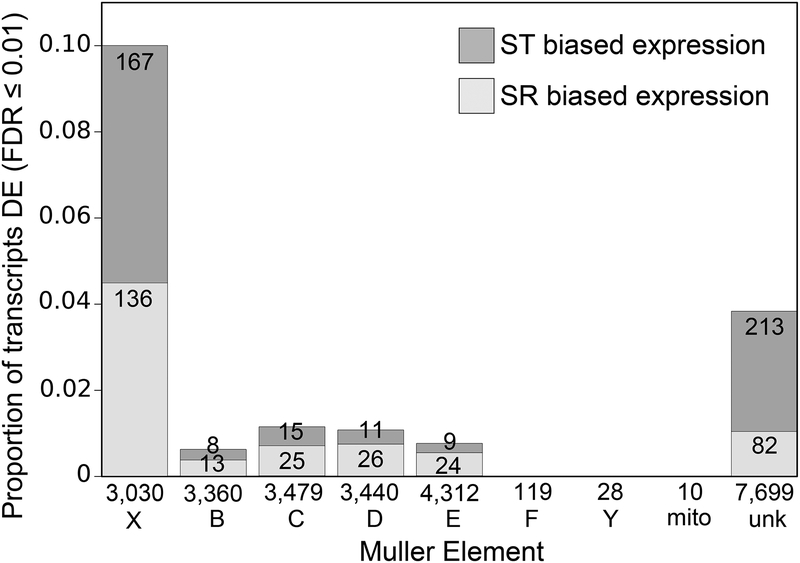
In total, the transcriptome contained 63,821 transcripts with an average contig length of 376.66 bp and a contig N50 value of 514 bp. The number of transcripts in this dataset overestimates the actual number of genes expressed in all tissues due to fragmented assembly. After filtering and normalization, 729 transcripts were identified as differentially expressed (DE) in the testes of ST and SR males (FDR ≤ 0.01 in both DESeq2 and edgeR). Of these, 306 had SR-biased expression patterns, and 423 were ST-biased (Figure 1). Additionally, 19 transcripts were expressed in SR but absent in ST, and 149 transcripts were expressed in ST but absent in SR. 42% (303 transcripts) of DE transcripts mapped to the X-chromosome (Figure 1); of these, transcripts with ST-biased expression about equaled those with SR-biased expression. In contrast, the other four large autosomes combined carry only 18% (131 transcripts) of DE transcripts, despite having a total number of mapped transcripts nearly quadruple that of the X-chromosome. These mapped relatively uniformly across the autosomes. No differentially expressed transcripts were found to map to the dot chromosome (Muller element F), the Y-chromosome, or the mitochondria (Figure 1a). The remaining 40.5% (295 transcripts) of DE transcripts were not able to be mapped to any region of the genome using homology with D. virilis and D. melanogaster or sequence differences between ST and SR. A chi-squared test comparing DE transcripts with total mapped transcripts for the five large Muller elements was highly significant (χ2=868.01, df=4, p<2.2e-16), indicating the X-chromosome is enriched for DE transcripts. This is expected as the ST and SR males used are genetically identical except for the X-chromosome; autosomal DE genes may be due to trans-acting X-linked genes.
Figure 1.
Differentially expressed (DE) transcripts are enriched on the X-chromosome. The total number of transcripts that mapped to each chromosome is listed beneath the bars. The large Muller elements are X, B, C, D, and E. F is the small, non-recombining dot chromosome, mito is the mitochondria, Y is the Y-chromosome, and unk stands for unknown location. Dark and light grey indicate the proportion of ST and SR biased transcripts, respectively, given the total number of transcripts that mapped to that chromosome. The number of DE transcripts in each category is printed within each bar.
Amount of sequence differences between ST and SR is as expected
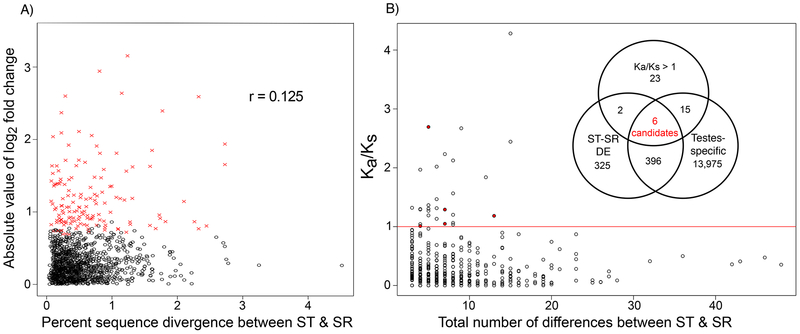
Of all 2,748 transcripts that initially mapped to the X-chromosome based on homology, 38.8% (1,067 transcripts) had at least one nucleotide difference between ST and SR (Table S1, Figure 2a). Because of the stringent requirements used to identify sequence differences, this likely underestimates the differentiation between ST and SR. Among transcripts with a sequence difference, the nucleotide sequence divergence ranged from 0.03 – 4.5% with a mean difference of 0.53% (median, 0.41%) (Figure 2a). As a comparison, the average percent sequence divergence in the coding regions of five arbitrarily chosen X-linked non-candidate genes in a large population genetic sample of ST males ranged from 0.16 to 1.25%, with an average of 0.56% (Pieper & Dyer, 2016). Among transcripts that contained at least one nucleotide difference between ST and SR, there was a very weak correlation between the absolute value of log2 fold change in expression between ST and SR and the percent sequence difference (r=0.125, p=0.017, Pearson’s correlation) (Figure 2a, Figure S1).
Figure 2.
Sequence differences between ST and SR in transcripts. A) There is weak relationship between differential expression between ST and SR (shown as the absolute value of log2 fold change) and sequence differences between ST and SR (Pearson’s correlation, r = 0.125). Percent sequence difference is calculated as the total number of differences divided by the length of the transcript times 100. Transcripts with significant differential expression are marked in red. Only transcripts meeting the minimum coverage criteria for detecting sequence differences and had at least one difference are included. The mean percent different of transcripts with nucleotide differences was 0.53%. B) Transcripts with the highest number of differences are not the same as those with the highest Ka/Ks values. Only transcripts with more than three differences and at least one synonymous difference are included in the figure. Transcripts with more than three differences but no synonymous differences were also included in the Ka/Ks > 1 set. The identified candidates are marked in red; one of these had no synonymous differences and is not pictured. The inset Venn diagram shows the criteria used to identify Ka/Ks candidates and the number of transcripts in each category. The total number of transcripts with Ka/Ks > 1 or more than three synonymous differences was 46, the total number of ST-SR DE transcripts was 729, and the total number of testes-specific transcripts was 14,392. Ka/Ks was calculated between ST and SR.
Identification of transcripts with elevated Ka/Ks between ST and SR
Ka/Ks values were calculated between ST and SR for each transcript to evaluate the potential functional consequences of these nucleotide differences. Of the 1,349 transcripts with sequence differences, 1,116 transcripts had annotated open reading frames containing nucleotide differences. The transcripts with at least one synonymous substitution (N=1,001) had an average Ka/Ks value of 0.23, and 31 of these transcripts had Ka/Ks >1 (Figure 2b). Combining these 31 transcripts with those that had at least three nucleotide differences but no synonymous differences (N=15) yielded a total of 46 transcripts with an elevated number of non-synonymous differences. Notably, these transcripts were not the transcripts with the most substitutions (Figure 2b). Using the transcripts with Ka/Ks < 1 as the expectation, transcripts with elevated Ka/Ks were not overrepresented among loci that were DE (χ2=2.407, p=0.121; Table 1). More of these transcripts than expected were testes specific (χ2=10.383, p=0.001; Table 1).
Table 1.
Chi-squared tests examining enrichment of positively selected transcripts for a) testes-specific expression, and b) differential expression (DE) between ST and SR. Each test had one degree of freedom. Values larger than expected are bolded. The set of Ka/Ks > 1 includes transcripts with Ka/Ks > 1 as well as those that have three or more nonsynonymous substitutions but no synonymous substitutions. Ka/Ks was calculated between ST and SR.
| Ka/Ks > 1 | Ka/Ks < 1 | χ2 | p-value | |
|---|---|---|---|---|
| testes specific | 21 | 241 | 10.383 | 0.001 |
| not testes specific | 27 | 829 | ||
| DE | 8 | 94 | 2.407 | 0.121 |
| not DE | 40 | 976 |
Identification of candidate genes
To narrow down our list of candidate transcripts for involvement in the drive mechanism, we focused on those that have Ka/Ks > 1 (or over 3 non-synonymous differences and no synonymous differences), are expressed only in the testes, and are differentially expressed between ST and SR (Figure 2b). While testis-specificity is not necessarily a requirement for participation in the mechanism of drive, we chose this criterion to help narrow down the field of candidates. Six transcripts matching these criteria were identified (Table S3). Two of them (TR24932 and TR5481) had no identifiable orthologs in D. melanogaster and D. virilis. Transcript TR6297 was identified as an ortholog of the D. melanogaster gene CG7366, which is located on an autosome in D. melanogaster and highly expressed in the testes with an unknown function (http://flybase.org/reports/FBgn0035855.html). Transcript TR23125 is a homolog of an X-linked gene lethal(1)1Bi, which is expressed during the mitosis stage of spermatogenesis (http://flybase.org/reports/FBgn0001341.html). This gene contains armadillo DNA-binding repeats, which often function in intracellular signaling and cytoskeletal regulation.
The final two candidate transcripts (TR10603 and TR2814) both mapped to an autosomal gene called importin-α2 (http://flybase.org/reports/FBgn0267727.html). Also known as Pendulin, this gene is involved in nuclear transport and also contains several armadillo DNA-binding repeats (Goldfarb, Corbett, Mason, Harreman, & Adam, 2004). Aligning these two transcripts to the D. melanogaster importin-α2 sequence revealed that they mapped to sequential sections of the protein (Figure S2). Searching the transcriptome data for other transcripts that mapped to importin-α2 revealed transcript TR37105 that aligned upstream and partially overlapped the other two transcripts. TR37105 also has sequence differences between ST and SR but has a Ka/Ks value of only 0.35 (Table S4). It also has SR-biased expression like the other two transcripts, but it was not considered significantly DE in the earlier analysis because the adjusted p-value is 0.013. To identify the 3’ end of the gene we queried the SR and ST specific assemblies using blastn (Camacho et al., 2009).
Searching for transcripts with homology to importin-α2 also revealed a full-length transcript (TR7043) that had no sequence differences between ST and SR and was not differentially expressed or testis-specific. This transcript also had much higher sequence similarity with the D. melanogaster and D. virilis importin-α2 sequences compared to the candidate transcripts (Figure 3, Figure S2). Therefore, TR7043 must represent the homolog of importin-α2, an autosomal gene in D. melanogaster, whereas the three candidate transcripts in D. neotestacea represent an X-linked duplicate copy of this gene. Supporting this, Sanger sequencing of the X-linked copy was never heterozygous in males, whereas this was not true for the autosomal importin-α2. We hereafter refer to the X-linked duplicate as X-importin-α2. The transcript is 2,106bp long in SR and 1,886bp long in ST; the 3’ UTR is longer in SR. The open reading frame of X-importin-α2 is the same length in ST and SR, but it is 16 amino acids shorter than the open reading frame of the autosomal importin-α2. This shortened end does not affect any of the protein domains (Figure S3). Visual inspection suggests the C-terminal end of X-importin-α2 is more diverged from the autosomal copy than the rest of the open reading frame (Figure S3).
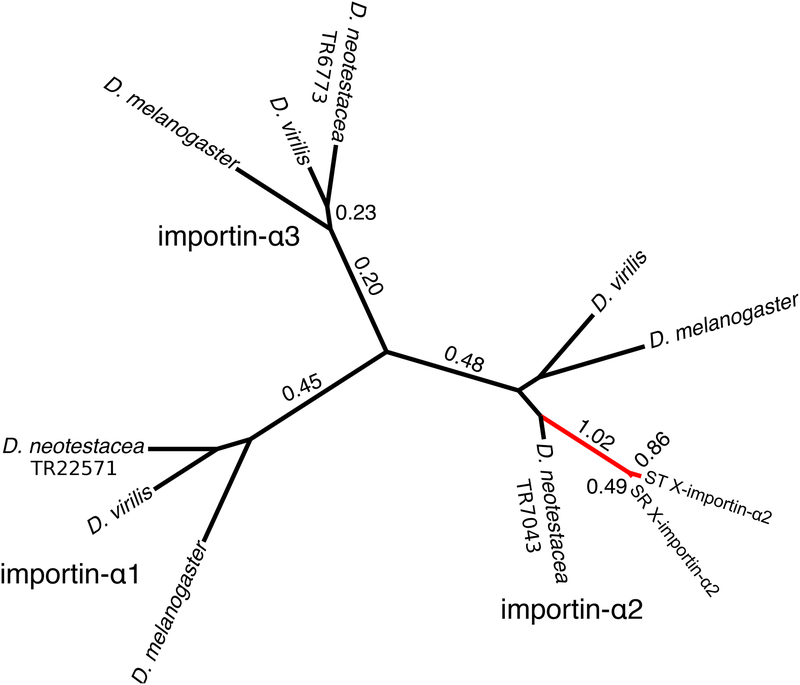
Figure 3.
X-importin-α2 is a fast-evolving X-linked duplicate of the autosomal gene importin-α2. Branch lengths were estimated with a neighbor-joining tree of 1,000 bootstraps. All nodes had bootstrap support > 99. Each of the three importin-α clades is labeled. X-importin-α2 includes TR10603, TR2814, TR37105. Estimated dN/dS values larger than 0.1 are labeled, and branches with dN/dS values > 0.60 are marked in red.
X-importin-α2 is a rapidly evolving member of the importin-α gene family
Importin-α2 is a member of the importin-α gene family along with importin-α1 and importin-α3. Full length transcripts of D. neotestacea importin-α1 (TR22571) and importin-α3 (TR6773) were identified from the transcriptome data. Neither of these transcripts were DE or had sequence differences between ST and SR, consistent with their autosomal location. A neighbor-joining unrooted phylogenetic tree was built from all members of the importin-α family in D. melanogaster, D. virilis, and D. neotestacea, including SR and ST X-importin-α2. Clearly, both X-importin-α2 sequences are most closely related to the importin-α2 copy in D. neotestacea, indicating the X-linked and autosomal copies are paralogs (Figure 3).
A likelihood ratio test supported a model where each branch of the tree has an independent dN/dS value over a model with one value across the entire phylogeny (χ2=485.5, df=18, p<0.0001). The estimated dN/dS values are uniformly quite low within the importin-α1 and importin-α3 clades, indicating strong purifying selection (Figure 3). The same is true of the importin-α2 branches for the two outgroups and the autosomal copy in D. neotestacea. However, the branch leading from the autosomal copy in D. neotestacea to the split between the X-linked copies has a dN/dS value of 1.03, and the branches leading to the individual ST and SR sequences have relatively high values of 0.86 and 0.49 respectively. Clearly, this section of the tree is evolving faster than the rest.
Sanger sequencing of X-importin-α2 in a selection of D. neotestacea, D. testacea, D. orientacea, D. putrida, and D. bizonata individuals showed that the X-linked duplication is specific to the testacea-species group. It was found in the very closely related D. neotestacea, D. testacea, and D. orientacea, but not in the more distantly related D. putrida or D. bizonata species (Dyer, White, Bray, Pique, & Betancourt, 2011). In the three testacea group species, some PCR primers designed for X-importin-α2 coincidentally captured variants from both X-importin-α2 and importin-α2. However, in D. putrida or D. bizonata these same primers only detected variants from the autosomal copy of the gene. It is possible that the X-linked copy is present in these species but too far diverged to be detectable with our primers, and more work is needed to pinpoint the origin of the duplication.
Top candidate loci have high non-synonymous variation on ST and low variation on SR
We examined the population genetic patterns of the entire open reading of X-importin-α2, the four other candidate loci described above, and five additional transcripts that had positive Ka/Ks values but did not otherwise meet the candidacy criteria (Tables 2, S3). Five of these 10 loci had no fixed differences between ST and SR in the population genetic sample (Table 2). These were thus excluded from further analyses, where the set of five “top candidates” were amended to include all sequenced loci with fixed differences between ST and SR (Table 2). The presence of loci with fixed differences between ST and SR is striking because previous work in this system found no fixed differences in 11 arbitrarily chosen X-linked loci (Pieper & Dyer, 2016). Population genetic data from five protein coding loci in this previously analyzed dataset was used as a comparison here – these loci are hereafter referred to as “non-candidates” (Pieper & Dyer, 2016)(Table S6). Additionally, it is notable that even though TR23135 had no fixed differences and is not a top candidate, it appears to be evolving non-neutrally as the ST samples have no synonymous polymorphisms but eight non-synonymous polymorphisms. Considering only the five top candidate loci that have fixed differences, it is striking that they also have a low number of shared mutations between ST and SR. This differentiation is reflected in the KST values between ST and SR of these top candidates, where the mean KST between ST and SR (0.43, sd=0.27) was significantly higher than in the set of non-candidate X-linked markers (mean=0.10, sd=0.10; t=2.74, df=6.62, p=0.03; Figure S4).
Table 2.
Population genetic summary statistics of sequenced loci. Top candidates for involvement in the driving mechanism are marked. Silent sites include noncoding and synonymous sites, and NS stands for nonsynonymous sites. Indicated is the number of samples (N); number of segregating sites (S); nucleotide diversity (π); Tajima’s D (D); number of fixed differences between ST and SR across all (total) and nonsynonymous sites; number of net nucleotide substitutions per site (Da) between ST and SR; and KST and Snn calculated between ST and SR. Values of π and Da are multiplied by 100.
| Top candidate | Transcript ID | Total sites | Silent sites | NS sites | X-type | N | S | Silent π (%) | NS π (%) | πa/πs | Silent D | NS D | Total fixed diffs | NS fixed diffs | Shared mutations | Da (%) | KST | Snn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| yes | X-importin-α2 | 1905 | 745 | 1156 | ST | 12 | 142 | 3.0 | 1.4 | 0.46 | −0.85 | −1.11 | 13 | 5 | 0 | 1.18 | 0.33 | 1.00 |
| yes | TR261 | 648 | 269 | 378 | ST | 10 | 25 | 1.0 | 1.4 | 1.39 | −0.78 | −0.47 | 2 | 2 | 8 | 0.94 | 0.34 | 0.97 |
| yes | TR11103 | 826 | 247 | 573 | ST | 9 | 15 | 1.3 | 0.3 | 0.22 | −1.28 | 0.39 | 1 | 0 | 0 | 0.26 | 0.33 | 0.95 |
| yes | TR24932 | 280 | 93 | 183 | ST | 8 | 10 | 1.3 | 1.1 | 0.83 | −1.60 | −1.48 | 5 | 4 | 0 | 2.05 | 0.67 | 1.00 |
| yes | TR37304 | 770 | 176 | 592 | ST | 10 | 4 | 0.1 | 0.1 | 1.12 | −1.11 | −1.03 | 4 | 3 | 0 | 0.60 | 0.84 | 1.00 |
| no | TR50351 | 649 | 268 | 375 | ST | 10 | 50 | 3.1 | 1.8 | 0.59 | −1.05 | −0.25 | 0 | 0 | 18 | 0.41 | 0.09 | 0.92 |
| no | TR23125 | 343 | 67 | 272 | ST | 9 | 8 | 0 | 1.0 | NA | NA | −0.26 | 0 | 0 | 2 | 0.37 | 0.19 | 0.76 |
| no | TR1778 | 477 | 116 | 355 | ST | 17 | 29 | 2.0 | 1.4 | 0.68 | −0.42 | −0.60 | 0 | 0 | 5 | 1.58 | 0.40 | 0.92 |
| no | TR5481 | 541 | 122 | 391 | ST | 12 | 31 | 1.4 | 1.3 | 0.90 | −1.91 | −1.69 | 0 | 0 | 6 | 0.51 | 0.16 | 0.86 |
| no | TR6297 | 394 | 99 | 294 | ST | 5 | 10 | 1.1 | 0.1 | 0.87 | −1.09 | −0.33 | 0 | 0 | 0 | 0.22 | 0.13 | 0.76 |
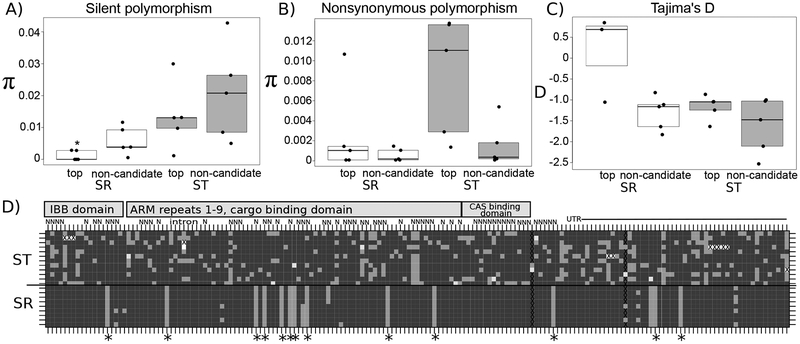
The top candidate markers, including X-importin-α2, showed a pattern characterized by very low diversity on SR, but high diversity on ST, particularly at non-synonymous sites (Figure 4; Table S6). Considering only SR chromosomes, the top candidate markers have significantly lower silent polymorphism relative to the non-candidates (U=2, n1=n2=5, p=0.034, two-tailed Mann-Whitney U-test; Figure 4a). There was no difference in non-synonymous polymorphism on SR between the top candidates and non-candidates (U=14, n1=n2=5, p=0.83, two-tailed Mann-Whitney U-test; Figure 4b). However, considering only ST chromosomes, the top candidate markers do not have reduced polymorphism relative to the non-candidates at silent sites (U=10, n1=n2=5, p=0.69, two-tailed Mann-Whitney U-test; Figure 4a), but they do have higher polymorphism at non-synonymous sites (U=22, n1=n2=5, p=0.056, two-tailed Mann-Whitney U-test; Figure 4b). Though not statistically significant, this elevated segregating non-synonymous variation in ST at X-importin-α2 and the other top candidates can be easily viewed in Figures 4a,d and Figure S5. Additionally, one top candidate had a non-synonymous variant that resulted in a premature stop codon (Figure S6).
Figure 4.
Molecular evolutionary patterns of top candidates are consistent with positive selection on SR and relaxed purifying selection on ST. A) Top candidates (top) on SR have significantly lower silent polymorphism than on ST. Silent sites include synonymous sites as well as non-coding sites. The star denotes statistical significance at p < 0.05, two-tailed Mann-Whitney U-test. B) The top candidates have higher non-synonymous polymorphism on ST than the non-candidates, though not significantly so (p = 0.056, two-tailed Mann-Whitney U-test). C) Tajima’s D is elevated in the top candidates on SR compared to the non-candidates, but not significant (p = 0.071, two-tailed Mann-Whitney U-test). In panels A through C, SR is represented by white boxes and ST by grey boxes. All data points are shown; the edges of the boxes are the first and third quartiles, and the middle line is the median. D) Haplotype structure of X-importin-α2. Each row is a chromosome, with ST phenotype males above the solid black line and SR phenotype males below. Each column is a single segregating site. Dark grey represents the individual carries the major allele, and light grey is the minor allele. Some sites have a third segregating allele, which is represented by white. Sites with a gap are marked with an X. Non-synonymous sites are denoted with an N; deletion polymorphisms are denoted with a G. Unlabeled sites are either synonymous or non-coding. Fixed differences between ST and SR are marked with a star. Sites in the intron and UTR are labeled. Sites located in specific protein domains are also labeled by grey blocks: the importin-ß binding (IBB) domain, the cargo binding domains (Armadillo [ARM] repeats 1 through 9), and the nuclear export factor (CAS) binding domain (ARM repeat 10) (Goldfarb et al., 2004).
Examining the polymorphism at X-importin-α2 also reveals the presence of two separate haplotypes at roughly equal frequency in SR (Figure 4d), whereas many singletons are observed in the ST samples. This pattern holds more generally across loci: within SR chromosomes, Tajima’s D is somewhat higher on the top candidates than non-candidate loci, though not statistically significant (U=14, n1=3, n2=5, p=0.071, two-tailed Mann-Whitney U-test; Figure 4c). However, the sample size is very small for this test because two of the top candidates lacked any segregating sites on SR and Tajima’s D could not be calculated. For the ST chromosomes Tajima’s D is similar between the candidates and non-candidates (U=16, n1 =n2=5, p=0.55, two-tailed Mann-Whitney U-test; Figure 4c). These trends also hold when only synonymous or nonsynonymous variation is considered (Figure S7).
We used HKA tests using divergence between ST and SR to ask if positive selection could explain the polymorphism differences between the top candidates and the non-candidate X-linked loci. When only considering SR samples, a maximum likelihood analysis supported a model where the five top candidate loci are under positive selection compared to the non-candidate loci (LRT, χ2=13.58, df=6, p=0.035). This result is likely due to the high divergence between ST and SR combined with the low diversity at the candidates on SR (Table S7). For ST samples, however, no model outperformed the one with all of the top candidate and non-candidate loci evolving neutrally (LRT, χ2=0.001, df=1, p=0.97; Table S7).
Discussion
Expected divergence and differential expression, with some exceptions
We identified differentially expressed transcripts between the testes of D. neotestacea males carrying a wild-type X-chromosome and those carrying the selfish sex-ratio X-chromosome. We found widespread differentiation between ST and SR, including differential expression and nucleotide sequence differences (Figures 1, 2). Nearly half of all transcripts that mapped to the X-chromosome had at least one sequence difference between ST and SR (Table S1), and some showed an elevated Ka/Ks suggestive of positive selection or relaxed purifying selection. Any two X-chromosomes from natural populations would be expected to have many sequence differences between them, and the average percent difference between ST and SR is very close to the average percent difference at five loci within a large sample of ST chromosomes (Figure 2a) (Pieper & Dyer, 2016). It is difficult to say how many transcripts have divergence that falls above a neutral expectation, but the tail of highly diverged transcripts likely indicates regions of particularly strong differentiation between SR and ST. Previous work on a smaller scale showed that differentiation between ST and SR was both widespread and variable across the chromosome, and this genome-scale investigation supports these conclusions (Pieper & Dyer, 2016).
There are a large number of DE transcripts between ST and SR, most of which are found on the X-chromosome (Figure 1). Though the presence of inversions on SR suggests the mechanism of drive may involve more than one gene, most of the 729 DE transcripts are probably not involved in the drive phenotype. Many of the DE genes in this study likely represent activation of pathways downstream of drive like apoptosis (see Table S8 for results of GO terms enrichment analysis). This is especially true for DE transcripts that map to the autosomes, as the ST and SR lines in this study have identical autosomes. In other cases, differential expression on the X-chromosome may represent gene expression levels that are diverging neutrally because of suppressed recombination with ST and their subsequent separate evolutionary histories (Harrison, Wright, & Mank, 2012). Though SR-ST nucleotide divergence is not correlated with expression differences (Figure 2a), differences may accumulate in cis-regulatory regions that are not captured in this analysis. Diverging expression due to drift is also interesting given that reduced male fertility is the only known phenotypic difference between individuals who carry SR versus ST. Similar widespread expression differences were seen in the stalk-eyed fly, though in that system there are clear pleiotropic consequences to carrying the driver (Cotton, Foldvari, Cotton, & Pomiankowski, 2014; Reinhardt et al., 2014). Similarly, it may also be the case that there has been selection for gene expression to mitigate any deleterious effects of SR in D. neotestacea. The high number of identified DE transcripts and the fact that many of them are likely not involved in drive makes the use of additional criteria (particularly elevated Ka/Ks and to a lesser degree testis-specificity) necessary for identifying candidates.
Molecular evolutionary patterns of candidate transcripts suggest involvement in drive
Previous work found that none of 11 arbitrarily chosen X-linked markers had fixed nucleotide differences between ST and SR (Pieper & Dyer, 2016). In contrast, of the 10 candidate loci we assayed for population genetic variation, five had fixed nucleotide differences between SR and ST (Table 2, Figure 4D). Population genetic patterns suggest these five “top candidate” loci are under positive selection on SR but under relaxed purifying selection on ST.
The relative location of the top candidates on the X-chromosome is unknown, but a possible reason for the low variation on SR at these loci is that they are involved in the driving mechanism or tightly linked to loci involved in drive. Involvement in drive or tight linkage to the driver is expected to result in molecular evolutionary patterns similar to a selective sweep (Derome et al., 2008; Kingan et al., 2010). Consistent with this, three of these five loci harbor little to no polymorphism on SR (Figure 4). However, the other two loci show evidence of multiple segregating haplotypes on SR; thus, any selective sweep that may have occurred at these loci was not recent. Between loci these SR haplotypes are not in phase with one another, suggesting recombination may be occurring between them (Figure S5, Figure 4D). This is consistent with previous findings of evidence for recombination in SR/SR females, which are fully fertile in this system and estimated to account for 2–3% of all females in the wild (Dyer, 2012; Pieper & Dyer, 2016; Pinzone & Dyer, 2013). The average population frequency of SR across North America is 15%; in some populations, as many as 30% of X-chromosomes are SR (Dyer, 2012). Cycling of drive haplotypes has been observed in the autosomal SD system of D. melanogaster (Brand, Larracuente, & Presgraves, 2015), and recombination in SR/SR females may eliminate deleterious mutations linked to drive loci.
In contrast to SR, ST chromosomes have a high level of segregating nonsynonymous variation at the top candidates (Figure 4B), suggestive of purifying selection failing to remove slightly deleterious variation. One sample in the top candidate TR261 contained a single base pair insertion that resulted in a premature stop codon, and another sample has an entire codon deleted in X-importin-α2 (Figure 4D, S6). If these loci are solely functioning as drivers, there may be no selection to maintain them on the ST X-chromosome and they are thus free to accumulate mutations. There are also numerous fixed nonsynonymous differences between ST and SR in the top candidate loci, further suggesting that these sequences may have divergent functions or phenotypic effects in SR vs ST (Table 2, Figure 4D, S5).
X-importin-α2 is a rapidly evolving X-linked duplicate
We argue that X-importin-α2 is a strong candidate for involvement in meiotic drive. X-importin-α2 is overexpressed in sex-ratio males, testes-specific, and highly differentiated between ST and SR (Table S4, Figure 4D, S5). Furthermore, it is evolving differently from the rest of the importin-α gene family (Figure 3). While most of the tree appears to be under strong purifying selection, the dN/dS value of 1.03 estimated for the X-importin-α2 branch is comparatively extremely high, suggesting selection for functional changes between the two copies of the gene. Phylogenetic analyses place the origin of the duplication between the split between the lineage leading to D. neotestacea, D. orientacea, and D. testeacea and that leading to D. putrida (Dyer et al., 2011). We note that sex-ratio meiotic drive is also present in D. orientacea (K. Dyer, unpublished data) and D. testacea (Keais, Hanson, Gowen, & Perlman, 2017); thus, the same three closely-related species that have sex-ratio drive in the testacea group also have X-importin-α2. The duplication event resulted from a retrotransposition event involving a poorly spliced transcript: in D. melanogaster there are three small introns in importin-α2, but only one of these in included in X-importin-α2 in D. neotestacea (Figure S8).
Duplication events have been implicated in the origin of multiple other meiotic drive systems. For instance, this includes the distorter locus of the SD system of D. melanogaster (Larracuente & Presgraves, 2012), both the distorter and an autosomal suppressor in the Winters sex-ratio system of D. simulans (Tao, Araripe, et al., 2007; Tao, Masly, et al., 2007), and a segmental duplication spanning six genes that includes the distorter in the Paris sex-ratio system of D. simulans (Fouvry, Ogereau, Berger, Gavory, & Montchamp-Moreau, 2011). Gene duplication is a potent force for genetic innovation as it allows new genes to evolve new functions while still maintaining the original function of the gene (Lynch & Walsh, 2007).
X-importin-α2 suggests nuclear transport may be a target of drive
We argue that while its molecular evolutionary patterns and status as a rapidly evolving duplicate make X-importin-α2 a good candidate for the mechanism of drive, the function of its parent gene, importin-α2, makes it an excellent candidate. All importin-α proteins play a key role in the nuclear transport pathway (Goldfarb et al., 2004; Matsuura & Stewart, 2005; Stewart, 2007; Sun, Fu, Ciziene, Stewart, & Musser, 2013). Nuclear transport has been previously implicated in genetic conflict in the male germline, most prominently in the autosomal SD meiotic driver of D. melanogaster (Larracuente & Presgraves, 2012). During spermatogenesis, the haploid nuclei of sperm share a cytoplasm until the individualization stage, when half of the sperm in SD males die (De Cuevas, Lilly, & Spradling, 1997; Fuller, 1993; Larracuente & Presgraves, 2012). The distorter locus of SD (sd-RanGAP) is a truncated, duplicated copy of RanGAP that mislocalizes to the nucleus instead of the cytoplasm (Ayumi Kusano et al., 2001; Larracuente & Presgraves, 2012). Though the exact mechanism is unknown, sd-RanGAP is enzymatically active and causes selective failure of sperm through disruption of the GTP-gradient required for nuclear transport (A. Kusano et al., 2002; Larracuente & Presgraves, 2012).
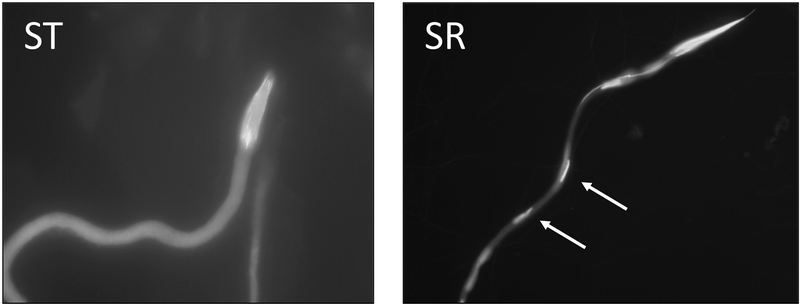
Of the three canonical importin-α genes in D. melanogaster, importin-α2 is primarily expressed in the testes and plays a critical role in spermatogenesis (Mason, Fleming, & Goldfarb, 2002). Male homozygous null importin-α2 flies are sterile and the sperm fail at the individualization checkpoint (Giarrè et al., 2002; Mason et al., 2002). In D. neotestacea, the autosomal copy of importin-α2 is under purifying selection and is expressed at the same high level in the testes of both ST and SR males, and thus likely retains the ancestral function as observed in D. melanogaster (Figure 3). The divergence of X-importin-α2 from the parent copy suggests that it make have taken on a new function. In D. neotestacea, microscopy visualizing the late stages of spermatogenesis shows that in SR males, a portion of spermatids begin to fail during the elongation phase prior to individualization (Figure 5), though a GO terms enrichment analysis of SR-biased, testis-specific transcripts found the biological process “sperm individualization” was significantly enriched (Table S8). More work is necessary to fully describe the cellular spermatogenesis phenotype of SR in D. neotestacea, but the evidence we have currently suggests the involvement of X-importin-α2 is highly plausible. We also examined several known interacting partners of importin-α2 (e.g., importin-ß, CAS, Ran, RanGTP, RanGEF, Nup153, Nup50) in our dataset, but we found no indication of DE between ST and SR, elevated Ka/Ks, or duplication (data not shown).
Figure 5.
Developing bundle of 64 spermatids in the testes of ST males (left) and SR males (right) at 650× magnification. The DNA is stained with DAPI, revealing the heads of the spermatids. In SR, roughly half of the sperm do not develop properly, which are presumably Y-bearing spermatids. Arrows point out the heads of sperm that are not maturing properly.
Duplication of an importin-α gene is not unique to the testacea group. At least three independent duplication events of importin-α2 and α3 have been identified in other Drosophila lineages, and like X-importin-α2, these duplicates are expressed primarily in the testes and at least one of them has signatures of positive selection in certain lineages (Phadnis, Hsieh, & Malik, 2011). Genes primarily expressed during spermatogenesis tend to be fast-evolving, and new duplicate genes often have testis-specific expression patterns (Betrán, Thornton, & Long, 2002; Emerson, Kaessmann, Betrán, & Long, 2004; Haerty et al., 2007). This is often attributed to sexual selection, but another hypothesis may be that spermatogenesis is frequently the site of genetic conflict over a fair meiosis (Haerty et al., 2007; Kleene, 2005). Phadnis et al. (2011) suggest that the repeated duplication and evolution of new importin-α family members is due to evolutionary pressure to maintain wild-type nuclear import against selfish genetic elements like SD. It may be the case that the ST copy of X-importin-α2 was once part of this defense system, but is no longer functional, resulting in the signatures of relaxed purifying selection we observed. If X-importin-α2 is involved in drive in D. neotestacea, this defense system may have been subverted and coopted. More work is needed to untangle the evolutionary relationship between the ST and SR copies of X-importin-α2 and the autosomal copy of importin-α2, including a larger and more complete gene tree that includes sequences from the autosomal paralog as well as both X-linked SR and ST alleles from D. orientacea and D. testacea (Keais et al., 2017).
In summary, we find a strong pattern of widespread expression and molecular evolutionary differences between SR and ST of D. neotestacea, supporting the results of previous work with a smaller set of loci (Pieper & Dyer, 2016). We also identified a set of candidates for involvement in the mechanism of drive based on their expression and unique molecular evolutionary patterns. Particularly notable is the X-linked, fast-evolving duplicate of importin-α2 called X-importin-α2 that is overexpressed in SR males. This appealing candidate is worthy of further investigation, including comparisons of expression in different SR lines and eventually knockdown experiments to functionally confirm its involvement in SR. If X-importin-α2 is involved in drive, it would be a remarkable example of convergent evolution with SD in D. melanogaster of manipulation of the nuclear transport pathway (Larracuente & Presgraves, 2012). The rapid sequence evolution of RanGAP and other associated genes in D. melanogaster suggests a long history of genetic conflict over nuclear transport (Presgraves, 2007). Further experiments with X-importin-α2 could provide evidence that certain pathways and processes are particularly susceptible to genetic conflict.
Supplementary Material
Acknowledgements:
This project was funded by a Rosemary Grant Award from the Society for the Study of Evolution, a Presidential Fellowship from the University of Georgia, and a National Institutes of Health (NIH) award under T32GM007103 to KEP; NIH award R00GM114714 to RLU; and National Science Foundation grants 1149350 and 1457707 and funds from the University of Georgia Research Foundation to KAD. We thank Yasir Ahmed-Braimah for providing us with the D. virilis genome assembly, Brooke White and Ariane Wong for laboratory assistance, and three anonymous reviewers and members of the Dyer, Hall, Bergman, and White labs at UGA for feedback on the manuscript.
Footnotes
Data Accessibility: The data presented in this paper can be accessed on Dryad with the accession number doi:10.5061/dryad.9b21271.
References
- Anders S, & Huber W (2012). Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL). [Google Scholar]
- Bauer H, Veron N, Willert J, & Herrmann BG (2007). The t-complex-encoded guanine nucleotide exchange factor Fgd2 reveals that two opposing signaling pathways promote transmission ratio distortion in the mouse. Genes & Development, 21(2), 143–147. doi: 10.1101/gad.414807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Willert JR, Koschorz B, & Herrmann BG (2005). The t complex-encoded GTPase-activating protein Tagap1 acts as a transmission ratio distorter in mice. Nature Genetics, 37(9), 969–973. doi: 10.1038/ng1617 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological), 289–300. [Google Scholar]
- Betrán E, Thornton K, & Long M (2002). Retroposed new genes out of the X in Drosophila. Genome Res, 12(12), 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Larracuente AM, & Presgraves DC (2015). Origin, evolution, and population genetics of the selfish Segregation Distorter gene duplication in European and African populations of Drosophila melanogaster. Evolution; international journal of organic evolution, 69(5), 1271–1283. doi: 10.1111/evo.12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, & Trivers R (2006). Genes in conflict: The biology of selfish genetic elements. Cambridge, Mass.: Belknap Press of Harvard University Press. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, & Madden TL (2009). BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, & Vaz SC (1999). Are Drosophila SR drive chromosomes always balanced? Heredity, 83, 221–228. doi: 10.1038/sj.hdy.6886100 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, & Hartl DL (1978). Population-dynamics of Segretation Distorter polymorphism of Drosophila melanogaster. Genetics, 89(1), 171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson SJ, Brand CL, & Wilkinson GS (2011). Reduced polymorphism associated with X chromosome meiotic drive in the stalk-eyed fly Teleopsis dalmanni. PLoS ONE, 6(11), 1–7. doi: 10.1371/journal.pone.0027254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AJ, Foldvari M, Cotton S, & Pomiankowski A (2014). Male eyespan size is associated with meiotic drive in wild stalk-eyed flies (Teleopsis dalmanni). Heredity, 112(4), 363–369. doi: 10.1038/hdy.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, … Durbin R (2011). The variant call format and VCFtools. Bioinformatics, 27(15), 2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cuevas M, Lilly M, & Spradling A (1997). Germline cyst formation in Drosophila. Annual review of genetics, 31(1), 405–428. [DOI] [PubMed] [Google Scholar]
- Derome N, Baudry E, Ogereau D, Veuille M, & Montchamp-Moreau C (2008). Selective sweeps in a 2-locus model for sex-ratio meiotic drive in Drosophila simulans. Mol. Biol. Evol, 25(2), 409–416. doi: 10.1093/molbev/msm269 [DOI] [PubMed] [Google Scholar]
- Dyer KA (2012). Local selection underlies the geographic distribution of sex-ratio drive in Drosophila neotestacea. Evolution; international journal of organic evolution, 66(4), 973–984. doi: 10.1111/j.1558-5646.2011.01497.x [DOI] [PubMed] [Google Scholar]
- Dyer KA, Bray MJ, & Lopez SJ (2013). Genomic conflict drives patterns of X-linked population structure in Drosophila neotestacea. Mol. Ecol, 22(1), 157–169. doi: 10.1111/mec.12097 [DOI] [PubMed] [Google Scholar]
- Dyer KA, White BE, Bray MJ, Pique DG, & Betancourt AJ (2011). Molecular evolution of a Y chromosome to autosome gene duplication in Drosophila. Mol. Biol. Evol, 28(3), 1293–1306. doi: 10.1093/molbev/msq334 [DOI] [PubMed] [Google Scholar]
- Emerson J, Kaessmann H, Betrán E, & Long M (2004). Extensive gene traffic on the mammalian X chromosome. Science, 303(5657), 537–540. [DOI] [PubMed] [Google Scholar]
- Fouvry L, Ogereau D, Berger A, Gavory F, & Montchamp-Moreau C (2011). Sequence analysis of the segmental duplication responsible for Paris sex-ratio drive in Drosophila simulans. G3: Genes, Genomes, Genetics, 1(5), 401–410. doi: 10.1534/g3.111.000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M (1993). Spermatogenesis. Plainview, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Giarrè M, Török I, Schmitt R, Gorjánácz M, Kiss I, & Mechler BM (2002). Patterns of importin-α expression during Drosophila spermatogenesis. Journal of structural biology, 140(1–3), 279–290. [DOI] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, & Adam SA (2004). Importin α: a multipurpose nuclear-transport receptor. Trends in cell biology, 14(9), 505–514. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, … Regev A (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc, 8(8), 1494–1512. doi: 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W, Jagadeeshan S, Kulathinal RJ, Wong A, Ram KR, Sirot LK, … Civetta A (2007). Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics, 177(3), 1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD (1967). Extraordinary sex ratios. Science, 156(3774), 477–488. [DOI] [PubMed] [Google Scholar]
- Harrison PW, Wright AE, & Mank JE (2012). The evolution of gene expression and the transcriptome–phenotype relationship. Paper presented at the Seminars in cell & developmental biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleu Q, Gérard PR, Dubruille R, Ogereau D, Prud’homme B, Loppin B, & Montchamp-Moreau C (2016). Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proceedings of the National Academy of Sciences, 113(15), 4110–4115. doi: 10.1073/pnas.1519332113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann BG, Koschorz B, Wertz K, McLaughlin KJ, & Kispert A (1999). A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature, 402(6758), 141–146. doi: 10.1038/45970 [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, … Svirskas R (2015). The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res, 25(3), 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, & Pomiankowski A (1991). Causes of sex-ratio bias may account for unisexual sterility in hybrids - a new explanation of Haldane’s rule and related phenomena. Genetics, 128(4), 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J (2001). Sex chromosome meiotic drive. Annual Review of Ecology and Systematics, 32, 25–49. doi: 10.1146/annurev.ecolsys.32.081501.113958 [DOI] [Google Scholar]
- James AC, & Jaenike J (1990). Sex-ratio meiotic drive in Drosophila testacea. Genetics, 126(3), 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keais GL, Hanson MA, Gowen BE, & Perlman SJ (2017). X-chromosome drive in a widespread Palearctic woodland fly, Drosophila testacea. Journal of Evolutionary Biology, 30(6), 1185–1194. doi: 10.1111/jeb.13089 [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, … Drummond A (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingan SB, Garrigan D, & Hartl DL (2010). Recurrent selection on the Winters sex-ratio genes in Drosophila simulans. Genetics, 184(1), 253–265. doi: 10.1534/genetics.109.109587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene KC (2005). Sexual selection, genetic conflict, selfish genes, and the atypical patterns of gene expression in spermatogenic cells. Developmental biology, 277(1), 16–26. [DOI] [PubMed] [Google Scholar]
- Kusano A, Staber C, & Ganetzky B (2001). Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Developmental cell, 1(3), 351–361. [DOI] [PubMed] [Google Scholar]
- Kusano A, Staber C, & Ganetzky B (2002). Segregation distortion induced by wild-type RanGAP in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 99(10), 6866–6870. doi: 10.1073/pnas.102165099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Meth, 9(4), 357–359. doi:10.1038/nmeth.1923 10.1038/nmeth.1923http://www.nature.com/nmeth/journal/v9/n4/abs/nmeth.1923.html#supplementary-informationhttp://www.nature.com/nmeth/journal/v9/n4/abs/nmeth.1923.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, & Presgraves DC (2012). The selfish Segregation Distorter gene complex of Drosophila melanogaster. Genetics, 192(1), 33–53. doi: 10.1534/genetics.112.141390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, & Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, … Durbin R (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, & Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11), 1451–1452. doi:Doi 10.1093/Bioinformatics/Btp187 [DOI] [PubMed] [Google Scholar]
- Lin CJ, Hu F, Dubruille R, Vedanayagam J, Wen J, Smibert P, … Lai EC (2018). The hpRNA/RNAi Pathway Is Essential to Resolve Intragenomic Conflict in the Drosophila Male Germline. Dev Cell, 46(3), 316–326.e315. doi: 10.1016/j.devcel.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L, … Price TAR (2016). The Ecology and Evolutionary Dynamics of Meiotic Drive. Trends Ecol Evol, 31(4), 315–326. doi: 10.1016/j.tree.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology, 15(12), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, & Walsh B (2007). The origins of genome architecture (Vol. 98): Sinauer Associates Sunderland (MA). [Google Scholar]
- Lyon MF (1991). The genetic basis of transmission-ratio distortion and male sterility due to the t complex. American Naturalist, 349–358. [Google Scholar]
- Lyon MF (2003). Transmission ratio distortion in mice. Annual review of genetics, 37, 393–408. doi: 10.1146/annurev.genet.37.110801.143030 [DOI] [PubMed] [Google Scholar]
- Mason DA, Fleming RJ, & Goldfarb DS (2002). Drosophila melanogaster importin α1 and α3 can replace importin α2 during spermatogenesis but not oogenesis. Genetics, 161(1), 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, & Stewart M (2005). Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. The EMBO journal, 24(21), 3681–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill C, Bayraktaroglu L, Kusano A, & Ganetzky B (1999). Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science, 283(5408), 1742–1745. doi: 10.1126/science.283.5408.1742 [DOI] [PubMed] [Google Scholar]
- Paczolt KA, Reinhardt JA, & Wilkinson GS (2017). Contrasting patterns of X-chromosome divergence underlie multiple sex-ratio polymorphisms in stalk-eyed flies. Journal of Evolutionary Biology, 30(9), 1772. doi: 10.1111/jeb.13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman SJ, Spicer GS, Shoemaker DD, & Jaenike J (2003). Associations between mycophagous Drosophila and their Howardula nematode parasites: a worldwide phylogenetic shuffle. Mol Ecol, 12(1), 237–249. doi: 10.1046/j.1365-294X.2003.01721.x [DOI] [PubMed] [Google Scholar]
- Phadnis N, Hsieh E, & Malik HS (2011). Birth, death, and replacement of karyopherins in Drosophila. Molecular Biology and Evolution, 29(5), 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper KE, & Dyer KA (2016). Occasional recombination of a selfish X-chromosome may permit its persistence at high frequencies in the wild. Journal of Evolutionary Biology, 29(11), 2229–2241. doi: 10.1111/jeb.12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzone CA, & Dyer KA (2013). Association of polyandry and sex-ratio drive prevalence in natural populations of Drosophila neotestacea. Proc. R. Soc. B Biol. Sci, 280(1769), 20131397. doi: 10.1098/rspb.2013.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC (2007). Does genetic conflict drive rapid molecular evolution of nuclear transport genes in Drosophila? Bioessays, 29(4), 386–391. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, & Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–842. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RCoreTeam. (2014). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; Retrieved from http://www.R-project.org [Google Scholar]
- Reinhardt JA, Brand CL, Paczolt KA, Johns PM, Baker RH, & Wilkinson GS (2014). Meiotic drive impacts expression and evolution of X-linked genes in Stalk-Eyed flies. Plos Genetics, 10(5), 14. doi: 10.1371/journal.pgen.1004362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR (2013). Nothing in genetics makes sense except in light of genomic conflict. Annual Review of Ecology and Systematics, 44(1), 217–237. doi:doi: 10.1146/annurev-ecolsys-110411-160242 [DOI] [Google Scholar]
- Risso D, Ngai J, Speed TP, & Dudoit S (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nature biotechnology, 32(9), 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, & Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L, & Novitski E (1957). Meiotic drive as an evolutionary force. American Naturalist, 91(857), 105–110. doi: 10.1086/281969 [DOI] [Google Scholar]
- Schaeffer SW, Bhutkar AU, McAllister BF, Matsuda M, Matzkin LM, O’Grady PM, … Kaufman TC (2008). Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics, 179(3), 1601–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M (2007). Molecular mechanism of the nuclear protein import cycle. Nature reviews Molecular cell biology, 8(3), 195. [DOI] [PubMed] [Google Scholar]
- Sun C, Fu G, Ciziene D, Stewart M, & Musser SM (2013). Choreography of importin-α/CAS complex assembly and disassembly at nuclear pores. Proceedings of the National Academy of Sciences, 110(17), E1584–E1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, & Hartl DL (2007). A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. Plos Biology, 5(11), e293. doi: 10.1371/journal.pbio.0050293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Masly JP, Araripe L, Ke Y, & Hartl DL (2007). A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. Plos Biology, 5(11), e292. doi: 10.1371/journal.pbio.0050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. (2014). UniProt: a hub for protein information. Nucleic Acids Research, 43(D1), D204–D212. doi: 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, & Rozen SG (2012). Primer3—new capabilities and interfaces. Nucleic Acids Research, 40(15), e115–e115. doi: 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, … DePristo MA (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics, 11(1110), 11 10 11–11 10 33. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, & Charlesworth B (2004). The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics, 168(2), 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (2007). PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24(8), 1586–1591. doi: 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, & Yu J (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics, 4(4), 259–263. doi: 10.1016/s1672-0229(07)60007-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.