Abstract
The aim of the study was to investigate the effects of miRNA-101 and miRNA-345 on HBV replication and liver cancer cell growth. qPCR was performed to detect the expression of miRNA-101 and miRNA-345. The expression of HBV RNA was detected by PCR. The expression of HbsAg was detected using ELISA. BEL-7404 cell line proliferation was detected by MTT assay. The expression levels of miR-101 and miR-345 in BEL-7404 pSUPER.neo-miR-101 group and BEL-7404 pSUPER.neo-miR-345 group were significantly higher than those in BEL-7404 pSUPER.neo group (P<0.05). The expression levels of miR-101 and miR-345 in MHCC97-L pSUPER.neo-miR-101 group and MHCC97-L pSUPER.neo-miR-345 group were significantly higher than those in MHCC97-L pSUPER.neo group (P<0.05). The expression of HBV DNA in MHCC97-L pSUPER.neo-miR-101 group was significantly lower than that in MHCC97-L pSUPER.neo group (P<0.05), and the expression of HBV DNA in MHCC97-L pSUPER.neo-miR-345 group was significantly higher than that in MHCC97-L pSUPER.neo group (P<0.05). The expression of HbsAg in MHCC97-L pSUPER.neo-miR-101 group was significantly lower than that in MHCC97-L pSUPER.neo group (P<0.05), and the expression of HbsAg in MHCC97-L pSUPER.neo-miR-345 group was significantly higher than that in MHCC97-L pSUPER.neo group (P<0.05). There was a significant difference in terms of HbsAg expression between the MHCC97-L pSUPER.neo-miR-101 and MHCC97-L pSUPER.neo-miR-345 groups (P<0.05). The proliferation of BEL-7404 cells in the BEL-7404 pSUPER.neo-miR-101 group was significantly lower than that in the BEL-7404 pSUPER.neo group (P<0.05). The proliferation of BEL-7404 cells in the BEL-7404 pSUPER.neo-miR-345 group was significantly higher than that in the BEL-7404 pSUPER.neo group (P<0.05). The proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-101 group was different from that in BEL-7404 pSUPER.neo-miR-345 group (P<0.05). miR-101 reduced the level of HBV replication, and inhibited the proliferation of liver cancer cells. miR-345 also upregulated the level of HBV replication, and promoted the proliferation of liver cancer cells.
Keywords: miR-101, miR-345, HBV replication, proliferation, expression vector
Introduction
Hepatitis B is a worldwide major infectious disease, mainly caused by hepatitis B virus (HBV) infection and manifested as liver damage (1). Previous findings have shown that there are approximately 400 million HBsAg-positive individuals in the world, of which China is a country with a high incidence of HBV, and approximately 120 million individuals are affected (2). Many liver cancers are associated with HBV infection. HBV enters the hepatocytes and destroys the DNA in the hepatocytes, causing cirrhosis of the liver and developing into liver cancer (3). Some studies have shown that the incidence of HBV-related liver cancer is approximately 80% of the incidence of liver cancer (4). Domestic and foreign scholars have conducted extensive and in-depth research on liver cancer, but the relationship between the development of liver cancer and HBV has not been studied clearly (5,6).
miRNAs are widely expressed in eukaryotic cells and regulate cell proliferation, differentiation and apoptosis, while abnormal changes in miRNA biosynthesis are involved in many pathophysiological processes (7,8). Previous findings have shown that many virus replication and proliferation are closely related to the expression level of miRNA. Wei et al reported that HBV protein can downregulate the expression of miR-101 (9). Shiu et al also found in the study that the expression of hepatitis C virus protein can enhance the expression of miR-345 (10). However, the correlation between HBV replication and miR-101 and miR-345, as well as the effects of miR-101 and miR-345 on the growth of hepatoma cells have yet to be fully elucidated.
Therefore, in this study, MHCC97-L and BEL-7404 cells were, respectively, used to replicate the HBV-associated liver cancer model and non-HBV replication liver cancer model, in order to explore the effect of miR-101 and miR-345 on HBV replication and hepatoma cell growth.
Materials and methods
BEL-7404 (STR is 5 ‘TTAGGG-3’) and MHCC97-L were both purchased from Shanghai Aolu Biotechnology Co., Ltd., Shanghai, China (cat nos. XFS3110, XFS3388, respectively) and proliferated in DEME medium (Shanghai Saily Biotechnology Co., Ltd., Shanghai, China) containing 15% fetal bovine serum. BEL-7404 and MHCC97-L cell culture condrated in DEME medium (Shanghai Saily Biotechnology Co., Ltd.; www.sailybio.com) containing 15% fetal bovine serum. BEL-7404 and MHCC97-L cell culture conditions were 37°C constant temperature, pH 6.8–7.4, and 5% CO2. The miR-101 and miR-345 expression vectors were constructed by Shanghai Genepharma Pharmaceutical Co., Ltd. (Shanghai, China), including pSUPER.neo-miR-101 and pSUPER.neo-miR-345. The constructed vector and pSUPER.neo and trypsinized BEL-7404 and MHCC97-L were added into DMEM medium, and replaced every 24 h for two weeks. The eight groups obtained were: BEL-7404 pSUPER.neo-miR-101, BEL-7404 pSUPER.neo-miR-345, BEL-7404 pSUPER.neo-101, BEL-7404 pSUPER.neo-345, MHCC97-L pSUPER.neo-miR-101, MHCC97-L pSUPER.neo-miR-345, MHCC97-L pSUPER.neo-101, and MHCC97-L pSUPER.neo-345 groups. Lipofectamine® 2000 transfection reagent was purchased from Shanghai Hengfei Biotechnology Co., Ltd., Shanghai, China (cat no. 11668019).
The study was approved by the Ethics Committee of Jinan Infectious Disease Hospital (Jinan, China).
Total cell miRNA extraction by TRIzol reagent
Total RNA was extracted from BEL-7404 and MHCC97-L cells using TRIzol reagent (Shanghai Mingjin Biotech Co., Ltd., Shanghai, China; www.mjswkj.cn). The procedure was performed according to the protocol. UV spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to analyze the concentration and purity of RNA extracted, and 3% agarose gel electrophoresis was used to analyze RNA integrity.
miRNA RT-qPCR reaction
Following extraction of total miRNA cDNA synthesis was performed by reverse transcription according to the TaqMan® MicroRNA reverse transcription kit [Thermo Fisher Scientific (China) Co., Ltd., Beijing, China]. Reaction conditions for the PCR were: 37°C for 45 min and 95°C for 5 min. The product was frozen at −20°C. A total of 20 µl cDNA was used for the amplification reaction system. Thermocycling conditions were as follows: pre-denaturation at 95°C and the results were analyzed by using the 2−∆∆Cq method (11).
Primers used for the study were: miR-101: F: 5′-TACAGTACTGTGATAACTGAA-3′, and R: 5′-CTCAACTGGTGTCGTGGA-3′; miR-345: F: 5′-GTCGTATCCAGTGCAGGGTCCGAGG-3′, and R: 5′-TATGCTGCTCGGGACCTGATCCTCA-3′; and the results were analyzed by using the 2−∆∆Cq method. Primers used for the study were: miR-101: F: 5′-TACAGTACTGTGATAACTGAA-3′, and R: 5′-CTCAACTGGTGTCGTGGA-3′; miR-345: F: 5′-GTCGTATCCAGTGCAGGGTCCGAGG-3′, and R: 5′-TATGCTGCTCGGGACCTGATCCTCA-3′; U6: F: 5′-CGCTTCGGCAGCACATATAC-3′, and R: 5′-TTCACGAATTTGCGTGTCAT-3′.
qPCR detection of HBV DNA
DNA extraction kit (Guangzhou Jianlun Biotechnology Co., Ltd.) was used to extract HBV DNA in MHCC97-L cells. The qPCR reaction system was amplified with 20 was used as an internal reference. All the samples were repeated in triplicate, and the results were analyzed by using the 2−∆Cq method. The DNA Amplification kit was purchased from Guangzhou Huayun Biotechnology Co., Ltd., Guangzhou, China was used as an internal reference. All the samples were repeated three times, and the results were analyzed by using the 2−∆∆Cq method (11). The DNA Amplification kit was purchased from Guangzhou Huayun Biotechnology Co., Ltd.
After 48 h MHCC97-L cells were transfected with Lipofectamine 2000, and the cell culture medium was collected and centrifuged at 3,200 × g for 5 min. The supernatant was taken as the ELISA test sample. The specific steps referred to the instructions of the HbsAg ELISA Test kit (Shanghai Jingkang Bioengineering Co., Ltd., Shanghai, China). Multi-functional microplate reader (Berthold LB943, Bad Wildbad, Germany) was used for readings, and the OD value of each well under the wavelength of 450 nm was measured three times.
MTT assay for the measurement of BEL-740 cells in vitro proliferation
The BEL-740 cells were prepared into singlearranged cell suspensions. The BEL-740 cells were prepared into single-arranged cell suspensions. The cells were routinely inoculated in 96-well cell culture plates with DEME+10 % FBS (fetal bovine serum). Part of the cultured cells were removed after 6 h, and the cells were added with 20 µl of MTT (5 mg/ml; Shanghai Lianmai Bioengineering Co., Ltd., Shanghai, China) and continued at 37°C. After incubation for 4 h, the supernatant containing impurities was exhausted and added with dimethylsulfoxide (DMSO) solution. After shaking for 15 min on a horizontal shaker, the absorbance at a wavelength of 570 nm was measured by an enzyme-linked immunosorbent assay (ELISA). The above steps were repeated at 12, 24, 48, and 72 h of the experiment, respectively.
Statistical analysis
SPSS 19.0 (Asia Analytics, formerly SPSS China) was used for statistical analysis. The measurement data were expressed as mean ± standard deviation. The non-parametric Kolmogorov-Smirnov test was used to compare the data that did not fit the normal distribution between the two groups. The t-test was used to compare the data with the normal distribution. P<0.05 was considered to indicate a statistically significant difference.
Results
RT-qPCR results
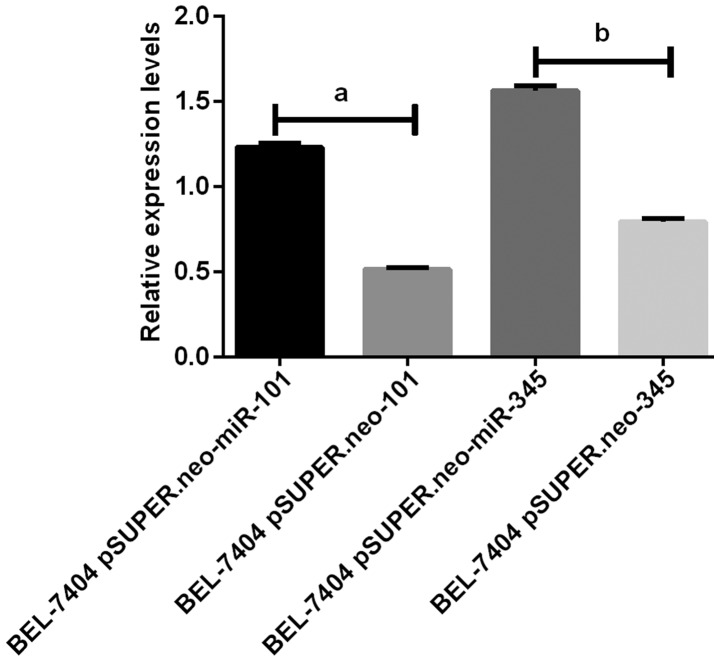
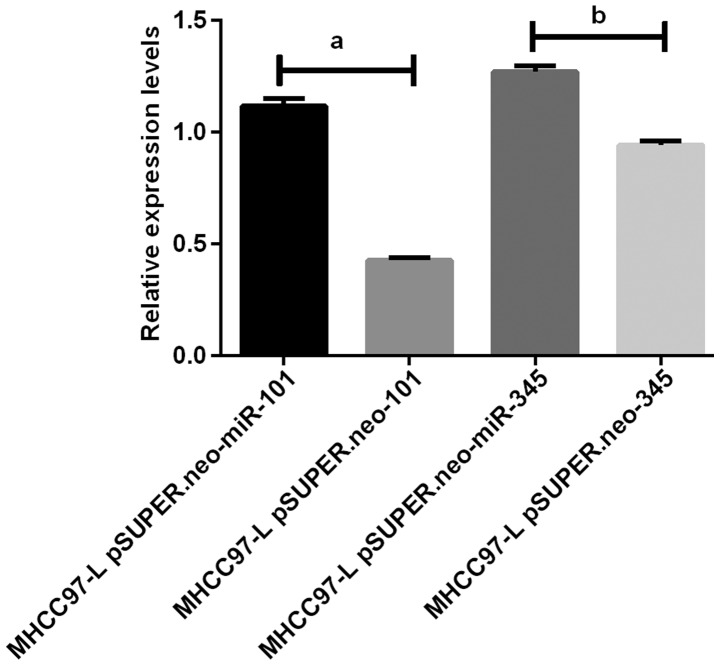
The results of RT-PCR amplification of miR-101 and miR-345 in BEL-740 and MHCC97-L cells showed that the expression of miR-101 and miR-345 in the BEL-740 pSUPER.neo-miR-101 and BEL-740 pSUPER.neo-miR-345 groups were significantly higher than those in the BEL-740 pSUPER.neo-101 and BEL-740 pSUPER.neo-345 groups (P<0.05). The expression levels of miR-101 and miR-345 in the MHCC97-L pSUPER.neo-miR-101 and MHCC97-L pSUPER.neo-miR-345 groups were significantly higher than those in the MHCC97-L pSUPER.neo-101 and MHCC97-L pSUPER.neo-345 groups (P<0.05) (Figs. 1 and 2).
Figure 1.
Results of RT-PCR amplification of BEL-7404 cells miR-101 and miR-345. The results of RT-PCR show that the expression levels of miR-101 and miR-345 in BEL-7404 pSUPER.neo-miR-101 group and BEL-7404 pSUPER.neo-miR-345 group were significantly higher than those in BEL-7404 pSUPER.neo group (P<0.05). aP<0.05. bP<0.05.
Figure 2.
Results of RT-PCR amplification of miR-101 and miR-345 in MHCC97-L cells. The results of RT-PCR show that the expression levels of miR-101 and miR-345 in MHCC97-L pSUPER.neo-miR-101 group and MHCC97-L pSUPER.neo-miR-345 group were significantly higher than those in MHCC97-L pSUPER.neo group (P<0.05). aP<0.05. bP<0.05.
Results of qPCR detection of HBV DNA
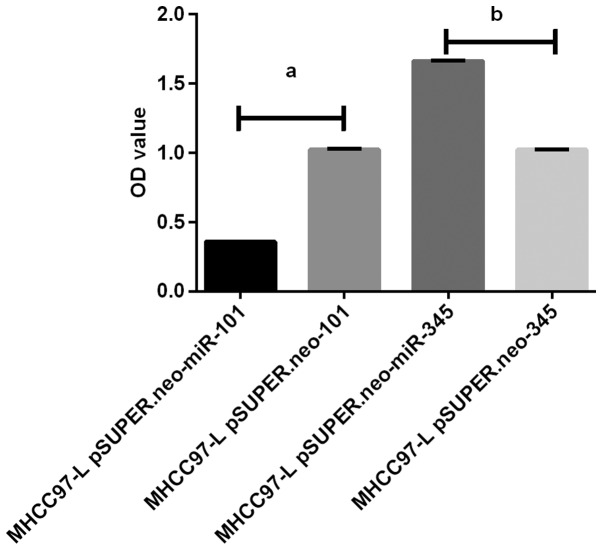
HBV DNA qPCR amplification results showed that HBV DNA expression level in MHCC97-L pSUPER.neo-miR-101 group was significantly lower than that in MHCC97-L pSUPER.neo-101 group (P<0.05). The HBV DNA level in MHCC97-L pSUPER.neo-miR-345 group was significantly higher than that in MHCC97-L pSUPER.neo-345 group (P<0.05). The HBV DNA level in MHCC97-L pSUPER.neo-miR-101 group was significantly different from that in MHCC97-L pSUPER.neo-miR-345 group (P<0.05) (Fig. 3).
Figure 3.
Results of PCR amplification of HBV DNA. The results of qPCR amplification showed that the expression of HBV DNA in MHCC97-L pSUPER.neo-miR-101 group was significantly lower than that in MHCC97-L pSUPER.neo group (P<0.05), and the expression level of HBV DNA in MHCC97-L pSUPER.neo-miR-345 group was significantly higher than that in MHCC97-L pSUPER.neo group (P<0.05). In addition, MHCC97-L pSUPER.neo-miR-101 group was significantly different from that in MHCC97-L pSUPER.neo-miR-345 group (P<0.05). aP<0.05. bP<0.05.
ELISA detection of HbsAg
The results of HbsAg ELISA showed that the expression level of HbsAg in MHCC97-L pSUPER.neo-miR-101 group was significantly lower than that in MHCC97-L pSUPER.neo-101 group (P<0.05). The expression of HbsAg in MHCC97-L pSUPER.neo-miR-345 group was significantly higher than that in MHCC97-L pSUPER.neo-345 group (P<0.05). The expression of HbsAg in MHCC97-L pSUPER.neo-miR-101 group was significantly different from that in MHCC97-L pSUPER.neo-345 group (P<0.05) (Fig. 4).
Figure 4.
Results of ELISA test for HbsAg level. The results of HbsAg ELISA showed that the expression level of HbsAg in MHCC97-L pSUPER.neo-miR-101 group was significantly lower than that in MHCC97-L pSUPER.neo group (P<0.05), and HbsAg expression in MHCC97-L pSUPER.neo-miR-345 group was significantly higher than that in MHCC97-L pSUPER.neo group (P<0.05). In addition, the expression level of HbsAg was different between in MHCC97-L pSUPER.neo-miR-101 group and in MHCC97-L pSUPER.neo-miR-345 group (P<0.05). aP<0.05. bP<0.05.
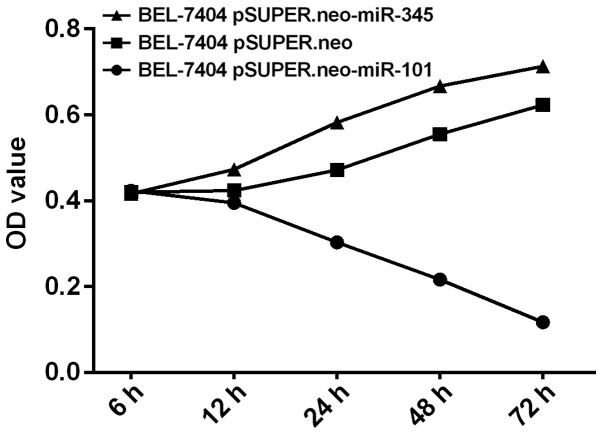
BEL-7404 cell proliferation in vitro results using MTT assay
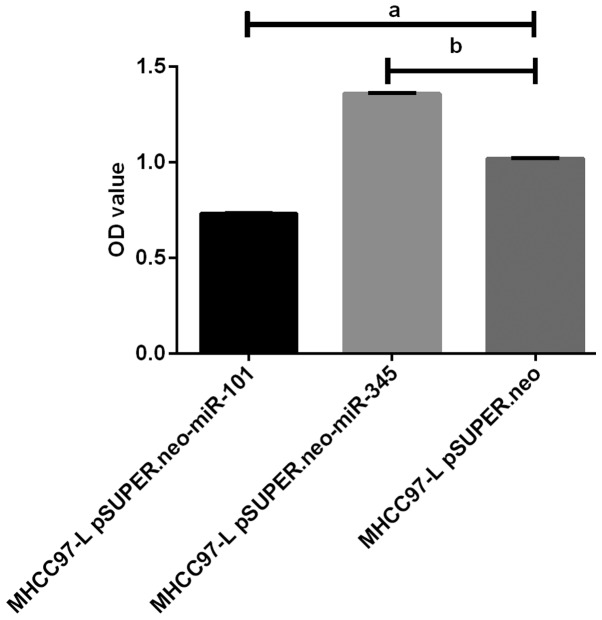
The proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-101 group was significantly lower than that in BEL-7404 pSUPER.neo-101 group (P<0.05) by MTT assay, and showed a gradual downward trend. The proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-345 group was significantly higher than that in BEL-7404 pSUPER.neo-345 group (P<0.05), and showed a gradual upward trend. The proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-101 group was different from that in BEL-7404 pSUPER.neo-miR-345 group (P<0.05) (Fig. 5).
Figure 5.
Results of MTT assay on BEL-7404 cell proliferation. The proliferation of BEL-7404 cells in vitro by MTT assay showed that the proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-101 group was significantly lower than that in BEL-7404 pSUPER.neo group (P<0.05). The proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-345 group was significantly higher than that in BEL-7404 pSUPER.neo group (P<0.05). The proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-101 group was different from that in BEL-7404 pSUPER.neo-miR-345 group (P<0.05).
Discussion
HBV is a type of hepadnavirus that has a very high mutation rate; although it can be controlled, it cannot be completely eliminated, and it is one of viruses which is the most difficult to cure under present medical technology (1). HBV infection is the main cause of liver cancer. Long-term HBV infection stimulates the body to produce a sustained immune response, causing liver immune damage, progressing into cirrhosis and eventually liver cancer (12,13). HBV DNA can also be integrated into the genome of hepatocytes by reverse transcription reactions, causing mutations in the gene and inducing liver cancer (14). Therefore, in this study, the effects of miR-101 and miR-345 on HBV replication and hepatoma cell proliferation were investigated, in order to provide clinical help for the treatment of HBV and HBV-related liver cancer.
In this study, MHCC97-L cells and BEL-7404 cells were respectively used to replicate the HBV-associated liver cancer model and HBV-free liver cancer model, in order to explore the relationship between miR-101, miR-345 and HBV replication and liver cancer cell proliferation.
In this experiment, the expression of miR-101 and miR-345 in MHCC97-L cells were regulated by the construction of pSUPER.neo-miR-101 and pSUPER.neo-miR-345 expression vectors. RT-qPCR results showed that the miR-101 expression in MHCC97-L pSUPER.neo-miR-101 group and the miR-345 expression in MHCC97-L pSUPER.neo-miR-345 groups was higher than that in MHCC97-L pSUPER.neo group (P<0.05), indicating that the expression vector was successfully constructed, and the vectors successfully regulated the expression level of miR-101 and miR-345 in MHCC97-L cells. We then examined the HBV DNA, and qPCR results showed that the expression of HBV DNA in MHCC97-L pSUPER.neo-miR-101 group was significantly lower than that in MHCC97-L pSUPER.neo group, but MHCC97-L pSUPER.neo-miR-345 group HBV DNA expression was significantly higher than that in MHCC97-L pSUPER.neo group, indicating that upregulation of miR-101 expression can reduce the expression of HBV DNA, thereby inhibiting HBV replication, and upregulation of miR-345 expression can increase the level of HBV DNA expression, thereby promoting HBV replication. The results of ELISA also showed that the expression level of HbsAg in MHCC97-L pSUPER.neo-miR-101 group was lower than that in MHCC97-L pSUPER.neo group, while the expression level of HbsAg in MHCC97-L pSUPER.neo-miR-345 group was higher than that in MHCC97-L pSUPER.neo group. Although there are some studies on the relationship between miR-101 and virus replication, the focus is the effect of virus replication on the expression level of miR-101.
As Sheng et al (15) reported in their study, HBV can downregulate the expression of miR-101, and promote the proliferation of liver cancer cells through Rab5a. Only Zheng et al (16) reported that miR-101 inhibits herpes simplex virus-1 replication by targeting ATP5B. This is in line with our findings. We did not examine ATP5B levels, but we speculate that the targeted regulation of miR-101 on ATP5B may also be a mechanism by which miR-101 regulates HBV replication. We hope to discuss this issue in future studies. However, most studies tend to examien hepatitis C virus (HCV) on the relationship between miR-345 and viral replication. No studies have yet been found on the relationship between miR-345 and HBV. Zhang et al (17) found that miR-345 expression levels in plasma of HCV patients was 2–3-fold higher than normal, which is similar to our result. Therefore, the expression levels of miR-101 and miR-345 are closely related to HBV replication.
This experiment also transfected pSUPER.neo-miR-101 and pSUPER.neo-miR-345 expression vector in BEL-7404 cells. RT-qPCR results showed that miR-101 level in BEL-7404 pSUPER.neo-miR-101 group and miR-345 expression level in BEL-7404 pSUPER.neo-miR-345 group were higher than those in BEL-7404 pSUPER.neo group (P<0.05); thus, miR-101 and miR-345 expression levels in BEL-7404 cells were successfully regulated. The results of MTT test showed that the proliferation of BEL-7404 cells in BEL-7404 pSUPER.neo-miR-101 group and BEL-7404 pSUPER.neo-miR-345 group was significantly lower than that in BEL-7404 pSUPER.neo group (P<0.05). The results showed that the upregulation of miR-101 and miR-345 could reduce the proliferation of BEL-7404 cells. Shen et al (18) showed that miR-101 acts as a tumor suppressor by directly targeting lipoprotein kinase in liver cancer. This is similar to our results, in which the upregulation of miR-101 can inhibit liver cancer cell proliferation. Srivastava et al (19) also reported that the downregulation of miR-345 expression level is conducive to pancreatic cancer cell apoptosis. Thus, the expression levels of miR-101 and miR-345 are closely related to the proliferation of hepatoma cells.
Du et al (20) showed that HBV replication levels were closely related to HBV-related hepatoma cell proliferation; reducing the level of HBV replication can reduce the proliferation of liver cancer cells. Therefore, through the transfection assay on MHCC97-L cell and BEL-7404 cells, we can speculate that upregulating the expression of miR-101 or downregulating the expression of miR-345 can reduce the HBV replication, and thus further reduce the level of liver cancer cells, which has a positive meaning on the treatment of HBV-related liver cancer. However, we do not know whether miR-101 and miR-345 have the same effect on human, thus we need to obtain further data in the clinic to verify our results.
In summary, miR-101 can reduce the level of HBV replication and inhibit the proliferation of hepatoma cells. In addition, miR-345 can upregulate the level of HBV replication and promote the proliferation of liver cancer cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
AD and LZ performed PCR and MTT assay. CW helped with total cell miRNA extraction and cell culture. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Jinan Infectious Disease Hospital (Jinan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liang X, Bi S, Yang W, Wang L, Cui G, Cui F, Zhang Y, Liu J, Gong X, Chen Y, et al. Reprint of: Epidemiological serosurvey of Hepatitis B in China - declining HBV prevalence due to Hepatitis B vaccination. Vaccine. 2013;31(Suppl 9):J21–J28. doi: 10.1016/j.vaccine.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, Washington MK, Germanidis G, Flaherty JF, Schall Aguilar R, et al. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: A 5-year open-label follow-up study. Lancet. 2013;381:468–475. doi: 10.1016/S0140-6736(12)61425-1. [DOI] [PubMed] [Google Scholar]
- 3.Ringelhan M, Heikenwalder M, Protzer U. Direct effects of hepatitis B virus-encoded proteins and chronic infection in liver cancer development. Dig Dis. 2013;31:138–151. doi: 10.1159/000347209. [DOI] [PubMed] [Google Scholar]
- 4.Singh S, Fujii LL, Murad MH, Wang Z, Asrani SK, Ehman RL, Kamath PS, Talwalkar JA. Liver stiffness is associated with risk of decompensation, liver cancer, and death in patients with chronic liver diseases: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2013;11(1573–1584):e1571–1572. doi: 10.1016/j.cgh.2013.07.034. quiz e1588-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ringelhan M, O'Connor T, Protzer U, Heikenwalder M. The direct and indirect roles of HBV in liver cancer: Prospective markers for HCC screening and potential therapeutic targets. J Pathol. 2015;235:355–367. doi: 10.1002/path.4434. [DOI] [PubMed] [Google Scholar]
- 6.Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, Co NN, Chan AW, Li PS, Lung RW, et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Zhang G, Wu Z, Lu B, Yuan D, Li X, Lu Z. MicoRNA-451 is a novel tumor suppressor via targeting c-myc in head and neck squamous cell carcinomas. J Cancer Res Ther. 2015;11(Suppl 2):C216–C221. doi: 10.4103/0973-1482.168189. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y, Mu N. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann Clin Biochem. 2017;54:127–133. doi: 10.1177/0004563216649377. [DOI] [PubMed] [Google Scholar]
- 9.Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, Wu Z. miR-101 is down-regulated by the hepatitis B virus × protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25:439–446. doi: 10.1016/j.cellsig.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Shiu TY, Huang SM, Shih YL, Chu HC, Chang WK, Hsieh TY. Hepatitis C virus core protein down-regulates p21(Waf1/Cip1) and inhibits curcumin-induced apoptosis through microRNA-345 targeting in human hepatoma cells. PLoS One. 2013;8:e61089. doi: 10.1371/journal.pone.0061089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−∆∆C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Ozen C, Yildiz G, Dagcan AT, Cevik D, Ors A, Keles U, Topel H, Ozturk M. Genetics and epigenetics of liver cancer. N Biotechnol. 2013;30:381–384. doi: 10.1016/j.nbt.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Chen WQ, Zheng RS, Zhang SW. Liver cancer incidence and mortality in China, 2009. Chin J Cancer. 2013;32:162–169. doi: 10.5732/cjc.013.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu C, Chen T, Fan C, Zhan Q, Wang Y, Lu J, Lu LL, Ni Z, Huang F, Yao H, et al. Efficacy of neonatal HBV vaccination on liver cancer and other liver diseases over 30-year follow-up of the Qidong hepatitis B intervention study: A cluster randomized controlled trial. PLoS Med. 2014;11:e1001774. doi: 10.1371/journal.pmed.1001774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng Y, Li J, Zou C, Wang S, Cao Y, Zhang J, Huang A, Tang H. Downregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5a. Arch Virol. 2014;159:2397–2410. doi: 10.1007/s00705-014-2084-5. [DOI] [PubMed] [Google Scholar]
- 16.Zheng SQ, Li YX, Zhang Y, Li X, Tang H. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antiviral Res. 2011;89:219–226. doi: 10.1016/j.antiviral.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Xu M, Qu Y, Li Z, Zhang Q, Cai X, Lu L. Analysis of the differential expression of circulating microRNAs during the progression of hepatic fibrosis in patients with chronic hepatitis B virus infection. Mol Med Rep. 2015;12:5647–5654. doi: 10.3892/mmr.2015.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Q, Bae HJ, Eun JW, Kim HS, Park SJ, Shin WC, Lee EK, Park S, Park WS, Lee JY, et al. MiR-101 functions as a tumor suppressor by directly targeting nemo-like kinase in liver cancer. Cancer Lett. 2014;344:204–211. doi: 10.1016/j.canlet.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava SK, Bhardwaj A, Arora S, Tyagi N, Singh S, Andrews J, McClellan S, Wang B, Singh AP. MicroRNA-345 induces apoptosis in pancreatic cancer cells through potentiation of caspase-dependent and -independent pathways. Br J Cancer. 2015;113:660–668. doi: 10.1038/bjc.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.