Abstract
Urocanic aciduria is caused by a deficiency in the enzyme urocanase (E.C. 4.2.1.49) encoded by the gene UROC1. In the past, deficiency of urocanase has been associated with intellectual disability in a few case studies with some suggestion that the enzyme deficiency was the causative etiology. Here, we describe two phenotypically normal siblings with compound heterozygous pathogenic variants in UROC1 and characteristic biochemical evidence of urocanase deficiency collected utilizing untargeted metabolomic analysis. These findings suggest that urocanic aciduria may represent an otherwise benign biochemical phenotype and that those individuals with concurrent developmental delay should continue to be evaluated for other underlying causes for their symptoms.
Keywords: UROC1, Urocanic aciduria, Untargeted metabolomics, Cis-urocanate, Trans-urocanate, Imidazole propionate
1. Introduction
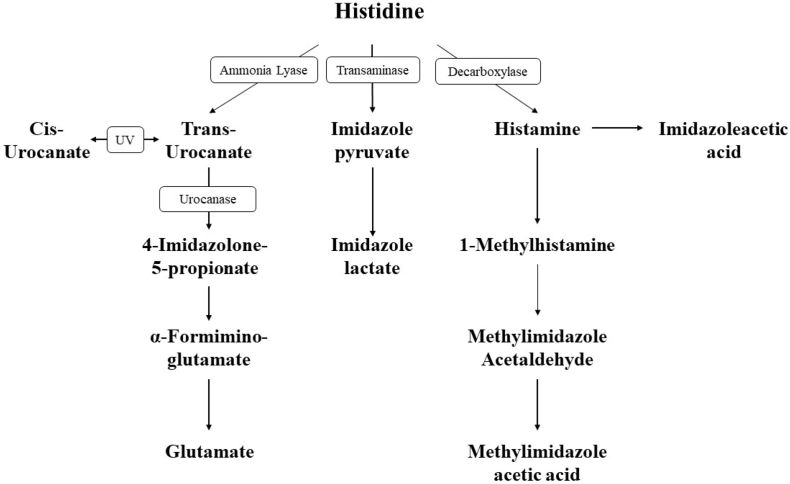
Inborn errors of histidine metabolism were first described in 1961 with the discovery of histidinuria in two siblings by Ghadimi et al. [1] and the identification of histidinemia as a distinct biochemical phenotype. Further studies on additional patients thereafter allowed for the complete elucidation of the histidine metabolic pathway and the discovery of urocanic aciduria as the second clinically significant error in this pathway [2]. Whereas histidinemia is caused by a deficiency in histidase, the enzyme that catalyzes the deamination of L-histidine to trans-urocanic acid, urocanic aciduria is caused by defects in urocanase, which catalyzes the conversion of urocanic acid to imidazolonepropionic acid (Fig. 1). Urocanase is encoded by the gene urocanate hydratase 1 (UROC1, MIM#613012), located on chromosome 3q21. The family of urocanases is highly conserved across multiple species and appears to be almost exclusively expressed in the liver [3].
Fig. 1.
Histidine metabolism.
Urocanic aciduria has been described in association with varying degrees of intellectual disability, though from very early on, this association has been somewhat tenuous [[4], [5], [6]]. Yoshide et al. for instance, first reported the case of a 16 year old young man with severe intellectual disability and significant urocanic aciduria in the setting of significantly decreased liver urocanase activity on biopsy [4]. While the patient did have some developmental delays from early infancy, he had also been diagnosed with tuberculous meningitis at 19 months with residual hemiplegia and seizures even after effective treatment. Similarly, Kalafatic et al. [5] identified two siblings with severe intellectual disability, mood disorder and biochemical evidence of urocanase deficiency. The siblings did have a significant family history of mood disorders and intellectual disability though at the time only their seemingly unaffected mother was available for biochemical testing.
It was not until 2009, however, that the first pathogenic variants in UROC1 were found in a 19 year old female with urocanic aciduria, intellectual disability and intermittent ataxia [6]. Prior to this study, patients with urocanic aciduria were diagnosed based upon biochemical data only, leaving open the possibility of other concurrent molecular diagnoses. This is particularly true given the vast advances in exome sequencing and the increased incidence of so-called “second diagnoses” upon comprehensive molecular testing [7]. While the exact molecular changes in prior individuals were unknown, we now report the case of two developmentally normal siblings with compound heterozygous pathogenic variants in UROC1 and urocanic aciduria. In addition, global untargeted metabolomic analysis is able to identify signature metabolic perturbations in both plasma and urine which may aid in its future diagnosis.
2. Subjects and methods
2.1. Subjects
We identified a family consisting of two adult children with biochemical evidence of urocanase deficiency. Each affected individual underwent sequencing of UROC1 to confirm the presence of biallelic pathogenic variants. The parents of the two patients also underwent targeted sequencing to confirm their presumed carrier status and the expected trans-configuration of variants. Untargeted metabolomics was also carried out on the affected subjects and revealed metabolic perturbations characteristic of urocanase deficiency. This study was conducted according to Baylor College of Medicine (BCM) Institutional Review Board (IRB) approved protocols.
2.2. Clinical subjects
Case 1 is a 27 year old female who was initially identified with a large amount of urocanic acid in her urine by routine urine screening at 8 weeks of age [8]. She was born to a 22 year old healthy mother after an uneventful primigravida pregnancy. The infant's birth, delivery and neonatal course were normal and she was breast fed. At the time of her birth, urine samples were being collected on every infant at 3–4 weeks of life as part of the Massachusetts State Newborn Screening Program primarily as a means of supplementing blood screening protocols. Following detection of urocanic acid in her urine, she was seen for metabolic evaluation at age 5 months. Examination revealed a normal infant with weight and length both 90-95th percentiles for age and appropriate developmental milestones. Family history was negative for metabolic disorders or any other familial diseases. Her red blood cell folate level was normal at 346 μgm/L (reference 200–270). Urine paper chromatography with special Pauly stain revealed a moderate amount of urocanic acid and unidentified related metabolites. Plasma amino acid analysis was normal. She was thus diagnosed with urocanic aciduria presumed to be due to urocanase deficiency. This was considered to be benign and she was allowed to continue breast feeding. A normal diet continued and her subsequent growth and development continued to remain normal. At age 3 years, psychological assessment by the McCarthy Scales of Children's Abilities revealed a general cognitive score of 115 placing her in the 83rd percentile for age. She was followed in the Metabolic Program at Boston Children's Hospital until age 5 years when the family relocated. Her subsequent clinical course was normal. She is now a registered nurse in an intensive care unit of a hospital.
Case 2 is the now 21 year old brother of Case 1. He was found to have urocanic aciduria by specific urine examination at age 7 months, performed because of the history of urocanic aciduria in his sister. He also has enjoyed an entirely normal clinical course with excellent growth and development. He is currently in nursing school to become a registered nurse. The family does have a third child who was found to be unaffected with an uneventful clinical course and no urocanic acid in urine.
2.3. Global metabolomics
Metabolomic profiling (Global MAPS®) was performed by Baylor Genetics (Houston, TX) (www.baylorgenetics.org) and Metabolon, Inc. (Durham, NC) (www.metabolon.com), as described previously [[9], [10], [11], [12], [13]]. Briefly, small molecules were extracted in an 80% methanol solution before being analyzed by four different mass spectrometry methods. The first method utilized acidic, positive ionization conditions chromatographically optimized for hydrophilic compounds (UPLC-MS/MS Pos polar). The second method used the same acidic positive ionization conditions but was chromatographically optimized for hydrophobic compounds (UPLC-MS/MS Pos lipid). The third method used negative ionization optimized conditions (UPLC-MS/MS Neg), and the final method utilized negative ionization optimized conditions with hydrophilic interaction liquid chromatography (UPLC-MS/MS Polar). All chromatographic separations were completed using an ACQUITY UPLC (Waters) equipped with either a Waters BEH C18 or Waters BEH Amide column followed by analysis with an Orbitrap Elite high-resolution mass spectrometer (Thermo-Finnigan).
The chemical structures of known metabolites were identified by matching the ions' chromatographic retention index, nominal mass, and mass spectral fragmentation signatures with reference library entries created from authentic standard metabolites under the identical analytical procedure as the experimental samples. Currently, the reference library contains entries for ~2500 unique human metabolites. Semi-quantitative analysis was achieved by comparing patient samples to a set of invariant anchor specimens included in each batch. Raw spectral intensity values were normalized to the anchor samples, log transformed, and compared to a normal reference population to generate z-score values. Median raw intensity values were calculated for all analytes identified in ≥2/3 of the anchor specimens, and these median values were then used to normalize corresponding analyte raw intensity values in the patient specimen. Analytes not identified in at least 2/3 of the anchor specimens were excluded from analysis. Z-scores were calculated using the mean and standard deviation of the entire median-scaled log- transformed dataset. Results were considered abnormally low if the z-score for a compound is equal or less than two standard deviations below the mean or abnormally high if the z-score is equal or greater than two standard deviations above the mean of the control reference population.
2.4. DNA sequencing
Sanger sequencing was performed both subjects as well as both parents as a service at Baylor Genetics (Houston, TX) (www.baylorgenetics.org). Genomic DNA was obtained from peripheral blood, and a PCR-based array was used to amplify the 21 coding exons of UROC1 based on reference sequences NM_144639.2 and NG_016286. Direct sequence analysis was then performed in both the forward and reverse directions using automated fluorescence dideoxy sequencing methods. Identified variants were then annotated using the recommended Human Genome Variation Society nomenclature.
3. Results
3.1. Metabolomic profiling
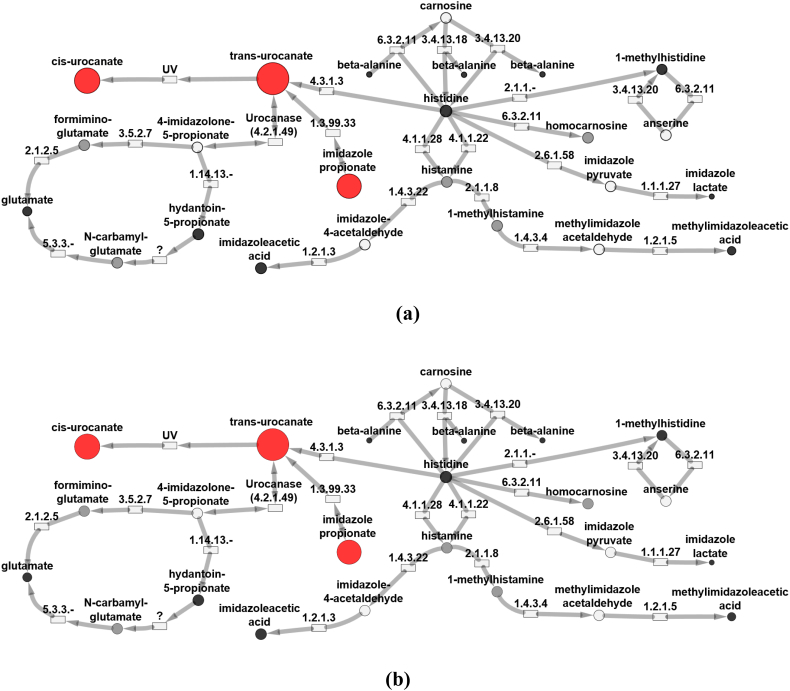
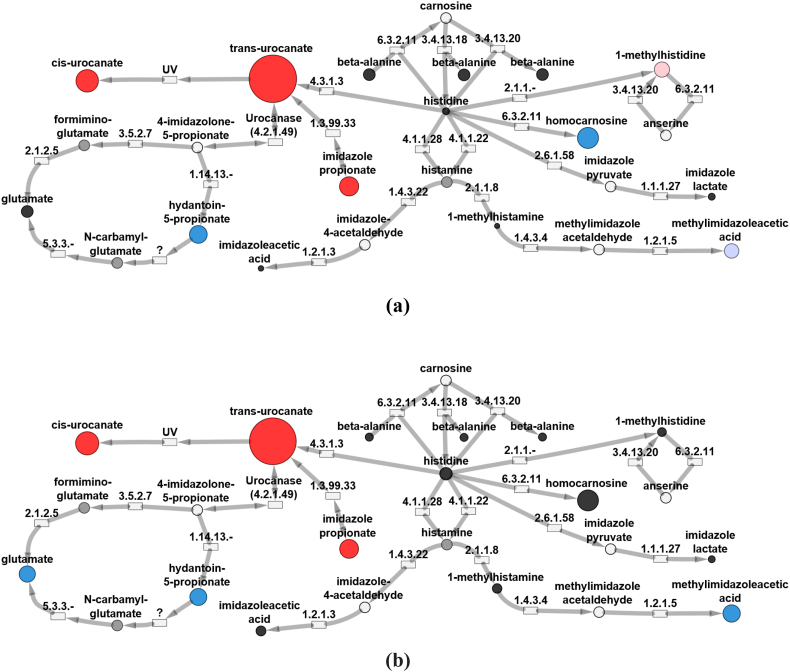
Untargeted metabolomic profiling was performed on both urine and plasma samples (Table 1). Both plasma samples from the subjects demonstrated significant elevations in cis-urocanate, trans-urocanate and imidazole propionate. Similarly, Z-scores for compounds in alternative histidine degradation pathways were found to be within normal limits, including 1-methylhisitidine, imidazole lactate, and 1-methylhistamine (Fig. 2, Fig. 3). Notably, histidine levels were found to be within normal limits in all samples. Some metabolic intermediates failed to be identified in either subject due to their inherent physical properties, relative levels in plasma or urine, or inherent limitations in the metabolomics platform. This pattern of abnormalities, in particular, significantly elevated trans-urocanic acid, cis-urocanic acid and imidazole propionate, was rather unique and was only found in a total of five patients in our internal database; the two patients mentioned here along with three other individuals. While we do not have molecular confirmation of UROC1 pathogenic variants in these additional patients, we do feel that this biochemical pattern is be diagnostic for this disorder (See Data in Brief).
Table 1.
Untargeted metabolomics identifies multiple intermediates in histidine metabolism in plasma and urine for patients with urocanic aciduria. z-Scores are shown.
| Compound | Plasma |
Urine |
||
|---|---|---|---|---|
| z-Score |
z-Score |
|||
| Subject 1 | Subject 2 | Subject 1 | Subject 2 | |
| trans-Urocanate | 7.0939 | 6.8137 | 7.7051 | 7.5464 |
| cis-Urocanate | 5.7492 | 5.2359 | 3.1651 | 3.2677 |
| Imidazole propionate | 3.7030 | 3.2626 | 2.5705 | 2.4829 |
| 1-methylhistidine | 1.7940 | 1.6233 | 1.8154 | 0.6895 |
| beta-Alanine | 1.0711 | 0.2461 | 1.2420 | 0.5525 |
| Imidazole lactate | 1.0099 | 0.1857 | 0.3640 | −0.3691 |
| 1-methylhistamine | NA | NA | 0.0476 | −0.8100 |
| 4-imidazoleacetate | 0.5374 | 1.0653 | −0.1010 | −0.8854 |
| Histidine | 0.1381 | −1.7535 | −0.3511 | −1.3456 |
| Glutamate | 0.929 | 0.123 | −1.2165 | −2.0166 |
| 1-methylimidazoleacetate | −1.4329 | −0.7770 | −1.6684 | −2.2551 |
| Hydantoin-5-propionic acid | NA | NA | −2.2781 | −2.1873 |
| Homocarnosine | NA | NA | −2.9191 | NA |
Fig. 2.
UROC1 pathogenic variants result in altered histidine metabolism. Biochemical pathway map showing metabolic perturbations in plasma from Patient 1 (a) and Patient 2 (b), both diagnosed with urocanic acidemia. Red circles indicate biochemicals with abnormally high z-scores (z-score ≥ 2) and blue circles indicate biochemicals with abnormally low (z-score ≤ −2) z-scores. The diameters of the circles indicate the magnitude of the z-scores. Pink circles represent biochemicals with z-score of 1.5 ≤ z < 2.0 and light blue circles represent biochemicals with −2.0 < z ≤ −1.5. Black circles represent other biochemicals in the pathway detected in the EDTA plasma samples but had z-scores of −1.5 < z < 1.5. Gray circles represent biochemicals in the library but not detected. White circles are pathways linked to the histidine metabolism pathway. Rectangles represent enzymes in the pathways with the associated EC number for identification. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Urine from patients with UROC1 pathogenic variants reveals altered histidine metabolism. Biochemical maps showing metabolic perturbations in urine from Patient 1 (a) and Patient 2 (b), both diagnosed with urocanic aciduria. Red circles indicate biochemicals with positive z-scores (z-score ≥ 2) and blue circles indicate biochemicals with negative z-scores (z-score ≤ −2). The diameters of the circles indicate the magnitude of the z-score. Pink circles represent biochemicals with z-score of 1.5 ≤ z < 2.0 and light blue circles represent biochemicals with −2.0 < z ≤ −1.5. Black circles represent other biochemicals in the pathway detected in the EDTA plasma samples but had z-scores of −1.5 < z < 1.5. Gray circles represent biochemicals in the library but not detected. White circles are pathways linked to the histidine metabolism pathway. Rectangles represent enzymes in the pathways with the associated EC number for identification. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. UROC1 sequencing
Both subjects were found to be compound heterozygotes for missense variants in UROC1: c.356C > G (p.P119R) and c.907G > C (p.A303P). Targeted sequence analysis of their parents indicated that the mother was heterozygous for the variant c.356C > G (p.P119R), while the father was heterozygous for c.907G > C (p.A303P). These data indicate that the two variants are in trans configuration in both children. The missense variant c.356C > G (p.P119R) is present in 12/61,216 of European (non-Finnish) alleles for an allele frequency of 1.797e-04 (ExAC Database [14]). Using publicly available prediction models (SIFT [15]/Polyphen [16]), the variant is predicted to be damaging and probably damaging, respectively. The second familial missense variant, c.907G > C (p.A303P), is also found exclusively in European (non-Finnish) individuals with an allele frequency approximately 7.502e-05 within that population. This variant is similarly predicted to be damaging (SIFT) and probably damaging (Polyphen) based on in silico calculations.
4. Discussion
In addition to urocanase, histidine catabolism may proceed through several different enzymatic pathways (Fig. 1). Alternative histidine pathways include conversion to histamine via enzymatic decarboxylation, transamination to produce imidazole pyruvic acid and minor conversions to carnosine, homocarnosine and anserine. In our study, both subjects demonstrated markedly elevated levels of trans-urocanic acid, cis-urocanic acid and imidazole propionate, while the products of alternative pathways were found to be only slightly decreased or otherwise normal (Fig. 2, Fig. 3). These results indicate clear biochemical evidence of isolated urocanase deficiency which was further confirmed by the presence of biallelic variants in UROC1 predicted to be deleterious by various platforms.
Urocanic acid has long been recognized to contribute to skin photo-protection and immune response regulation [17]. However, it remains unclear whether or not elevated levels of the compound impact these functions, and most studies linking urocanic aciduria to developmental delays have focused instead on altered folate metabolism as a potential cause. This explanation, however, is somewhat problematic given that other enzyme deficiencies in folate metabolism also similarly cause a complex and sometimes contradictory phenotype. Glutamate formiminotransferase, in particular, is known to play an important role in tetrahydrofolate utilization and indeed, patients with deficiencies in this enzyme do exhibit varying degrees of developmental and physical delays in the setting of megaloblastic anemia [18,19]. Conversely, however, there have also been reported patients with massive amounts of formiminoglutamic acid (FIGLU) in urine but no signs of developmental or physical delays [20]. This variation may be due to the relatively minor contribution of glutamate formiminotransferase to overall folate metabolism along with, once again, a level of ascertainment bias and previously limited molecular testing.
As with many rare diseases, both urocanase deficiency and glutamate formiminotransferase deficiency remain poorly understood. While their particular biochemical signatures in urine made their initial diagnosis relatively simple, our understanding of these abnormalities remains limited making the issue of counseling and ongoing management difficult. One notable limitation of the present study is that we only describe two related individuals, both with the same two missense variants. It is possible, for instance, that there may be significant differences between these two individuals and others with more severe changes like truncating or frameshift variants. While further studies on genotype-phenotype correlations may help alleviate some of these challenges, we would argue that the finding of significant urocanic aciduria in a child with developmental delays is benign and should not preclude standard, comprehensive testing [21]. Untargeted metabolomics may be considered as part of this work-up, as this method is capable of screening for multiple metabolic aberrations and key features of urocanase deficiency simultaneously. As more children with this disorder obtain such testing, it is hoped that more information can be gained as to the exact clinical consequences of different UROC1 variants.
5. Conclusion
Significant urocanic aciduria may exist as an otherwise benign phenotype, as observed in two healthy, developmentally appropriate patients. The diagnosis was initially made based on biochemical data, though was later confirmed by the presence of two heterozygous pathogenic/likely pathogenic variants. Using untargeted metabolomics, we were able to identify pathway aberrations specific to this disease which served to support this as a single, isolated defect without significant inhibition or enrichment of alternate pathways. Based on these observations, the finding of urocanic aciduria in patients with developmental delay, should prompt a complete and comprehensive evaluation for other molecular or biochemical causes.
Acknowledgments
Acknowledgements
The authors would like to thank the patients and their families for their participation and openness. The authors also like to thank Jing Xiao and Charul Gijavanekar for their kind assistance with data management and processing.
Conflicts of interest
KG and SHE are employees of Baylor College of Medicine. Baylor Genetics generates genetic testing revenue in a partnership with Baylor College of Medicine. ADK and KLP are employees of Metabolon, Inc. and, as such, have affiliations with or financial involvement with Metabolon, Inc. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the article apart from those disclosed.
Funding
This work was supported by the BCM Department of Molecular & Human Genetics.
References
- 1.Ghadimi H., Partington M.W., Hunter A. A familial disturbance of histidine metabolism. N. Engl. J. Med. 1961;265(5):221–224. doi: 10.1056/NEJM196108032650504. [DOI] [PubMed] [Google Scholar]
- 2.Levy Harvey L. In: "Disorders of Histidine Metabolism." The Online Metabolic and Molecular Bases of Inherited Disease. Valle David., editor. McGraw-Hill; New York, NY: 2014. http://ommbid.mhmedical.com/content.aspx?bookid=971§ionid=62674206 [Google Scholar]
- 3.Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., Asplund A. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida T., Tada K., Honda Y., Arakawa T. Urocanic aciduria: a defect in the urocanase activity in the liver of a mentally retarded. Tohoku J. Exp. Med. 1971;104(4):305–312. doi: 10.1620/tjem.104.305. [DOI] [PubMed] [Google Scholar]
- 5.Kalafatic Z., Lipovac K., Jezerinac Z., Juretic D., Dumic M., Zurga B. A liver urocanase deficiency. Metabol. Clin. Exp. 1980;29(11):1013–1019. doi: 10.1016/0026-0495(80)90209-7. [DOI] [PubMed] [Google Scholar]
- 6.Espinós C., Pineda M., Martinez-Rubio D., Lupo V., Ormazabal A., Vilaseca M.A., Spaapen L.J., Palau F., Artuch R. Mutations in the urocanase gene UROC1 are associated with urocanic aciduria. J. Med. Genet. 2009;46(6):407–411. doi: 10.1136/jmg.2008.060632. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z., Hardison M. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy H., Coulombe J.T., Shih V.E. Newborn urine screening. In: Bickel H., Guthrie R., Hammersen G., editors. Neonatal Screening for Inborn Errors of Metabolism. 1980. pp. 89–103. Berlin. [Google Scholar]
- 9.Miller M.J., Kennedy A.D., Eckhart A.D., Burrage L.C., Wulff J.E., Miller L.A., Milburn M.V., Ryals J.A., Beaudet A.L., Sun Q., Sutton V.R. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. J. Inherit. Metab. Dis. 2015;38(6):1029–1039. doi: 10.1007/s10545-015-9843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans A.M., DeHaven C.D., Barrett T., Mitchell M., Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009;81(16):6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 11.DeHaven C.D., Evans A.M., Dai H., Lawton K.A. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminforma. 2010;2(1):9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atwal P.S., Donti T.R., Cardon A.L., Bacino C.A., Sun Q., Emrick L., Sutton V.R., Elsea S.H. Aromatic L-amino acid decarboxylase deficiency diagnosed by clinical metabolomic profiling of plasma. Mol. Genet. Metab. 2015;115(2):91–94. doi: 10.1016/j.ymgme.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy A.D., Miller M.J., Beebe K., Wulff J.E., Evans A.M., Miller L.A., Sutton V.R., Sun Q., Elsea S.H. Metabolomic profiling of human urine as a screen for multiple inborn errors of metabolism. Genet. Test. Mol. Biomarkers. 2016;20(9):485–495. doi: 10.1089/gtmb.2015.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Tukiainen T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4(7):1073. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 16.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7(4):248. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko K., Walker S.L., Lai-Cheong J., Matsui M.S., Norval M., Young A.R. cis-Urocanic acid enhances prostaglandin E2 release and apoptotic cell death via reactive oxygen species in human keratinocytes. J. Investig. Dermatol. 2011;131(6):1262–1271. doi: 10.1038/jid.2011.37. [DOI] [PubMed] [Google Scholar]
- 18.Erbe R.W. Inborn errors of folate metabolism. N. Engl. J. Med. 1975;293(15):753–757. doi: 10.1056/NEJM197510092931506. [DOI] [PubMed] [Google Scholar]
- 19.Hilton J.F., Christensen K.E., Watkins D., Raby B.A., Renaud Y., de la Luna S., Estivill X., MacKenzie R.E., Hudson T.J., Rosenblatt D.S. The molecular basis of glutamate formiminotransferase deficiency. Hum. Mutat. 2003;22(1):67–73. doi: 10.1002/humu.10236. [DOI] [PubMed] [Google Scholar]
- 20.Perry T.L., Applegarth D.A., Evans M.E., Hansen S., Jellum E. Metabolic studies of a family with massive formiminoglutamic aciduria. Pediatr. Res. 1975;9(3):117. doi: 10.1203/00006450-197503000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Moeschler J.B., Shevell M. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014;134(3):e903–e918. doi: 10.1542/peds.2014-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]