Abstract
Background
The aim of the current study was to investigate the prevalence and clinicopathologic characteristics of ROS1‐rearranged non‐small cell lung cancer (NSCLC) in routine genotypic screening in conjunction with the study of PD‐L1 expression, a biomarker for first‐line treatment decisions.
Methods
Reflex simultaneous genotypic screening for EGFR by peptide nucleic acid clamping, and ALK and ROS1 by fluorescence in situ hybridization (FISH) was performed on consecutive NSCLC cases at the time of initial pathologic diagnosis. We evaluated genetic aberrations, clinicopathologic characteristics, and PD‐L1 tumor proportion score (TPS) using a PD‐L1 22C3 assay kit.
Results
In 407 consecutive NSCLC patients, simultaneous genotyping identified 14 (3.4%) ROS1 and 19 (4.7%) ALK rearrangements, as well as 106 (26%) EGFR mutations. These mutations were mutually exclusive and were found in patients with similar clinical features, including younger age, a prevalence in women, adenocarcinoma, and advanced stage. The PD‐L1 assay was performed on 130 consecutive NSCLC samples. High PD‐L1 expression (TPS ≥ 50%) was observed in 29 (22.3%) tumors. PD‐L1 expression (TPS ≥ 1%) was significantly associated with wild type EGFR, while ROS1 rearrangement was associated with high PD‐L1 expression. Of the 14 cases with ROS1 rearrangement, 12 (85.7%) showed PD‐L1 expression and 5 (35.7%) showed high PD‐L1 expression.
Conclusion
In the largest consecutive routine Asian NSCLC cohort analyzed to date, we found that high PD‐L1 expression frequently overlapped with ROS1 rearrangement, while it negatively correlated with EGFR mutations.
Keywords: Biomarker, epidermal growth factor receptor, human ROS1 protein, non‐small cell lung carcinoma, programmed death 1 ligand 1
Introduction
Non‐small cell lung cancer (NSCLC) is one of the leading causes of cancer death worldwide.1 Recent advances in the molecular characterization of NSCLC, together with personalized therapies against NSCLC molecular targets, have improved patient prognosis. Testing for activating mutations and rearrangements in the kinase domain of EGFR, ALK, and ROS1 is now recommended as part of the routine evaluation of patients with advanced lung adenocarcinoma.2, 3 Recently, immune checkpoint inhibitors against PD‐1 have shown remarkable therapeutic activity in NSCLC:4, 5, 6 for example, pembrolizumab has been approved by the United States Food and Drug Administration (USFDA) for first‐line systemic therapy in advanced NSCLC with a PD‐L1 tumor proportion score (TPS) of > 50% by PD‐L1 22C3 assay.7 However, testing of both genotype changes and PD‐L1 biomarker levels in routine practice has raised questions about first‐line treatment options when it comes to a PD‐L1 TPS > 50% combined with targetable oncogenic driver mutations.3 Notably, accumulating evidence has revealed a relationship between PD‐L1 expression and the presence of driver mutations in EGFR, ALK, and KRAS. 8, 9
ROS1 is a transmembrane tyrosine kinase receptor that shares a significant amino acid homology to the kinase domain of ALK.10 Previous studies have reported that chromosomal rearrangements involving ROS1 occur in 1–2% of NSCLC and are clinically associated with a history of never‐smoking, younger age, and adenocarcinoma histologic type.11, 12 There has been debate over the prevalence of ROS1 abnormality and its mutual exclusiveness to other mutations. Previous studies using fluorescence in situ hybridization (FISH), next‐generation sequencing, or quantitative PCR (qPCR) have reported that the rearrangements in ROS1 are generally distinct from and mutually exclusive to EGFR and ALK mutational status.13, 14, 15 In contrast, other studies using immunohistochemistry (IHC) screening alone, or with FISH, have reported that ROS1‐expressing NSCLCs frequently harbor concomitant oncogenic driver mutations.16, 17 The low prevalence of ROS1 aberrations and the diversity in the methods used to detect them are the main reasons for the difficulties in defining the clinicopathologic characteristics of ROS1‐aberrant NSCLCs. For example, recent studies assessing PD‐L1 expression in ROS1‐aberrant cases included no more than three cases and showed no statistical association.3, 18 PD‐L1 expression shows limited immunoreactivity in older tissue samples19 and changes after treatment.20, 21 The assessment of PD‐L1 expression in routine clinical practice at the time of the initial pathologic diagnosis, before any antitumor treatment, is the best way to evaluate PD‐L1 expression in ROS1‐rearranged NSCLCs, especially during first line treatment.
Therefore, we aimed to examine PD‐L1 expression using a USFDA‐approved companion diagnostic, the 22C3 pharmDx kit (Agilent/Dako, Santa Clara, CA, USA), in a ROS1‐rearranged population, by simultaneously genotyping EGFR, ALK, and ROS1 in unselected consecutive patients with NSCLC.
Methods
Patients
From 2011, the certified Molecular Pathology Center of the Catholic University of Korea Yeouido St. Mary Hospital, South Korea, has been conducting comprehensive genotyping screening for targeted therapy of patients with NSCLC from 36 hospitals in South Korea. From January 2014, ROS1 FISH has been integrated into the screening platform: a total of 1110 ROS1 FISH tests have been performed on samples from multiple institutions and 35 (3.2%) cases with ROS1 rearrangement were found up to December 2017. A total of 407 patients with NSCLC at the Yeouido St. Mary Hospital who consecutively underwent non‐sequential, simultaneous genotyping for EGFR, ALK, and ROS1 using tissue samples from the initial pathologic diagnosis or from the immediate subsequent surgery (no previous antitumor treatment) were selected for this study. The patients’ clinical data were retrieved from the electronic medical record system of the Yeouido St. Mary Hospital. NSCLC tumor pathology was classified according to World Health Organization criteria.22 A never‐smoker was defined as a patient who had never smoked or who had smoked < 100 cigarettes in their lifetime; pack‐years of smoking were calculated as packs‐day multiplied by the years of smoking. Processing and data analysis were performed on pseudonymized data sets. This study was conducted in accordance with the relevant legislation and approved by the institutional review board of Yeouido St. Mary's Hospital (SC18RNSI0037) and complied with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all participants included in the study.
Simultaneous diagnostic platform for genotyping and immunohistochemistry
We adhered to guidelines previously described for simultaneous genotypic testing and IHC.23 The sample with the largest tumor proportion and least stroma in each patient was selected for analysis.24 Additionally, the sample needed to have >100 tumor cells for the PD‐L1 test25 and > 50 tumors cells for ALK and ROS1 FISH tests.11, 26 In a tissue‐sparing manner, we also investigated an IHC panel including thyroid transcription factor 1 (TTF‐1), p63, p40, and cytokeratin 7; mucin staining using Periodic acid‐Schiff stain and Alcian blue for histological confirmation; ALK IHC to confirm the FISH results; and PD‐L1 IHC (using the PD‐L1 IHC 22C3 pharmDx, Agilent/Dako) for therapeutic purposes.23, 27 PD‐L1 IHC was performed on 4 μm thick formalin‐fixed paraffin‐embedded (FFPE) tissue sections using the PD‐L1 clone 22C3 pharmDx kit and Dako Automated Link 48 platform (Agilent/Dako). Immunohistochemical staining for TTF‐1 (clone SP141, Ventana Medical Systems, Tucson, AZ, USA), p63 (clone 4A4), p40 (polyclonal anti p40; Diagnostic BioSystems, Pleasanton, CA, USA), cytokeratin 7 (clone SP52, Ventana), and ALK (clone 5A4, dilution 1:50, Leica Biosystems, Newcastle, UK) was performed using the Benchmark Ultra Autostainer (Roche Tissue Diagnostics, Tucson, AZ, USA) with an OptiView Universal DAB detection kit for ALK IHC and UltraView universal DAB detection kit for the other proteins (Ventana Medical Systems).
Detection of EGFR mutations
DNA was isolated from three 5 μm thick FFPE tissue sections using the High Pure PCR Template Preparation Kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions.23 The extracted DNA was stored at −20 °C until analysis. The PNAClamp EGFR Mutation Detection Kit (PANAGENE Inc., Daejeon, Korea) was used to detect mutations by qPCR, as previously described.28 Briefly, qPCR was performed in total volume of 20 μL that included template DNA, a primer and peptide nucleic acid (PNA) probe set, and a SYBR Green PCR master mix. The control qPCR lacked the PNA probe and contained the wild‐type (WT) template. The qPCR was performed using a CFX96 PCR detection system (Bio‐Rad, Philadelphia, PA, USA). PCR cycling conditions were as follows: five minutes at 94°C and 40 cycles of 94°C for 30 seconds, 70°C for 20 seconds, 63°C for 30 seconds, and 72°C for 30 seconds. We used the ΔCt method to quantify the amplification: for this purpose, we used threshold cycle (Ct) values, which were automatically calculated from the PCR amplification plots of fluorescence against the number of cycles. ΔCt values were calculated as the Ct value of the standard minus the Ct value of the sample. Higher ΔCt values indicate more efficient amplification of the mutant. A cutoff value of 2.0 was used to indicate the presence of mutant DNA.
ALK and ROS1 rearrangements
ALK and ROS1 rearrangements were detected using FISH without prior screening by IHC. FISH is the preferred method used to investigate ALK and ROS1 mutations and is a prerequisite for crizotinib treatment.29, 30 Additionally, FISH allows the co‐detection of ALK and ROS1 rearrangements as opposed to sequential staining in IHC. Briefly, FISH was performed on unstained 4 mm thick FFPE tumor samples using the Vysis ALK FISH Break Apart kit and Vysis ROS1 FISH Break Apart kit (Abbott Molecular, Abbot Park, IL, USA), following the manufacturer's instructions.31 FISH images were captured using the automated BioView Duet scanning system (BioView, Rehovot, Israel) and scored by either one or two experienced pathologists. The procedure consisted of the following steps: (i) proper tumor tissue sections were selected for automated imaging and analysis using a 10× objective to locate the nuclei; (ii) the system automatically captured and analyzed the nuclei found in those regions using a 60× oil immersion objective and a single band 4′,6‐diamidino‐2‐phenylindole (DAPI)/Spectrum Green/Spectrum Orange filter; and (iii) the target nuclei were selected and scored by a pathologist using a specific algorithm for positive or negative signal patterns based upon the classifications described in the Vysis ALK FISH Break Apart kit instructions.
A minimum of 50 tumor cell nuclei was counted according to the manufacturer's instructions. ALK and ROS1 FISH‐positive cases were defined as those with > 25 nuclei (50%) with break‐apart (BA) signals or isolated red signals (IRS) for ALK and isolated green signals (IGS) for ROS1. ALK and ROS1 FISH‐negative samples were defined as those with < 5 nuclei (< 10%) with BA signals or IRS (for ALK) and IGS (for ROS1). ALK and ROS1 FISH cases were considered borderline if 5–25 cells (10–50%) were positive. In the case of borderline results, a second reader evaluated the slide: 50 additional tumor cell nuclei were counted, and a percentage was calculated from a total of 100 cells. If < 15% of the cells were positive, the sample was considered negative. If ≥ 15% of the cells were positive, the sample was considered positive.26
PD‐L1 tumor proportion score analysis
An experienced pathologist interpreted the expression of PD‐L1. PD‐L1 protein expression was determined by using the TPS, which is the percentage of viable tumor cells showing partial or complete membranous staining at any intensity. The specimen was considered to be negative or positive for PD‐L1 expression at TPS < 1% and TPS > 1%, respectively. PD‐L1 expression was subclassified as low PD‐L1 expression with TPS ≥ 1% and TPS < 50% and high PD‐L1 expression with TPS ≥ 50%.
Statistical analysis
Statistically significant differences between groups were analyzed using the Fisher's exact test. Data are expressed as means ± standard deviations. A two‐tailed P value < 0.05 was considered statistically significant. All statistical analyses were performed using R software, version 3.4.1 (R Foundation, Vienna, Austria).
Results
Baseline clinical and pathologic characteristics of patients
Among the 407 patients with NSCLC, 238 (58.6%) were male, and the mean patient age was 66.9 years (Table 1). The predominant histologic type was adenocarcinoma (306 cases, 75.4%), and 183 (46.4%) patients were never‐smokers. A high percentage of the study population (56.6%) had advanced stage cancer (stages IIIB and IV). Stage information was not available for 12 patients. Most of the study specimens (96.1%) were obtained by non‐surgical biopsy (bronchoscopic biopsy, 10.3%; percutaneous needle aspiration biopsy, 73.4%; metastatic site biopsy, 8.7%; cell block from pleural effusion, 1.2%; and endobronchial ultrasound‐guided transbronchial needle aspiration, 2.5%); the remainder was obtained by surgical resection (lobectomy, 3.2%; wedge resection, 0.7%).
Table 1.
Demographics of EGFR, ALK, and ROS1 status in 407 NSCLC consecutive patients
| EGFR (n = 106) | ALK (n = 19) | ROS1 (n = 14) | |||||
|---|---|---|---|---|---|---|---|
| Variables | Total | N (%) | P † | N (%) | P † | N (%) | P † |
| Age, mean ± SD | 66.9 ± 12.07 | 64.8 ± 12.0 | 0.033 ‡ | 55.7 ± 15.7 | < 0.001 † | 61.2 ± 15.5 | < 0.001 † |
| Gender, N (%) | |||||||
| Male | 238 (58.6) | 29 (27.4) | < 0.001 | 7 (36.8) | 0.058 | 4 (28.6) | 0.026 |
| Female | 168 (41.4) | 77 (72.6) | 12 (63.2) | 10 (71.4) | |||
| Smoking history | |||||||
| Never smoker | 183 (46.4) | 76 (73.8) | < 0.001 | 14 (73.7) | 0.018 | 10 (71.4) | 0.098 |
| Ever smoker | 211 (53.6) | 27 (26.2) | 5 (26.3) | 4 (28.6) | |||
| Packyears | 19.5 ± 23.6 | 5.5 ± 11.6 | < 0.001 ‡ | 6.9 ± 14.8 | 0.001 ‡ | 12.1 ± 24.3 | 0.237‡ |
| Histology | |||||||
| ADC | 306 (75.4) | 101 (95.3) | < 0.001 § | 17 (89.5) | 0.179 § | 14 (100) | 0.026 § |
| SCC | 74 (18.2) | 4 (3.8) | 1 (5.3) | 0 (0.0) | |||
| ADSQ | 9 (2.2) | 1 (0.9) | 0 (0.0) | 0 (0.0) | |||
| Other NSCLCs | 17 (4.2) | 0 (0.0) | 1 (5.3) | 0 (0.0) | |||
| Stage | |||||||
| IA | 60 (15.2) | 20 (19.6) | < 0.001 ¶ | 2 (10.5) | 0.006 ¶ | 1 (7.1) | 0.001 ¶ |
| IB | 37 (9.4) | 10 (9.8) | 0 (0.0) | 1 (7.1) | |||
| IIA | 18 (4.6) | 7 (6.9) | 1 (5.3) | 0 (0.0) | |||
| IIB | 18 (4.6) | 2 (2.0) | 1 (5.3) | 1 (7.1) | |||
| IIIA | 38 (9.6) | 7 (6.9) | 2 (10.5) | 0 (0.0) | |||
| IIIB | 32 (8.1) | 6 (5.9) | 0 (0.) | 0 (0.0) | |||
| IV | 191 (48.5) | 50 (49.0) | 13 (68.4) | 11 (78.6) | |||
| Type of sample | |||||||
| Biopsy | 0.258†† | 1.000†† | 1.000†† | ||||
| Bronchoscopy | 42 (10.3) | 6 (5.7) | 3 (15.8) | 1 (7.1) | |||
| PCNB | 298 (73.4) | 87 (82.1) | 13 (68.4) | 9 (64.3) | |||
| Metastatic site | 35 (8.7) | 7 (6.6) | 3 (15.8) | 3 (21.4) | |||
| Cell block | 5 (1.2) | 3 (2.8) | 0 (0.0) | 1 (7.1) | |||
| EBUS‐TBNA | 10 (2.5) | 1 (0.9) | 0 (0.0) | 0 (0.0) | |||
| Surgical resection | |||||||
| Lobectomy | 13 (3.2) | 2 (1.9) | 0 (0.0) | 0 (0.0) | |||
| Wedge resection | 3 (0.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||
P value was evaluated compared to wild type for the gene.
Evaluated by t‐test.
Adenocarcinoma versus all others.
Stage I–IIIa versus IIIb–IV.
Biopsy versus surgical resection.
Values in bold are statistically significant.
ADC, adenocarcinoma; SCC, squamous cell carcinoma; ADSQ, adeno‐squamous carcinoma; NSCLC, non‐small cell lung cancer; PCNB, percutaneous needle aspiration biopsy; EBUS‐TBNA, endobronchial ultrasound‐transbronchial needle aspiration biopsy.
Genetic alterations of EGFR, ALK, and ROS1
As shown in Table 1, among the 407 patients, we identified 106 (26.0%) EGFR mutations, and 19 (4.7%) ALK and 14 (3.4%) ROS1 rearrangements. Among the squamous cell carcinoma cases, none had a ROS1 rearrangement, one had an ALK mutation, and four had an EGFR mutation. In this cohort, coexisting genetic alterations were detected in two patients (0.7%): one had an EGFR mutation with an ALK rearrangement, the other had an EGFR mutation with a ROS1 rearrangement.
When comparing patients with WT or mutated oncogenes, we found that younger age and advanced stage (IIIB and IV) were significantly associated with the three oncogene mutations. EGFR mutations were significantly more frequent in patients with the following characteristics: younger age (P = 0.033), female gender (P < 0.001), never‐smoker (P < 0.001), shorter smoking timeframe (pack‐years, P < 0.001), adenocarcinoma (P < 0.001), and advanced stage (IIIB and IV; P < 0.001). A similar pattern was observed in patients with ALK and ROS1 rearrangements. Specifically, younger age (P < 0.001), never‐smoker (P = 0.018), shorter smoking timeframe (P < 0.001), and advanced tumor stage (P = 0.006) were also significantly associated with ALK rearrangements, while ROS1 rearrangements were significantly associated with younger age (P < 0.001), female gender (P = 0.026), adenocarcinoma (P = 0.026), and advanced tumor stage (P < 0.001).
Association of EGFR, ALK, and ROS1 gene mutation with PD‐L1 expression
Table 2 shows the relationship between EGFR, ALK, and ROS1 gene mutations and PD‐L1 expression. From September 2016 onwards, the PD‐L1 22C3 test was performed on 130 consecutive patients with NSCLC; we identified 29 cases (22.3%) with high PD‐L1 expression, 59 cases (45.4%) with low PD‐L1 expression, and 42 cases (32.3%) with no PD‐L1 expression. EGFR WT status was significantly associated with PD‐L1 expression (TPS > 1%, P = 0.031). ALK rearrangement showed no association with PD‐L1 expression, while ROS1 rearrangement was significantly associated with high PD‐L1 expression (P = 0.031).
Table 2.
Oncogenic aberration and PD‐L1 expression in 130 consecutive NSCLC patients
| PD‐L1 expression | ||||
|---|---|---|---|---|
| Type of mutation | PD‐L1 < 1% (n = 42) |
1% ≤ PD‐L1 < 50% (n = 59) |
PD‐L1 ≥ 50% (n = 29) |
P † |
| EGFR, N (%) | ||||
| Negative | 26 (61.9) | 45 (76.3) | 26 (89.7) | 0.051‡
0.031 § |
| Positive | 16 (38.1) | 14 (23.7) | 3 (10.3) | |
| Exon 19 deletion | 5 (11.9) | 6 (9.0) | 2 (10.5) | |
| Exon 21 L858R | 7 (16.7) | 4 (2.6) | 0 (0.0) | |
| Others | 4 (9.5) | 4 (0.7) | 1 (5.3) | |
| ALK, N (%) | ||||
| Negative | 41 (97.6) | 55 (93.2) | 27 (93.1) | 0.652‡
0.427§ |
| Positive | 1 (2.4) | 4 (6.8) | 2 (6.9) | |
| ROS1, N (%) | ||||
| Negative | 42 (100.0) | 58 (98.3) | 26 (89.7) |
0.031
‡
0.304§ |
| Positive | 0 (0.0) | 1 (1.7) | 3 (10.3) | |
P was evaluated by comparison with the wild type gene.
PD‐L1 < 50% versus PD‐L1 ≥ 50%.
PD‐L1 < 1% versus PD‐L1 ≥ 1%.
Values in bold are statistically significant.
Clinicopathologic profiles of patients with ROS1 gene mutations
The characteristics of the patients with ROS1 rearrangements are shown in Table 3 and Figure 1. Most of the patients were female (71.4%), never‐smokers (71.4%), at advanced tumor stage (IV, 78.6%). All patients had adenocarcinoma. Two coexisting mutations (EGFR and ROS1 mutations) were detected in one patient. Because of the unexpectedly strong association between high PD‐L1 expression and ROS1 mutation in four ROS1‐rearranged cases, we additionally performed PD‐L1 assay on the 10 earlier cases with ROS1 rearrangement that had not been tested for PD‐L1 expression. Out of the 14 ROS1 mutated patients, five (35.7%) had high PD‐L1 expression, 7 (50.0%) had low PD‐L1 expression, and 2 (14.2%) were negative for PD‐L1 expression.
Table 3.
Clinicopathologic profiles of patients with ROS1 rearrangement
| Patient | Age | Gender | Smoking | Pack‐years | Cell type | Sample | Stage | EGFR | ALK | PD‐L1 (%) | ROS1 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 66 | Male | Y | 50 | Adenocarcinoma | Bronchoscopy | IV | WT | Negative | 10 | 28 |
| 37 | 79 | Female | N | 0 | Adenocarcinoma | Cell block | IV | WT | Negative | 80 | 54 |
| 46 | 59 | Female | N | 0 | Adenocarcinoma | PCNB | IV | WT | Negative | 5 | 29 |
| 70 | 76 | Male | Y | 75 | Adenocarcinoma | Metastatic site | IV | WT | Negative | 0 | 59 |
| 275 | 72 | Male | Y | 40 | Adenocarcinoma | PCNB | IB | WT | Negative | 30 | 58 |
| 276 | 79 | Female | N | 0 | Adenocarcinoma | PCNB | IV | WT | Negative | 95 | 27 |
| 282 | 43 | Female | N | 0 | Adenocarcinoma | PCNB | IA | WT | Negative | 25 | 88 |
| 300 | 80 | Female | N | 0 | Adenocarcinoma | PCNB | IV | Exon 21 L858R | Negative | 25 | 49 |
| 360 | 69 | Female | N | 0 | Adenocarcinoma | PCNB | IV | WT | Negative | 2 | 31 |
| 375 | 57 | Female | N | 0 | Adenocarcinoma | PCNB | IV | WT | Negative | 0 | 86 |
| 396 | 47 | Female | N | 0 | Adenocarcinoma | PCNB | IIB | WT | Negative | 1 | 62 |
| 404 | 44 | Female | N | 0 | Adenocarcinoma | Metastatic site | IV | WT | Negative | 66 | 88 |
| 406 | 33 | Male | Y | 5 | Adenocarcinoma | Metastatic site | IV | WT | Negative | 90 | 62 |
| 407 | 53 | Female | N | 0 | Adenocarcinoma | PCNB | IV | WT | Negative | 88 | 86 |
WT, wild type; PCNB, percutaneous needle aspiration biopsy.
Figure 1.
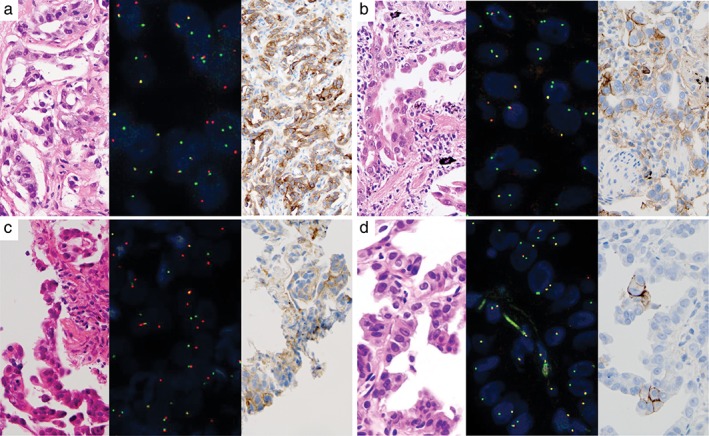
PD‐L1 tumor proportion score (TPS) in several cases of adenocarcinoma with positive fluorescence in situ hybridization (FISH) ROS1 split signal. (a) Case no. 407 showed adenocarcinoma histology (left panel), positive ROS1 FISH, mainly break‐apart signal (middle panel), and high PD‐L1 expression (right panel). (b) Case no. 404 showed adenocarcinoma histology (left panel), positive ROS1 FISH, mainly isolated green signal (middle panel) and high PD‐L1 expression (right panel). (c) Case no. 282 showed adenocarcinoma histology (left panel), positive ROS1 FISH, mainly break‐apart signal (middle panel), and low PD‐L1 expression (right panel). (d) Case no. 396 showed adenocarcinoma histology (left panel), positive ROS1 FISH, mainly isolated green signal (middle panel), and low PD‐L1 expression (right panel).
Discussion
To the best of our knowledge, our study is the first to simultaneously diagnose genetic alterations in EGFR, ALK, and ROS1 in an unselected Asian population. Previous studies have reported that the frequency of ROS1 rearrangement in EGFR/KRAS/ALK WT and never‐smoking patients with lung adenocarcinoma is 5.7–8.3%.32, 33 In an unselected NSCLC population, the rate of ROS1 mutation is 0.6–4.5%.11, 14, 34, 35, 36 Consistently, in this study, the rate of ROS1 rearrangement was 3.4% (14/407).
There were no cases of co‐occurring ROS1 and ALK mutation, but one case of co‐occurring ROS1 and EGFR mutation was found. These findings are similar to those of previous studies suggesting that ROS1 rearrangements are mutually exclusive to other oncogenic changes.11, 14, 36, 37 However, others have reported that ROS1 rearrangements occur in conjunction with other driver mutations.16, 17 This inconsistency may be a result of differences in ROS1 testing techniques. Updated molecular testing guidelines recommend that, while ROS1 IHC may be used as a screening test, any finding should be confirmed by molecular (qPCR) or cytogenetic (FISH) testing.2 Wiesweg et al. reported that 36% of ROS1 IHC‐positive cases have a concomitant EGFR mutation; however, only about half of the cases showed ROS1 rearrangement by FISH.17 This result suggests that IHC results include a high number of false‐positives. The significance of ROS1 IHC differs from that of ALK IHC: ALK expression is highly specific to tumors with ALK rearrangement,31 whereas ROS1 is expressed at typically low and occasionally high levels in tumors lacking ROS1 rearrangement.15, 38, 39 Therefore, we chose ROS1 FISH testing as the initial screening test.
In this study, ROS1‐rearranged NSCLCs were exclusively adenocarcinomas and were predominantly observed in women and younger patients, in line with previous studies.11, 14, 36, 40
As a result of the use of pembrolizumab in first‐line therapy, its companion diagnostic PD‐L1 22C3 assay has become a major tool in the routine clinical testing of NSCLC.3 Increasing costs and medical complexity are significant challenges in organizing the diagnostic platform of biomarkers in NSCLCs, especially in relation to the prevalence and mutual exclusiveness of these biomarkers.41, 42, 43 Accumulating evidence has revealed a relationship between PD‐L1 expression and driver mutations. Previous studies have shown that activating EGFR mutations induce PD‐L1 expression in cell lines and animal models.8, 44 Moreover, some studies have shown more frequent PD‐L1 expression in NSCLC with EGFR mutations.8, 45 These observations support the theory of an immunologic evasion mechanism by EGFR‐mutated lung cancer and suggest that therapeutic blockade of the PD‐1/PD‐L1 pathway could improve the treatment of patients with NSCLC carrying an EGFR mutation. On the contrary, a previously published study revealed the lack of high PD‐L1 expression in EGFR, ALK, or ROS1‐mutated lung adenocarcinoma and proposed therapeutic subgrouping as EGFR/ALK/ROS1 affected, high PD‐L1 expressing, and biomarker negative groups.3 Another large meta‐analysis reported that PD‐L1 expression is associated with gender, smoking status, histology, and tumor size; EGFR WT status was associated with PD‐L1 expression, while ALK or KRAS mutations were not.8 These data are consistent with the correlation between PD‐L1 expression and EGFR WT status we found. PD‐L1 expression in ROS1‐rearranged NSCLC has not been assessed prior to our study. Interestingly, 5 (35.7%) of the 14 patients with ROS1 rearrangement had high PD‐L1expression, and high PD‐L1 expression was also significantly associated with ROS1 rearrangement in our consecutive cohort, which is the largest PD‐L1 study cohort so far. Based on our results, ROS1 rearrangement and high PD‐L1 expression (≥ 50%) are not mutually exclusive but correlate. Further studies are necessary to establish the exact relationship between PD‐L1 expression and ROS1 rearrangement.
There are some limitations to this study. First, we have no data on the immune checkpoint inhibitor treatment response in patients with oncogenic driver mutations because our national insurance only covers pembrolizumab as a second‐line NSCLC treatment, while it covers other tyrosine kinase inhibitors as first‐line treatment. Second, PD‐L1 expression was only tested in 130 consecutive patients and the PD‐L1 test in ROS1 rearrangement cases was partly performed on a simultaneous platform because PD‐L1 has been only recently integrated into the routine diagnostic platform. Third, the number of ROS1 rearrangements was small even though this was the largest cohort analyzed so far. Thus, further research is needed to generalize our findings. Despite these limitations, this study is a large‐scale screen using simultaneous panel genotyping of EGFR, ALK, and ROS1 in unselected patients, which show mutual exclusiveness with other targetable mutations and demonstrates a strong association between ROS1 rearrangement and high PD‐L1 expression.
Disclosure
No authors report any conflict of interest.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT, grant No. 2017R1E1A1A01078335) and by a grant from the Institute of Clinical Medicine Research in the Yeouido St. Mary's Hospital, Catholic University of Korea.
Contributor Information
Jongmin Lee, Email: kimecho@catholic.ac.kr.
Tae‐Jung Kim, Email: kimecho@catholic.ac.kr.
References
- 1. Cersosimo RJ. Lung cancer: A review. Am J Health Syst Pharm 2002; 59: 611–42. [DOI] [PubMed] [Google Scholar]
- 2. Lindeman NI, Cagle PT, Aisner DL et al Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018; 142: 321–46. [DOI] [PubMed] [Google Scholar]
- 3. Rangachari D, VanderLaan PA, Shea M et al Correlation between classic driver oncogene mutations in EGFR, ALK, or ROS1 and 22C3‐PD‐L1 50% expression in lung adenocarcinoma. J Thorac Oncol 2017; 12: 878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borghaei H, Paz‐Ares L, Horn L et al Nivolumab versus docetaxel in advanced nonsquamous non‐small‐cell lung cancer. N Engl J Med 2015; 373: 1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garon EB, Rizvi NA, Hui R et al Pembrolizumab for the treatment of non‐small‐cell lung cancer. N Engl J Med 2015; 372: 2018–28. [DOI] [PubMed] [Google Scholar]
- 6. Herbst RS, Baas P, Kim DW et al Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer KEYNOTE‐010: A randomised controlled trial. Lancet 2016; 387: 1540–50. [DOI] [PubMed] [Google Scholar]
- 7. Reck M, Rodriguez‐Abreu D, Robinson AG et al Pembrolizumab versus chemotherapy for PD‐L1‐positive non‐small‐cell lung cancer. N Engl J Med 2016; 375: 1823–33. [DOI] [PubMed] [Google Scholar]
- 8. Azuma K, Ota K, Kawahara A et al Association of PD‐L1 overexpression with activating EGFR mutations in surgically resected nonsmall‐cell lung cancer. Ann Oncol 2014; 25: 1935–40. [DOI] [PubMed] [Google Scholar]
- 9. Zhang M, Li G, Wang Y et al PD‐L1 expression in lung cancer and its correlation with driver mutations: A meta‐analysis. Sci Rep 2017; 7: 10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chin LP, Soo RA, Soong R, Ou SHI. Targeting ROS1 with anaplastic lymphoma kinase inhibitors: A promising therapeutic strategy for a newly defined molecular subset of non‐small‐cell lung cancer. J Thorac Oncol 2012; 7: 1625–30. [DOI] [PubMed] [Google Scholar]
- 11. Bergethon K, Shaw AT, Ou SH et al ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012; 30: 863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non‐small‐cell lung cancer. Lancet 2016; 388: 1012–24. [DOI] [PubMed] [Google Scholar]
- 13. Lin JJ, Ritterhouse LL, Ali SM et al ROS1 fusions rarely overlap with other oncogenic drivers in non‐small cell lung cancer. J Thorac Oncol 2017; 12: 872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takeuchi K, Soda M, Togashi Y et al RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012; 18: 378–81. [DOI] [PubMed] [Google Scholar]
- 15. Yoshida A, Tsuta K, Wakai S et al Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol 2014; 27: 711–20. [DOI] [PubMed] [Google Scholar]
- 16. Warth A, Muley T, Dienemann H et al ROS1 expression and translocations in non‐small‐cell lung cancer: Clinicopathological analysis of 1478 cases. Histopathology 2014; 65: 187–94. [DOI] [PubMed] [Google Scholar]
- 17. Wiesweg M, Eberhardt WEE, Reis H et al High prevalence of concomitant oncogene mutations in prospectively identified patients with ROS1‐positive metastatic lung cancer. J Thorac Oncol 2017; 12: 54–64. [DOI] [PubMed] [Google Scholar]
- 18. Jiang L, Su X, Zhang T et al PD‐L1 expression and its relationship with oncogenic drivers in non‐small cell lung cancer (NSCLC). Oncotarget 2017; 8: 26845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giunchi F, Degiovanni A, Daddi N et al Fading with time of PD‐L1 Immunoreactivity in non‐small cells lung cancer tissues: A methodological study. Appl Immunohistochem Mol Morphol 2018; 26: 489–94. [DOI] [PubMed] [Google Scholar]
- 20. Han JJ, Kim DW, Koh J et al Change in PD‐L1 expression after acquiring resistance to Gefitinib in EGFR‐mutant non‐small‐cell lung cancer. Clin Lung Cancer 2016; 17: 263–70. [DOI] [PubMed] [Google Scholar]
- 21. Kim TJ, Hong SA, Kim O et al Changes in PD‐L1 expression according to tumor infiltrating lymphocytes of acquired EGFR‐TKI resistant EGFR‐mutant non‐small‐cell lung cancer. Oncotarget 2017; 8: 107630–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Travis WD, Brambilla E, Nicholson AG et al The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015; 10: 1243–60. [DOI] [PubMed] [Google Scholar]
- 23. Kim TJ, Park CK, Yeo CD et al Simultaneous diagnostic platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients with non‐small‐cell lung cancer highlights significantly higher ALK rearrangement rate in advanced stage. J Surg Oncol 2014; 110: 245–51. [DOI] [PubMed] [Google Scholar]
- 24. Ku BM, Heo MH, Kim JH et al Molecular screening of small biopsy samples using next‐generation sequencing in Korean patients with advanced non‐small cell lung cancer: Korean lung cancer consortium KLCC‐13‐01. J Pathol Transl Med 2018; 52: 148–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heymann JJ, Bulman WA, Swinarski D et al PD‐L1 expression in non‐small cell lung carcinoma: Comparison among cytology, small biopsy, and surgical resection specimens. Cancer Cytopathol 2017; 125: 896–907. [DOI] [PubMed] [Google Scholar]
- 26. Gainor JF, Varghese AM, Ou SH et al ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: An analysis of 1,683 patients with non‐small cell lung cancer. Clin Cancer Res 2013; 19: 4273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sterlacci W, Savic S, Schmid T et al Tissue‐sparing application of the newly proposed IASLC/ATS/ERS classification of adenocarcinoma of the lung shows practical diagnostic and prognostic impact. Am J Clin Pathol 2012; 137: 946–56. [DOI] [PubMed] [Google Scholar]
- 28. Yeo CD, Park KH, Park CK et al Expression of insulin‐like growth factor 1 receptor IGF‐1R predicts poor responses to epidermal growth factor receptor EGFR tyrosine kinase inhibitors in non‐small cell lung cancer patients harboring activating EGFR mutations. Lung Cancer 2015; 87: 311–7. [DOI] [PubMed] [Google Scholar]
- 29. Ou SH, Bazhenova L, Camidge DR et al Rapid and dramatic radiographic and clinical response to an ALK inhibitor crizotinib, PF02341066 in an ALK translocation‐positive patient with non‐small cell lung cancer. J Thorac Oncol 2010; 5: 2044–6. [DOI] [PubMed] [Google Scholar]
- 30. Shaw AT, Solomon BJ. Crizotinib in ROS1‐rearranged non‐small‐cell lung cancer. N Engl J Med 2015; 372: 683–4. [DOI] [PubMed] [Google Scholar]
- 31. Thunnissen E, Bubendorf L, Dietel M et al EML4‐ALK testing in non‐small cell carcinomas of the lung: A review with recommendations. Virchows Arch 2012; 46: 245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim HR, Lim SM, Kim HJ et al The frequency and impact of ROS1 rearrangement on clinical outcomes in never smokers with lung adenocarcinoma. Ann Oncol 2013; 24: 2364–70. [DOI] [PubMed] [Google Scholar]
- 33. Kim MH, Shim HS, Kang DR et al Clinical and prognostic implications of ALK and ROS1 rearrangements in never‐smokers with surgically resected lung adenocarcinoma. Lung Cancer 2014; 83: 389–95. [DOI] [PubMed] [Google Scholar]
- 34. Davies KD, Le AT, Theodoro MF et al Identifying and targeting ROS1 gene fusions in non‐small cell lung cancer. Clin Cancer Res 2012; 18: 4570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rimkunas VM, Crosby KE, Li D et al Analysis of receptor tyrosine kinase ROS1‐positive tumors in non‐small cell lung cancer: Identification of a FIG‐ROS1 fusion. Clin Cancer Res 2012; 18: 4449–57. [DOI] [PubMed] [Google Scholar]
- 36. Yoshida A, Kohno T, Tsuta K et al ROS1‐rearranged lung cancer: A clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol 2013; 37: 554–62. [DOI] [PubMed] [Google Scholar]
- 37. Shaw AT, Ou SHI, Bang YJ et al Crizotinib in ROS1‐rearranged non‐small‐cell lung cancer. N Engl J Med 2014; 371: 1963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mescam‐Mancini L, Lantuéjoul S, Moro‐Sibilot D et al On the relevance of a testing algorithm for the detection of ROS1‐rearranged lung adenocarcinomas. Lung Cancer 2014; 83: 168–73. [DOI] [PubMed] [Google Scholar]
- 39. Li C, Fang R, Sun Y et al Spectrum of oncogenic driver mutations in lung adenocarcinomas from east Asian never smokers. PLoS ONE 2011; 6: e28204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Go H, Kim DW, Kim D et al Clinicopathologic analysis of ROS1‐rearranged non‐small‐cell lung cancer and proposal of a diagnostic algorithm. J Thorac Oncol 2013; 8: 445–50. [DOI] [PubMed] [Google Scholar]
- 41. Jackman DM, Zhang Y, Dalby C et al Cost and survival analysis before and after implementation of Dana‐Farber clinical pathways for patients with stage IV non‐small‐cell lung cancer. J Oncol Pract 2017; 13: e346–52. [DOI] [PubMed] [Google Scholar]
- 42. Shim HS, Choi YL, Kim L et al Molecular testing of lung cancers. J Pathol Transl Med 2017; 51: 242–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wong DWS, Leung ELH, So KKT et al The EML4‐ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild‐type EGFR and KRAS. Cancer 2009; 115: 1723–33. [DOI] [PubMed] [Google Scholar]
- 44. Akbay EA, Koyama S, Carretero J et al Activation of the PD‐1 pathway contributes to immune escape in EGFR‐driven lung tumors. Cancer Discov 2013; 3: 1355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. D'Incecco A, Andreozzi M, Ludovini V et al PD‐1 and PD‐L1 expression in molecularly selected non‐small‐cell lung cancer patients. Br J Cancer 2015; 112: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]


