Abstract
Ubiquitin-specific protease 10 (USP10) is involved in a number of biological processes by stabilizing several proteins, which have been implicated in multiple stages of tumorigenesis and progression. Previous studies have indicated that USP10 stabilizes and deubiquitinates MutS homolog 2 (MSH2) in in vitro and in vivo models. The level of MSH2 protein has been positively correlated with that of the USP10 protein in a panel of lung cancer cell lines. Furthermore, depletion of USP10 in lung cancer cells causes decreased apoptosis and increased cell survival upon treatment with DNA-damaging agents. However, the expression and clinical implication of USP10 protein in lung cancer tissues is not clear. Additionally, whether the level of MSH2 protein is positively correlated with that of the USP10 protein in lung cancer tissues also remains unresolved. Therefore, USP10 protein expression was detected in 148 human non-small cell lung cancer (NSCLC) and 139 non-cancerous lung tissues using immunohistochemistry, whereas mRNA was investigated by Gene Expression Omnibus dataset and The Cancer Genome Atlas database analyses. It was identified that USP10 protein expression was significantly downregulated in NSCLC tissues compared with in normal lung tissues (P<0.05). However, no significant difference in USP10 mRNA expression between the two tissues was identified. In addition, a positive correlation was observed between the USP10 and MSH2 proteins in NSCLC tissues (P<0.05). However, the clinicopathological features and survival analysis indicated that the USP10 and MSH2 proteins were not associated with clinical features, including age, sex, tumor size, Tumor-Node-Metastasis stage and tumor cell differentiation, along with the prognosis of NSCLC. Collectively, these results suggest that downregulation of USP10 protein serves an important function in the tumorigenesis of NSCLC, and the level of USP10 protein is positively correlated with that of MSH2 protein in NSCLC tissues, which may indicate that USP10 also stabilizes the MSH2 protein in patients with lung cancer.
Keywords: non-small cell lung cancer, MutS homolog 2, human ubiquitin-specific protease 10, clinical implication
Introduction
The ubiquitination pathway is an important way to regulate protein levels in eukaryotic cells and serves an important function in the post-translational modification of proteins (1,2). It is well-documented that the ubiquitination of a number of proteins can be reversed by deubiquitinases (DUBs), thereby affecting cell viability, signal transduction, and a number of physiological and pathological processes (3). In total, ~100 DUBs of five associated classes have been identified in the human genome (4). The human ubiquitin-specific protease 10 (USP10), also known as UBPO and located on chromosome 16q24.1, belongs to the DUB family (5). An increasing number of studies into the USP10 protein and its tumor-associated molecular mechanism have been performed. It has been identified that USP10 is associated with the deubiquitination of certain proteins, including tumor protein p53, T-box protein 21, proliferating-cell nuclear antigen (PCNA), Beclin1, sirtuin 6 (SIRT6) and inhibitor of nuclear factor κB kinase subunit γ proteins (6–10), thus participating in cell proliferation, differentiation and autophagy.
It has been identified that USP10 protein expression was significantly decreased in human gastric cancer tissues and cell lines compared with that in the normal gastric mucosa and immortalized epithelial cell lines, which indicated that USP10 is a tumor suppressor gene involved in the tumorigenesis of gastric cancer (11). Furthermore, USP10 protein expression was negatively associated with the depth of invasion, lymph node metastasis and Tumor-Node-Metastasis (TNM) staging (12) of gastric carcinoma. Survival analysis demonstrated that USP10 may serve as a novel prognostic indicator predicting the outcome of gastric carcinoma (11). It has been reported that knockdown of USP10 in lung cancer cells causes increased cell survival and decreased apoptosis upon treatment with a DNA-methylating and antimetabolite agent (13). Thereafter, the aforementioned phenotypes may be rescued by ectopic expression of MutS homolog 2 (MSH2). A recent study demonstrated that 50% (9/18) of patients with non-small cell lung cancer (NSCLC) exhibited decreased expression of USP10 in tumor tissues compared with that in the respective adjacent normal lung tissues (14). However, the clinical sample size was not large and the functional role of USP10 in NSCLC remains unresolved. Therefore, it is necessary to use a large cohort of patients with NSCLC to validate whether USP10 serves critical functions in the tumorigenesis and progression of NSCLC.
MSH2, a crucial element of the highly conserved DNA mismatch repair system, maintains genetic integrity by correcting DNA replication errors (15). The function of MSH2 is to recognize DNA mismatches and then assist in recruiting DNA repair proteins to the mismatched site. A previous study indicated that MSH2 is a promising marker of the benefit of adjuvant cisplatin-based chemotherapy for NSCLC (16). Inhibition of protein kinase B activity and MSH2 expression increased the tamoxifen-mediated cytotoxicity in lung cancer A549 and H1703 cells (17). A previous study indicated that USP10 interacts with MSH2, and the major region of MSH2 responsible for the interaction with USP10 is located in the N-terminal region, whereas for USP10, the major region is a C-terminal hydrolase domain, which interacts with MSH2 (13). Further study indicated that USP10-mediated deubiquitination of MSH2 may partially account for MSH2 stability. Furthermore, the researchers identified that the protein level of MSH2 is positively associated with the USP10 protein level in a panel of lung cancer cell lines (17). However, whether the protein level of MSH2 is positively correlated with the USP10 protein level in lung cancer tissue samples also requires clarification. Therefore, the aim of the present study was to investigate the potential correlation between the USP10 and MSH2 proteins in NSCLC tissues. In addition, USP10 expression levels in NSCLC and normal tissues were investigated to determine whether USP10 is associated with tumorigenesis and progression of NSCLC. Furthermore, the association between USP10 or MSH2 expression and clinicopathological features as well as prognosis was also evaluated in patients with NSCLC.
Materials and methods
Tissue sample and data collection
A total of 148 patients with NSCLC (101 men and 47 women; age range, 40–76 years; mean age, 60.0±8.2 years) were included in the present study. All cases were identified through chest surgery and the Pathology Department (Remin Hospital of Wuhan University, Wuhan, China), as well as through cancer registries at Renmin Hospital of Wuhan University, between August 2013 and September 2014. Following provision of written informed consent, demographic and clinicopathological data (including age, sex, tumor size, tumor differentiation and TNM stage) were collected at the Renmin Hospital of Wuhan University, using a standard interviewer-administered questionnaire and/or medical records. In the present study, the TNM stage was confirmed according to the Union for International Cancer Control (UICC) classification (8th edition) (12). A total of 148 formalin-fixed paraffin-embedded NSCLC tumor tissues (54 cases of squamous cell carcinoma, 82 cases of adenocarcinoma, 4 cases of adenosquamous carcinoma, 3 cases of sarcomatoid carcinoma and 5 cases of large cell carcinoma) and 139 non-cancerous tissues (45 cases of bronchial epithelium and 94 cases of alveolar epithelium) were selected. The tissue microarrays were prepared by Wuhan Iwill Biological Technology Co., Ltd. (Wuhan, China).
For the survival analysis, a total of 56 patients, who received the same surgical therapy excluding preoperative chemotherapy or radiotherapy, were followed up. There were 26 cases of squamous cell carcinoma, 27 cases of adenocarcinoma, 2 cases of adenosquamous carcinoma and 1 case of large cell carcinoma. The last follow-up day was set for March 2018, and the survival status was confirmed by means of clinical records and patient or family contact. The duration of overall survival was defined as extending from the date of surgical treatment to the date of mortality or last known date alive. The present study was approved by the Ethics Committee of Renmin Hospital of Wuhan University.
Immunohistochemistry (IHC)
IHC staining was performed on formalin-fixed paraffin-embedded sections (4 µm) according to a standard protocol. Briefly, sections were deparaffinized with xylene twice for 10 min and rehydrated with 100% ethanol twice for 5 min, 95% ethanol twice for 2 min and 85% ethanol for 2 min, then washed in deionized H2O for 1 min at room temperature with stirring. Subsequently, sections were treated with 3% H2O2 at room temperature for 10 min and subjected to antigen retrieval by citrate buffer (pH 6.0) at 98°C for 15 min. Following blocking with 5% bovine serum albumin for 20 min at room temperature, the sections were incubated at 4°C overnight with anti-USP10 (1:250; cat. no. ab109219; Abcam, Cambridge, UK) and anti-MSH2, (cat. no. ZA-0622; ready-to-use; OriGene Technologies, Inc., Beijing, China) primary antibodies. The sections were incubated with biotinylated antibodies and horseradish peroxidase-labeled streptavidin [Ready-to-use; UltraSensitive™ SP (Mouse/Rabbit) IHC kit-9710; Fuzhou Maixin Biotech Co., Ltd., China] for 15 min each at room temperature. The reaction products were visualized with 3,3′-diaminobenzidine as a chromogen followed by counterstaining with hematoxylin at room temperature for 1 min. The sections without primary antibody served as a negative control (18).
Evaluation of IHC staining
The IHC staining results of USP10 and MSH2 in all sections were evaluated by two pathologists (Dr Zhi Zeng and Dr Yabing Huang, Department of Pathology, Renmin Hospital of Wuhan University) unaware of the disease outcome. Expression levels were ascertained according to the average score of the two observers' evaluations. If the difference of score was ≥2, the final score was determined by a third pathologist (Dr Jingping Yuan; Department of Pathology, Renmin Hospital of Wuhan University). USP10 in the tumor cells was mainly located in the cytoplasm and was present at a different staining intensity; therefore, the USP10 expression information in the tumor cells, scored by staining intensity was analyzed according to a four-tier grading system (scored as 0, absent; 1, weak; 2, moderate; and 3, strong staining intensity), as in a previous study (11). The MSH2 protein was located in the cell nucleus, therefore the percentages of nucleus-stain-positive cells were analyzed in the tumor tissues as the IHC score, and were divided into four groups according to the percentage of positive cells: 0 group, <10%; 1+ group, 10–25%; 2+ group, 26–50%; and 3+ group, >50%. For the analysis, USP10 and MSH2 expression levels were divided into two groups: Negative group (score, ≤1 or 1+) and positive group (score, >1 or 1+).
Analysis of publicly available data
Datasets of patients with lung cancer and corresponding clinical data were downloaded from the publicly available Gene Expression Omnibus (GEO) datasets (www.ncbi.nlm.nih.gov/gds). The data were retrieved from different GEO Series (GSE) and GEO Platform (GPL) datasets. A total of nine independent datasets from GSE (GSE36471, GSE44077, GSE32867, GSE31210, GSE46539, GSE67061, GSE33479, GSE19804 and GSE43458) were used to analyze the expression level of USP10 mRNA in lung cancer. The log2 intensity of different probes was first transformed into original data used to represent the expression level of USP10, and the y-axis of the figures of different GSE datasets represents the unlogged data of the gene expression intensity. P<0.05 was considered to indicate a statistically significant difference, as presented in Table I. Furthermore, UALCAN (ualcan.path.uab.edu), an interactive web portal for performing in-depth analyses of The Cancer Genome Atlas (TCGA) gene expression data, was utilized to mine USP10 mRNA expression between NSCLC and normal lung tissues. Kaplan-Meier plots presenting the association of USP10/MSH2 mRNA expression and patient survival were also obtained from TCGA database.
Table I.
USP10 mRNA expression in normal compared with cancer tissues of different Gene Expression Omnibus datasets from different gene expression platforms.
| GSE | Normal cases | Cancer cases | P-value | GPL | Probe |
|---|---|---|---|---|---|
| 36471 | 29 | 89 | 0.581 | 3720 | SNP A-1828377 |
| 44077 | 65 | 57 | 4.43×10−7 | 6244 | 7997633 |
| 32867 | 86 | 86 | 0.66 | 8490 | cg00200063 |
| 31210 | 20 | 226 | 0.001 | 570 | 209136_s_at |
| 46539 | 92 | 92 | 0.41 | 6883 | ILMN_1721116 |
| 67061 | 8 | 69 | 0.0028 | 6480 | A_23_P100196 |
| 33479 | 13 | 14 | 0.082 | 6480 | A_23_P100196 |
| 19804 | 60 | 59 | 0.5 | 570 | 209136_s_at |
| 43458 | 30 | 80 | 0.7 | 6244 | 7997633 |
USP10, ubiquitin-specific protease 10; GSE, Gene Expression Omnibus Series; GPL, Gene Expression Omnibus Platform.
Statistical analysis
All experimental data were analyzed using SPSS statistical software (version 15.0; SPSS, Inc., Chicago, IL, USA). χ2 and Fisher's exact tests were used to analyze the statistical significance of the association between USP10 expression and the clinicopathological features. Student's t-test was used to compare patients' age or tumor diameter between two groups. The correlation between USP10 and MSH2 expression was analyzed using Spearman's rank correlation analysis. For the survival analysis, survival curves were obtained with the Kaplan-Meier method and compared using the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of USP10 protein between NSCLC tissues and non-cancerous lung tissues
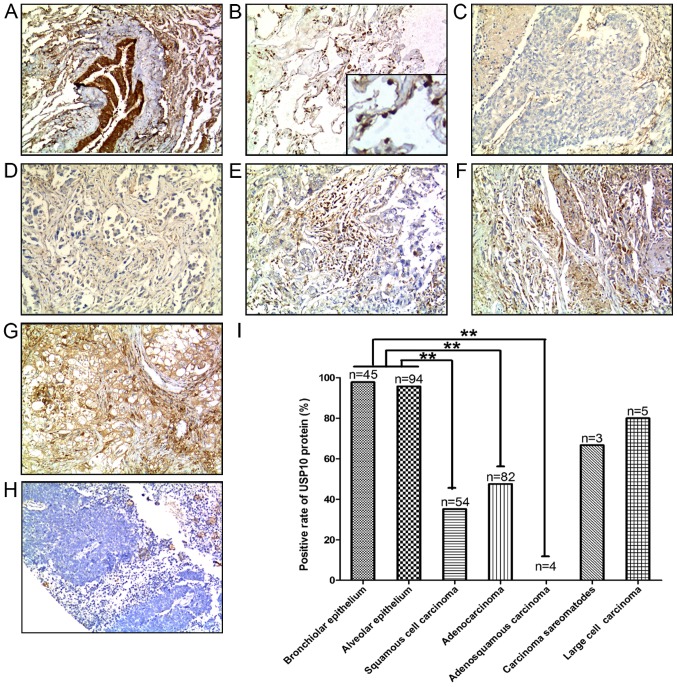
A total of 148 human NSCLC tissue samples were analyzed, including 54 cases of squamous cell carcinoma, 82 cases of adenocarcinoma, 4 cases of adenosquamous carcinoma, 3 cases of sarcomatoid carcinoma and 5 cases of large cell carcinoma, and a corresponding 139 non-cancerous lung tissues containing 45 cases of bronchial epithelium and 94 cases of alveolar epithelium were also analyzed. Representative IHC staining for USP10 protein expression in different types of the aforementioned tissues are presented in Fig. 1A-H. The positive rate of USP10 protein in different tissues, including bronchial epithelium, alveolar epithelium, squamous cell carcinoma, adenocarcinoma, adenosquamous carcinoma, sarcomatoid carcinoma and large cell carcinoma was 97.78, 95.74, 35.18, 45.76, 0, 66.67 and 80%, respectively (Fig. 1I). USP10 protein expression was significantly decreased in the NSCLC tumor tissues, including lung squamous cell carcinoma (P<0.001), lung adenocarcinoma (P<0.001) and adenosquamous carcinoma (P<0.001), compared with USP10 expression in normal lung tissues (Fig. 1I).
Figure 1.
Positive expression of USP10 protein in representative normal and NSCLC tissues: (A) Bronchial epithelium, (B) alveolar epithelium (C) squamous cell carcinoma, (D) adenocarcinoma, (E) adenosquamous carcinoma, (F) sarcomatoid carcinoma, (G) large cell carcinoma and (H) negative control. 3,3′-Diaminobenzidene staining (brown), nuclear counterstaining (hematoxylin). Original magnification, ×100. (I) Positive rate of USP10 protein between normal and lung cancer tissues. Statistical analysis was performed by χ2 test. **P<0.01. NSCLC, non-small cell lung cancer; USP10, ubiquitin-specific protease 10.
Expression of USP10 mRNA between NSCLC tissues and non-cancerous lung tissues
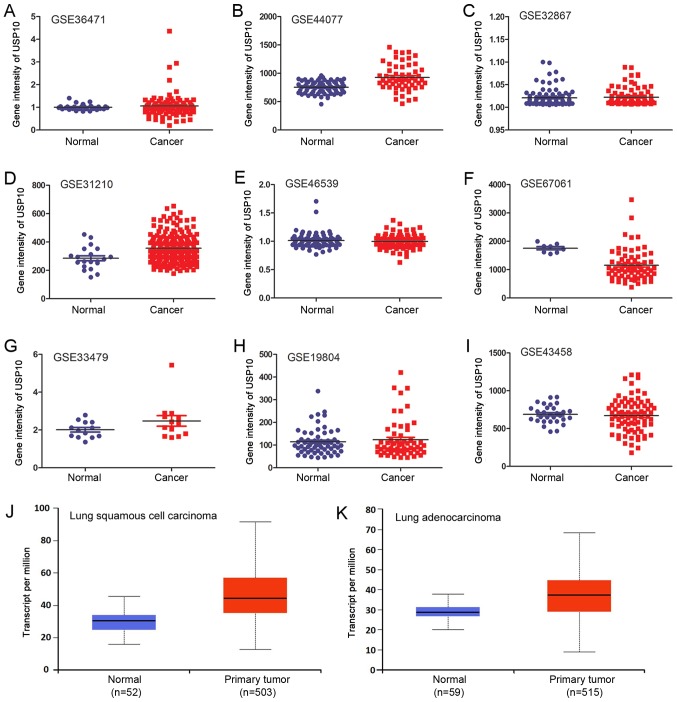
USP10 mRNA expression was investigated using bioinformatics analysis of the GEO datasets and TCGA database. A total of nine independent GSE datasets was selected (Fig. 2A-I). In total, six of the GSE datasets indicated that there was no significant difference in USP10 mRNA expression between NSCLC and normal lung tissues. Of the GSE datasets, two indicated significantly increased expression of USP10 in NSCLC tissues (P<0.05; Fig. 2B and Fig. 2D), and one indicated significantly decreased expression in NSCLC tissues (P<0.05; Fig. 2F); however, the increase or decrease was <1.5-fold in these three GSE datasets. The different probes and GPL used for the corresponding GSE are presented in Table I. Furthermore, the P-values and scatter plots between normal and NSCLC groups and the total cases in each group are presented in Table I and Fig. 2A-I, respectively. There was no significant difference in USP10 mRNA expression between the lung squamous cell carcinoma or adenocarcinoma NSCLC subtypes and normal lung tissues from TCGA database (Fig. 2J and K). Collectively, bioinformatics analysis did not reveal a significant difference in USP10 mRNA expression between NSCLC tissues and normal lung tissues.
Figure 2.
USP10 mRNA expression in normal and NSCLC tissues of (A) GSE36471, (B) GSE44077, (C) GSE32867, (D) GSE31210, (E) GSE46539, (F) GSE67061, (G) GSE33479, (H) GSE19804 and (I) GSE43458 datasets from the GEO database. Relative expression of USP10 between normal lung tissue and (J) lung squamous cell carcinoma or (K) lung adenocarcinoma from The Cancer Genome Atlas database. NSCLC, non-small cell lung cancer; USP10, ubiquitin-specific protease 10; GEO, Gene Expression Omnibus; GSE, GEO Series.
Correlation between USP10 and MSH2 protein expression
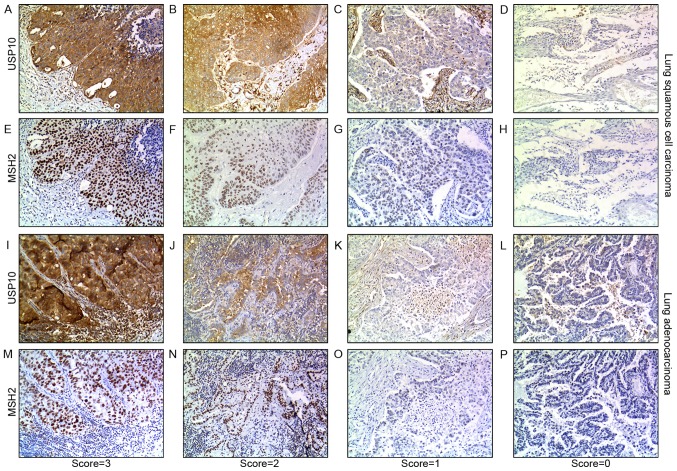
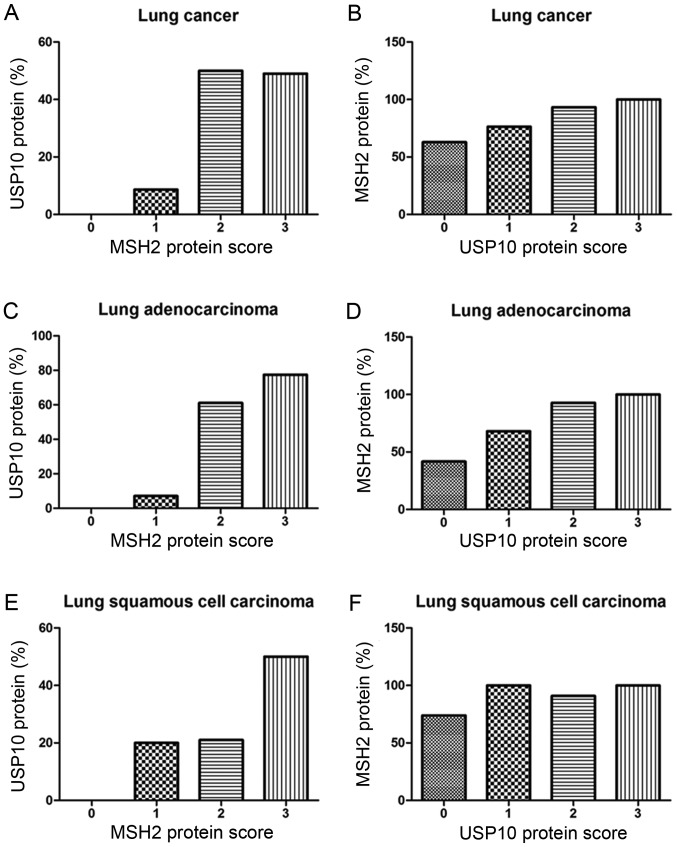
From the IHC staining of NSCLC cells, positive expression of USP10 was observed primarily in the cytoplasm, whereas the MSH2 protein was present primarily in the nucleus. Representative IHC staining specimens for different scores of USP10 or MSH2 protein expression are presented in Fig. 3, of which IHC staining specimens in Fig. 3A and E, B and F, C and G, D and H, I and M, J and N, K and O, and L and P were in the same field from the same slice. As presented in Fig. 4, the expression of MSH2 was positively correlated with the expression of USP10 (r=0.25, P=0.004). The positive expression rate of MSH2 in NSCLC was 61.90, 76.32, 93.33 and 100% with a USP10 protein score of 0, 1 2 and 3, respectively, whereas the positive expression rate of USP10 in NSCLC was 0, 8.67, 50.00 and 49.02% with an MSH2 protein score of 0, 1+, 2+ to 3+ (Fig. 4A and B). Similarly, further systematic analysis of the association between USP10 and MSH2 revealed the same trend in squamous cell carcinoma and adenocarcinoma, respectively (Fig. 4C-F). In lung adenocarcinoma, the positive expression rate of MSH2 was 41.67, 68.00, 92.86 and 100% with a USP10 protein score of 0, 1, 2 and 3, whereas the positive expression rate of USP10 was 0, 7.14, 61.11 and 77.42% with an MSH2 protein score of 0, 1+, 2+ and 3+ (Fig. 4C and D). In lung squamous cell carcinoma, the positive expression rate of MSH2 was 73.91, 100.00, 90.91 and 100.00% with a USP10 protein score of 0, 1, 2 and 3, whereas the positive expression rate of USP10 was 0, 20.00, 21.05 and 50.00% with an MSH2 protein score of 0, 1+, 2+ and 3+ (Fig. 4E and F).
Figure 3.
Immunohistochemical staining of USP10 and MSH2 proteins in representative lung squamous cell carcinoma and lung adenocarcinoma tissue specimens. (A-D) The expression of USP10 in representative tumor cells of lung squamous cell carcinoma. (A) Score=3. (B) Score=2. (C) Score=1. (D) Score=0. (E-H) The expression of MSH2 in representative tumor cells of lung squamous cell carcinoma. (E) Score=3+. (F) Score=2+. (G) Score=1+. (H) Score=0. (I-L) The expression of USP10 in representative tumor cells of lung adenocarcinoma. (I) Score=3. (J) Score=2. (K) Score=1. (L) Score=0. (M-P) The expression of MSH2 in representative tumor cells of lung adenocarcinoma (M) Score=3+. (N) Score=2+. (O) Score=1+. (P) Score=0. 3,3′-Diaminobenzidine staining (brown); nuclear counterstaining (hematoxylin). Original magnification, ×100. Score ≤1 or 1+ was defined as negative expression of USP10 or MSH2, score >1 or 1+ was defined as positive expression of USP10 or MSH2. MSH2, MutS homolog 2; USP10, ubiquitin-specific protease 10.
Figure 4.
Correlation between USP10 and MSH2 protein expression in (A and B) lung cancer, (C and D) lung adenocarcinoma and (E and F) lung squamous cell carcinoma tissues. The x-axis represents the score of USP10 or MSH2, and the y-axis represents the corresponding positive rate of MSH2 or USP10. MSH2, MutS homolog 2; USP10, ubiquitin-specific protease 10.
Association of the USP10 or MSH2 proteins in tumor cells with clinicopathological features of NSCLC
The association between USP10 protein cytoplasmic staining in tumor cells and the traditional clinicopathological features for the 148 NSCLC samples was analyzed further, as presented in Table II. There was no significant association of USP10 protein expression with the clinicopathological features, including age, sex, tumor size, TNM stage and tumor cell differentiation (P>0.05; Table II). Previous studies indicated that MSH2 was positively associated with the expression of USP10 (13); thus, the association between MSH2 protein nuclear staining in tumor cells and the traditional clinicopathological features for the 148 NSCLC samples was statistically analyzed further (Table II). Additionally, there was no significant association of MSH2 protein expression with the clinicopathological features, including age, sex, tumor size, TNM stage and tumor cell differentiation (P>0.05; Table II).
Table II.
Correlation between USP10/MSH2 expression and clinicopathological features of NSCLC.
| USP10 protein expression | MSH2 protein expression | |||||
|---|---|---|---|---|---|---|
| Clinicopathological feature | Negative, n | Positive, n (%) | P-valuea | Negative, n | Positive, n (%) | P-valuea |
| Sex | ||||||
| Male | 58 | 43 (42.57) | 0.81 | 13 | 77 (85.56) | 0.085 |
| Female | 26 | 21 (44.68) | 12 | 33 (73.33) | ||
| T stage | ||||||
| T1 | 31 | 29 (48.33) | 0.601 | 9 | 43 (82.69) | 0.553 |
| T2 | 35 | 23 (39.66) | 15 | 39 (72.22) | ||
| T3 | 9 | 4 (30.77) | 2 | 9 (81.82) | ||
| T4 | 9 | 8 (47.06) | 3 | 15 (83.33) | ||
| N stage | ||||||
| N0 | 58 | 45 (43.69) | 0.868 | 17 | 79 (82.29) | 0.974 |
| N1/N2/N3 | 26 | 19 (42.22) | 7 | 32 (82.05) | ||
| M stage | ||||||
| M0 | 82 | 64 (43.84) | − | 29 | 103 (78.03) | − |
| M1/M2/M3 | 2 | 0 (0) | 0 | 3 (100) | ||
| TNM | ||||||
| I | 47 | 35 (42.68) | 0.394 | 14 | 62 (81.58) | 0.186 |
| II | 13 | 15 (53.57) | 3 | 18 (85.71) | ||
| III/IV | 24 | 14 (36.84) | 12 | 26 (68.42) | ||
| Differentiationb | ||||||
| Well | 18 | 13 (41.94) | 0.739 | 8 | 19 (70.37) | 0.287 |
| Moderate | 35 | 30 (46.15) | 11 | 48 (81.36) | ||
| Poor | 24 | 15 (46.15) | 5 | 31 (86.11) | ||
| Age, years | 59.76±8.34 | 60.22±8.04 | 0.738c | 58.59±8.2 | 59.86±8.06 | 0.454c |
| Diameter, cm | 4.47±2.03 | 4.09±2.32 | 0.29c | 4.38±1.93 | 4.44±2.35 | 0.895c |
χ2 or Fisher's exact test.
For USP10 protein, there were 31 well-differentiated cases, 65 cases of moderate differentiation, 39 cases of poor differentiation, and 13 other cases among the 148 patients with NSCLC. For MSH2 protein, there were 27 well-differentiated cases, 59 cases of moderate differentiation, 36 cases of poor differentiation, and 13 other cases among 135 cases of patients with NSCLC.
Student's t-test. MSH2, MutS homolog 2; USP10, ubiquitin-specific protease 10; NSCLC, non-small cell lung cancer; TCGA, The Cancer Genome Atlas; TNM, Tumor-Node-Metastasis.
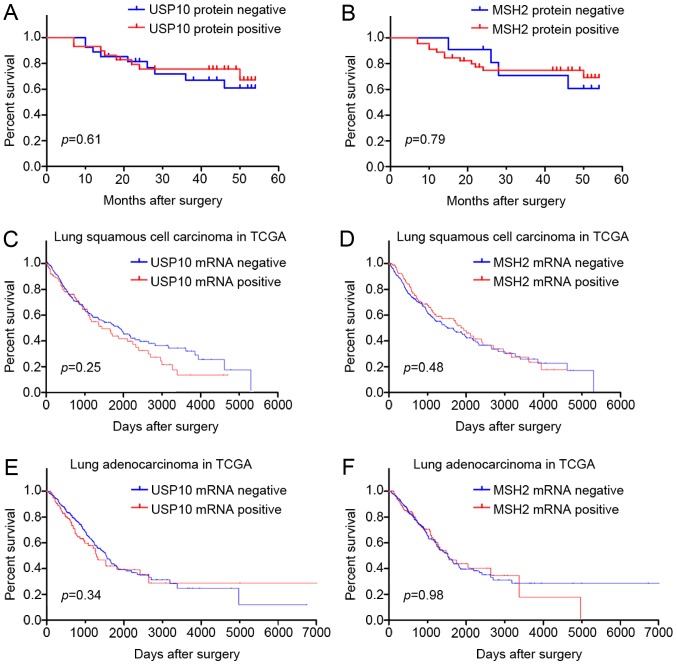
Correlation between USP10 or MSH2 protein expression and survival of patients with NSCLC
The prognostic value of USP10 or MSH2 protein for NSCLC in 56 patients was assessed using a postoperative follow-up. Kaplan-Meier analysis and log-rank testing revealed no significant difference between the USP10 or MSH2 protein negative expression group and the positive expression group (P>0.05; Fig. 5A and B). Furthermore, survival analysis from TCGA database also indicated that USP10 and MSH2 mRNA expression was not associated with survival of patients with lung squamous cell carcinoma and adenocarcinoma (Fig. 5C-F). Therefore, the survival analysis indicated that expression of USP10 and MSH2 may not serve as a biomarker for the tumor progression of NSCLC.
Figure 5.
Kaplan-Meier survival analysis of USP10 and MSH2 protein and mRNA expression in tumor cells of NSCLC (log-rank test). (A) Association between USP10 protein expression in NSCLC and overall survival (P>0.05). USP10 negative expression (n=27); USP10 positive expression (n=29). (B) Association between MSH2 protein expression in NSCLC and overall survival (P>0.05). MSH2 negative expression (n=10), MSH2 positive expression (n=46). (C) Association between USP10 mRNA expression in lung squamous cell carcinoma and overall survival, from TCGA database (P>0.05). USP10 negative expression (n=369); USP10 positive expression (n=125). (D) Association between MSH2 mRNA expression in lung squamous cell carcinoma and overall survival, from TCGA database (P>0.05). MSH2 negative expression (n=370); MSH2 positive expression (n=124). (E) Association between USP10 mRNA expression in lung adenocarcinoma and overall survival, from TCGA database (P>0.05). USP10 negative expression (n=376); USP10 positive expression (n=126). (F) Association between MSH2 mRNA expression in lung adenocarcinoma and overall survival (P>0.05), from TCGA database. MSH2 negative expression (n=375); MSH2 positive expression (n=127). MSH2, MutS homolog 2; USP10, ubiquitin-specific protease 10; NSCLC, non-small cell lung cancer; TCGA, The Cancer Genome Atlas.
Discussion
Lung cancer is a common malignant disease and the leading cause of cancer-associated mortality globally (19). NSCLC is the primary type of lung cancer, accounting for between 80 and 85% of cases (20), and is the leading cause of mortality caused by lung cancer encompassing lung adenocarcinomas, squamous cell lung carcinomas, large cell carcinomas, sarcomatoid carcinoma and adenosquamous carcinoma (21). Surgical treatment is generally considered curative for early-stage NSCLC, however, most patients present with advanced stages at diagnosis (22). Furthermore, the 5-year survival rate is only 16%, and patients have a high incidence of recurrence (23). Therefore, it is important to identify novel biomarkers for the early stages of NSCLC.
USP10 is involved in a number of biological processes by stabilizing several proteins (6,7) that serve direct or indirect functions in tumorigenesis and in the progression of several tumors. Yuan et al (3) reported that USP10, as a novel regulator of p53, provides an alternative mechanism of p53 inhibition in cancer. A number of studies also suggested increased USP10 expression in certain types of breast cancer (24) and glioblastoma (25). Lin et al (9) identified that the protein expression levels of two interacting proteins (USP10 and SIRT6) were downregulated in human colon cancer, which also indicated that gene-dysregulated USP10 function promotes tumorigenesis through SIRT6 degradation. Yang et al (8) reported that PCNA stability was regulated by USP10 and promoted cell viability by upregulating melanoma antigen family D1 in esophageal tumor tissues. A previous study indicated that a positive USP10 immunoreaction may be useful in distinguishing adrenal cortical tumors from pheochromocytomas (18). It was further demonstrated that USP10 could be used as an independent prognostic predictor for patients with gastric carcinoma (11). However, there is limited information about the functions of USP10 in human patients with NSCLC. Therefore, the aim of the present study was to clarify whether USP10 is involved in the tumorigenesis and progression of NSCLC.
The majority of lung squamous cell carcinoma originates from the bronchial epithelium, whereas type II alveolar cells and Clara cells are regarded as the possible origin of lung adenocarcinoma (26–28). Therefore, bronchial epithelium and alveolar epithelium samples were used in the present study as well-directed normal lung tissue controls.
In 2004, the World Health Organization published a new classification of lung tumors, of which the incidence of the four main types of lung cancer are as follows: 31.5% for lung adenocarcinoma, 29.4% for lung squamous carcinoma, 17.8% for small cell lung cancer, and 9.2% for large-cell lung cancer (29). Therefore, the clinical NSCLC samples enrolled in the present study were predominantly from lung adenocarcinoma and lung squamous cell carcinoma, and only these types of NSCLC sample were analyzed to determine the significant differences between NSCLC and non-cancerous lung tissues. In the IHC staining, USP10 was localized primarily to the cytoplasm in NSCLC tissues as well as in gastric carcinoma tissues (11). In the 148 patients with NSCLC enrolled in the present study, the positive rate of USP10 was 35.18 and 45.76% in squamous cell carcinoma and adenocarcinoma, respectively. By contrast, USP10 was highly expressed in bronchial epithelium and alveolar epithelium, with a positive rate of 97.78 and 95.74% (P<0.01), indicating that the expression of USP10 protein is significantly downregulated in the majority of NSCLC tumor tissues compared with non-cancerous lung tissues. This result was consistent with that of a recent study performed by Cao et al (14), which demonstrated that 50% (9/18) of patients have decreased expression of USP10 in NSCLC tumor tissue compared with that in the respective adjacent normal lung tissue. Several differences between the present study and the study of Cao et al (14) must be emphasized. First, an IHC assay was utilized in the present study to detect the USP10 protein expression, whereas a western blot assay was used in the study of Cao et al (14). In the present study, the protein expression level could be determined and the protein localization was also identified. Secondly, in the present study, the clinical sample size was enlarged to analyze the USP10 expression in NSCLC tissues. A total of 148 human NSCLC tissue samples and 139 non-cancerous lung tissues were analyzed in the present study, whereas only 18 cases were analyzed in the study of Cao et al (14). Finally, the present study emphasized elucidating the critical roles of USP10 in NSCLC; however, Cao et al (14) focused on the roles of eukaryotic translation initiation factor 4γ 1 (EIF4G1) in NSCLC, and identified that USP10 could act as a negative regulator of EIF4G1. Therefore, combining the two investigations, it is more convincing to conclude that USP10 is downregulated in NSCLC clinical tissue samples and may participate in the tumorigenesis of NSCLC.
To explore whether the mRNA level of USP10 is downregulated in human NSCLC tissues, the expression of USP10 mRNA was further analyzed in human normal lung and NSCLC tissues using bioinformatics analysis in the GEO datasets and TCGA database. A total of nine independent GSE datasets with different numbers of clinical samples and supplied by different investigators were selected to evaluate the difference in USP10 mRNA expression between normal lung tissues and NSCLC tissues. Different GPLs and the corresponding probe substrates were selected for the USP10 gene, which guaranteed the accuracy of the comprehensive results. Generally, the result of bioinformatics analysis in the GEO datasets and TCGA database revealed no significant difference in USP10 mRNA expression between normal lung tissues and NSCLC tissues. However, 3/9 GSE results exhibited a significant difference. Nevertheless, the increase and decrease were <1.5-fold in these GSE datasets. Therefore, bioinformatics analysis did not identify a significant difference in USP10 mRNA expression between normal lung tissues and NSCLC tissues, which indicated that the mechanism of downregulation of USP10 protein in the NSCLC tissues may occur at the post-transcriptional level, not at the transcriptional level. It is speculated that microRNA (miR) is involved in this process, as it was reported that miR-191 may inhibit protein levels of USP10 in pancreatic cancer (30). Additionally, it was further verified that H19-derived miR-675 is targeted at the 3′-untranslated region (UTR) of USP10 in C-kit(+) cardiac progenitor cells (31). Thus, the expression of USP10 protein will be downregulated if one of the microRNAs targets the 3′-UTR of USP10 mRNA, which requires investigation in a future study.
Previous research indicated that USP10 stabilizes and deubiquitinates MSH2 in vitro and in vivo (13). Similarly, Lin et al (9) also reported that USP10, as one of the SIRT6-interacting proteins, suppresses SIRT6 ubiquitination to protect SIRT6 from proteasomal degradation. Guturi et al (32) demonstrated that USP10 functions as a DUB that negatively regulates DNA topoisomerase II α ubiquitylation and limits its chromatin association in human breast cancer cell lines. A positive correlation was previously identified between S100A12 and USP10 protein in gastric carcinoma (33). In the present study, on the basis of previous research of the underlying molecular mechanism of interaction between USP10 and MSH2 in NSCLC cell lines (13), human clinical NSCLC tissue samples were investigated to clarify the association of these two proteins. A cohort of human NSCLC tissue samples was assessed to clarify whether there was a positive correlation between USP10 and MSH2 by evaluating the positive staining of USP10 and MSH2 in human NSCLC tissue samples utilizing IHC assays, which was consistent with the previous in vitro results (13). This result may indicate that USP10 stabilizes MSH2 in lung cancer tissues.
USP10 serves crucial functions in the tumor biological process, which is critically involved in the control of cell viability, differentiation and apoptosis (3,13). Increased USP10 expression has been detected in certain breast cancer (24) and glioblastoma (25) samples, and USP10 overexpression has been identified to be associated with a poor prognosis in patients with glioblastoma. However, USP10 has been identified to be underexpressed in a number of types of cancer, including gastric carcinoma (11). Furthermore, it was also demonstrated that the downregulated expression of USP10 indicates a worse outcome and decreased survival time in patients with gastric carcinoma, which may be used as an independent predictor of the prognosis of gastric carcinoma (11). Therefore, whether USP10 is irregularly expressed in the development and progression of NSCLC was the primary focus of the present study. The results of the present study failed to validate the clinical outcome and prognostic value of USP10 in the patients with NSCLC. No correlation was identified between USP10 protein expression and clinicopathological features, including age, sex, tumor size, TNM stage and tumor cell differentiation, indicating that USP10 may not participate in the tumor progression of NSCLC.
Multiple DNA repair pathways have previously been confirmed to be associated with tumor prognosis, drug efficacy or chemotherapeutic resistance. MSH2 in the mismatch repair pathway is associated with DNA repair following platinum insult. However, this protein was not identified to be associated with clinicopathological features of lung cancer, including smoking history, sex, age and TNM stage (34). A previous study failed to identify a correlation between MSH2 expression and prognosis in either lung squamous cell carcinoma or adenocarcinoma, which may be attributed to the limited number of clinical samples analyzed (34). Consistent with this study, the present study also failed to reveal a correlation between NSCLC prognostic value and MSH2 protein expression. Notably, a recent study indicated that MSH2/breast cancer early onset 1 (BRCA1) expression may act as a DNA-repair signature predicting survival in patients with early-stage lung cancer (35). The difference from that study is that the patients enrolled were all post-chemotherapy and in early Stage I and II. Furthermore, the two proteins were used together to predict the outcome, and only high BRCA1 and low MSH2 expression significantly predicted an improved overall survival time (35). In summary, more data are needed to support the use of MSH2 to predict NSCLC prognoses.
In conclusion, USP10 protein expression was downregulated in clinical NSCLC tissue samples compared with non-cancerous lung tissues, whereas USP10 mRNA exhibited no significant difference between normal lung tissues and NSCLC tissues. It was identified that USP10 protein expression was positively correlated with MSH2 in a large cohort of clinical NSCLC tissue samples. However, USP10 and MSH2 were not associated with clinicopathological features of NSCLC, including age, sex, tumor size, TNM stage and tumor cell differentiation. Furthermore, USP10 and MSH2 mRNA and protein expression were not associated with the prognosis of NSCLC. Thus, the detection of USP10 may be used as a biomarker to distinguish the tumorigenesis of NSCLC.
Acknowledgements
The authors would like to thank Dr Xu Zhang and Dr Liwen Yao (Renmin Hospital of Wuhan University) for collecting the samples.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81602535, 81704023 and 81803789), the scientific research project of Integrated Traditional Chinese and Western Medicine from the Health and Family Planning Commission of Hubei Province (2017–23) and the guidance funding of Renmin Hosipital of Wuhan University (RMYD2018M79).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZZ and DL performed the experiments and wrote the paper; HY and TY collected the samples and performed statistical analysis; YH and JY performed bioinformatics analysis; LG and JY designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Renmin Hospital of Wuhan University (Wuhan, China). Patients provided written informed consent for the use of their tissues in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 2.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 3.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Soncini C, Berdo I, Draetta G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene. 2001;20:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan L, Chen Z, Wang L, Chen C, Li D, Wan H, Li B, Shi G. Deubiquitination and stabilization of T-bet by USP10. Biochem Biophys Res Commun. 2014;449:289–294. doi: 10.1016/j.bbrc.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Ou C, Liu M, Xiao W, Wen C, Sun F. NRAGE promotes cell proliferation by stabilizing PCNA in a ubiquitin-proteasome pathway in esophageal carcinomas. Carcinogenesis. 2014;35:1643–1651. doi: 10.1093/carcin/bgu084. [DOI] [PubMed] [Google Scholar]
- 9.Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, Kong S, Ye J, Gao B, Fang D. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 2013;5:1639–1649. doi: 10.1016/j.celrep.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niu J, Shi Y, Xue J, Miao R, Huang S, Wang T, Wu J, Fu M, Wu ZH. USP10 inhibits genotoxic NF-κB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 2013;32:3206–3219. doi: 10.1038/emboj.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Z, Wu HX, Zhan N, Huang YB, Wang ZS, Yang GF, Wang P, Fu GH. Prognostic significance of USP10 as a tumor-associated marker in gastric carcinoma. Tumour Biol. 2014;35:3845–3853. doi: 10.1007/s13277-013-1509-1. [DOI] [PubMed] [Google Scholar]
- 12.Koul R, Rathod S, Dubey A, Bashir B, Chowdhury A. Comparison of 7th and 8th editions of the UICC/AJCC TNM staging for non-small cell lung cancer in a non-metastatic North American cohort undergoing primary radiation treatment. Lung Cancer. 2018;123:116–120. doi: 10.1016/j.lungcan.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Hu C, Tong D, Xiang S, Williams K, Bai W, Li GM, Bepler G, Zhang X. Ubiquitin-specific peptidase 10 (USP10) deubiquitinates and stabilizes MutS Homolog 2 (MSH2) to regulate cellular sensitivity to DNA damage. J Biol Chem. 2016;291:10783–10791. doi: 10.1074/jbc.M115.700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y, Wei M, Li B, Liu Y, Lu Y, Tang Z, Lu T, Yin Y, Qin Z, Xu Z. Functional role of eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) in NSCLC. Oncotarget. 2016;7:24242–24251. doi: 10.18632/oncotarget.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinen CD. Translating mismatch repair mechanism into cancer care. Curr Drug Targets. 2014;15:53–64. doi: 10.2174/1389450114666140106100128. [DOI] [PubMed] [Google Scholar]
- 16.Kamal NS, Soria JC, Mendiboure J, Planchard D, Olaussen KA, Rousseau V, Popper H, Pirker R, Bertrand P, Dunant A, et al. MutS homologue 2 and the long-term benefit of adjuvant chemotherapy in lung cancer. Clin Cancer Res. 2010;16:1206–1215. doi: 10.1158/1078-0432.CCR-09-2204. [DOI] [PubMed] [Google Scholar]
- 17.Ko JC, Chiu HC, Syu JJ, Chen CY, Jian YT, Huang YJ, Wo TY, Jian YJ, Chang PY, Wang TJ, Lin YW. Down-regulation of MSH2 expression by Hsp90 inhibition enhances cytotoxicity affected by tamoxifen in human lung cancer cells. Biochem Biophys Res Commun. 2015;456:506–512. doi: 10.1016/j.bbrc.2014.11.116. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Z, Zhou Z, Zhan N, Yuan J, Ye B, Gu L, Wang J, Jian Z, Xiong X. USP10 expression in normal adrenal gland and various adrenal tumors. Endocr Pathol. 2015;26:302–308. doi: 10.1007/s12022-015-9406-3. [DOI] [PubMed] [Google Scholar]
- 19.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 20.Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21:2213–2220. doi: 10.1158/1078-0432.CCR-14-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang H, Shrager JB. CRISPR/Cas-mediated genome editing to treat EGFR-mutant lung cancer: A personalized molecular surgical therapy. EMBO Mol Med. 2016;8:83–85. doi: 10.15252/emmm.201506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCloskey P, Balduyck B, Van Schil PE, Faivre-Finn C, O'Brien M. Radical treatment of non-small cell lung cancer during the last 5 years. Eur J Cancer. 2013;49:1555–1564. doi: 10.1016/j.ejca.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 23.Santarpia M, Rolfo C, Peters GJ, Leon LG, Giovannetti E. On the pharmacogenetics of non-small cell lung cancer treatment. Expert Opin Drug Metab Toxicol. 2016;12:307–317. doi: 10.1517/17425255.2016.1141894. [DOI] [PubMed] [Google Scholar]
- 24.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, Dong L, Tang E, Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- 25.Grunda JM, Nabors LB, Palmer CA, Chhieng DC, Steg A, Mikkelsen T, Diasio RB, Zhang K, Allison D, Grizzle WE, et al. Increased expression of thymidylate synthetase (TS), ubiquitin specific protease 10 (USP10) and survivin is associated with poor survival in glioblastoma multiforme (GBM) J Neurooncol. 2006;80:261–274. doi: 10.1007/s11060-006-9191-4. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura H, Kameda Y, Ito T, Hayashi H. Atypical adenomatous hyperplasia of the lung. Implications for the pathogenesis of peripheral lung adenocarcinoma. Am J Clin Pathol. 1999;111:610–622. doi: 10.1093/ajcp/111.5.610. [DOI] [PubMed] [Google Scholar]
- 27.Mori M, Kaji M, Tezuka F, Takahashi T. Comparative ultrastructural study of atypical adenomatous hyperplasia and adenocarcinoma of the human lung. Ultrastruct Pathol. 1998;22:459–466. doi: 10.3109/01913129809032282. [DOI] [PubMed] [Google Scholar]
- 28.Osanai M, Igarashi T, Yoshida Y. Unique cellular features in atypical adenomatous hyperplasia of the lung: Ultrastructural evidence of its cytodifferentiation. Ultrastruct Pathol. 2001;25:367–373. doi: 10.1080/019131201317101243. [DOI] [PubMed] [Google Scholar]
- 29.Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization classification of lung tumors. Semin Roentgenol. 2005;40:90–97. doi: 10.1053/j.ro.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Xu XF, Zhao Y, Tang MC, Zhou YQ, Lu J, Gao FH. MicroRNA-191 promotes pancreatic cancer progression by targeting USP10. Tumour Biol. 2014;35:12157–12163. doi: 10.1007/s13277-014-2521-9. [DOI] [PubMed] [Google Scholar]
- 31.Cai B, Ma W, Bi C, Yang F, Zhang L, Han Z, Huang Q, Ding F, Li Y, Yan G, et al. Long noncoding RNA H19 mediates melatonin inhibition of premature senescence of c-kit(+) cardiac progenitor cells by promoting miR-675. J Pineal Res. 2016;61:82–95. doi: 10.1111/jpi.12331. [DOI] [PubMed] [Google Scholar]
- 32.Guturi KK, Bohgaki M, Bohgaki T, Srikumar T, Ng D, Kumareswaran R, El Ghamrasni S, Jeon J, Patel P, Eldin MS, et al. RNF168 and USP10 regulate topoisomerase IIα function via opposing effects on its ubiquitylation. Nat Commun. 2016;7:12638. doi: 10.1038/ncomms12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Zeng Z, Yu T, Qin J, Wu J, Song JC, Zhou ZY, Yuan JP. Expression and clinical implication of S100A12 in gastric carcinoma. Tumour Biol. 2016;37:6551–6559. doi: 10.1007/s13277-015-4460-5. [DOI] [PubMed] [Google Scholar]
- 34.Xie KJ, He HE, Sun AJ, Liu XB, Sun LP, Dong XJ. Expression of ERCC1, MSH2 and PARP1 in non-small cell lung cancer and prognostic value in patients treated with platinum-based chemotherapy. Asian Pac J Cancer Prev. 2014;15:2591–2596. doi: 10.7314/APJCP.2014.15.6.2591. [DOI] [PubMed] [Google Scholar]
- 35.Levallet G, Dubois F, Fouret P, Antoine M, Brosseau S, Bergot E, Beau-Faller M, Gounant V, Brambilla E, Debieuvre D, et al. MSH2/BRCA1 expression as a DNA-repair signature predicting survival in early-stage lung cancer patients from the IFCT-0002 phase 3 trial. Oncotarget. 2017;8:4313–4329. doi: 10.18632/oncotarget.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.