Abstract
Resveratrol (RES) is a polyphenol produced by certain plant species that has been well studied due to its ability to slow the growth of cancer cells. In numerous cell types and tissues, RES has been demonstrated to promote mitochondrial biogenesis, fusion, and oxidative phosphorylation. The present study investigated the interaction between RES's effects on growth and metabolism in PC3 prostate cancer cells, and demonstrated that RES-mediated growth inhibition is only observed under conditions in which a metabolic shift from glucose fermentation to mitochondrial respiration can occur. When this shift was prevented by growing cells in galactose medium or by pharmacologically inhibiting prolyl hydroxylase (PHD) in order to stabilize hypoxia inducible factor-1α, RES did not effect mitochondrial fusion, biogenesis, respiration or cell growth. Similar results were observed in PC3 cells expressing a mutant HIF-1α lacking the prolines that are hydroxylated by PHD to promote its degradation. Thus, RES appears to slow PC3 cell growth by interfering with glucose fermentation and promoting respiration. Consistent with this, RES was observed to be particularly effective at inhibiting PC3 cell growth under hypoxic conditions that precluded increased reliance on oxidative phosphorylation. These observations are important in understanding how RES may affect cancer cell growth in vivo where hypoxia is common in growing tumours.
Keywords: resveratrol, mitochondria, cancer, hypoxia-inducible factor, HIF-1, fusion, fission, bioenergetics, mitochondrial dynamics
Introduction
Cancer cells use exceptionally high rates of glucose catabolism to produce ATP, NADPH, and anabolic building blocks needed for the production of daughter cells, a metabolic phenotype termed ‘the Warburg effect’ (1–3). Although the Warburg effect was originally attributed to mitochondrial defects compromising oxidative phosphorylation, it is now appreciated to occur in cells with competent oxidative phosphorylation (4). Strategies targeting the Warburg effect by inhibiting glucose fermentation and shifting cellular metabolism toward oxidative phosphorylation can slow the growth of cancer cells (5,6). Since hypoxia inducible factor-1 plays a key role in establishing the Warburg phenotype, this protein in particular is an important target for inhibiting cancer growth (7,8).
The plant polyphenol resveratrol (RES) inhibits cell cycle progression (9) and thus proliferative growth in many cancer cell lines (10). Coincident with these growth inhibitory effects, RES promotes mitochondria network fusion (11), mitochondrial biogenesis, and oxidative phosphorylation (12) in a variety of cancer cell lines. Thus, RES appears to reverse the Warburg effect in many cancer cells. RES has also been reported to reduce HIF-1α expression in vitro (13) and in vivo (14), suggesting that this could underlie the metabolic phenotype.
Here we used PC3 prostate cancer cells, in which RES inhibits growth and reduces HIF-1α levels to investigate the interactions between RES's effects on the metabolic phenotype and growth. We show that the inhibition of PC3 cell growth by treatment with 10 µM RES is coincident with increased mitochondrial network fusion, biogenesis, and respiration. We then prevented RES-induced metabolic switching from the glycolytic to oxidative metabolism by growing cells in galactose medium, which does not support glucose fermentation (15) or stabilizing HIF-1α using pharmacological or genetic approaches. Under these conditions, the effects of RES on growth and metabolism were either attenuated or abolished. These observations suggest that the metabolic and growth effects of RES on PC3 cells are inter-dependent. The ability of RES to reduce HIF-1α levels suggests that it could be particularly effective in hypoxic conditions, as are common in growing tumours. Indeed, RES inhibited PC3 cell growth more strongly and at lower concentrations under hypoxic conditions. These observations have important implications for understanding how RES may inhibit cancer growth under conditions prevailing in vivo.
Materials and methods
Materials
Dulbecco's modified Eagle's medium (DMEM) high glucose (4,500 mg/l) containing L-glutamine, sodium pyruvate and sodium bicarbonate (cat. no. D6429), supplement-free DMEM powdered media (cat. no. D5030), fetal bovine serum (cat. no. F1051), non-essential amino acids, penicillin/streptomycin solution, 0.25% trypsin/EDTA solution, bovine serum albumin (BSA), Glucose Oxidase from Aspergillus niger (cat. no. G2133), Horseradish Peroxidase (2KU; cat. no. P6140), Bradford reagent (cat. no. B6916), IOX2 (cat. no. SML0652), MG132 (cat. no. C2211) and deferoxamine mesylate salt (DFO; cat. no. D9533) were obtained from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). trans-Resveratrol (Product no. 70675) and Amplex Red reagent were purchased from Cayman Chemical (Ann Arbor, MI, USA). Dimethylsufoxide (DMSO), DL-dithiothreitol (DTT), Bradford reagent, D-galactose, L-glutamine, HEPES [(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] and Trypan blue were obtained from BioShop (Burlington, ON, Canada). Tissue culture dishes (100×20 mm and 60×15 mm) and cell scrapers were obtained from Sarstedt, Inc (Newton, SC, USA). MitoTracker Red CMXRos and Lipofectamine 2000 transfection reagent was purchased from Life Technologies Incorporated (Burlington, ON, Canada). Mouse anti-human HIF-1α (Product no. 610958; Lot no. 5174837 and Product no. 610959; Lot no. 4073775) was obtained from BD Biosciences (Franklin Lakes, NJ, USA). Rabbit polyclonal Lamin B1 antibody (Product no. ab16048; Lot no. GR-263244-1) was obtained from Abcam (Cambridge, UK). Ham's F12 Nutrient Mix powdered media (cat. no. 21700-075) and Alexa Fluor® 647-conjugated secondary (IgG) antibody against mouse (cat. no. A-31571; Lot no. 1069838) were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). HA-HIF1α-pcDNA3 (plasmid no. 18949), and HA-HIF1α P402A/P564A-pcDNA3 (plasmid no. 18955) were purchased from Addgene (Cambridge, MA, USA). Human HIF-1α recombinant protein (cat. no. GWB-184E1F) was purchased from GenWay Biotech, Inc. (San Diego, CA, USA).
Cell lines and culture conditions
PC3, LNCaP, C2C12, and SHSY5Y cell lines were acquired from ATCC and cultured in high glucose DMEM unless otherwise indicated. For experiments performed in galactose medium, cells grown in glucose were harvested and then seeded in glucose-free DMEM supplemented with 10 mM galactose, 6 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate for at least 3 days before starting experiments. Cells were cultured at 37°C in humidified 5% CO2 atmosphere, 18% O2 atmosphere, unless otherwise indicated. Cell density and population doubling time were determined by hemocytometer counting using trypan blue exclusion to identify live cells. Treatments were added directly to the culture media for 48 h. Media and treatments were refreshed every day.
Hypoxia conditions
For atmospheric hypoxia experiments, cells were grown within a humidified hypoxia modulator incubator chamber (MIC-101; Billups-Rothenberg, Del Mar, CA, USA) maintained at ≤0.4 O2%, 5% CO2 with the aid of a Roxy-1 Universal O2 Controller (Sable Systems, North Las Vegas, NV, USA). Culture media was refreshed daily with pre-conditioned hypoxic media kept in the same chamber. Alternatively, in some experiments (where indicated) ‘pseudo-hypoxia’ was induced through chemical means via treatment with the PHD inhibitor IOX2 (25 µM). Cells were incubated overnight with IOX2 prior to commencing experiments of interest.
Preparation of whole cell lysates
Cells were harvested and washed in PBS then lysed in ice cold lysis buffer (10 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM DTT, 40% glycerol (v/v), 0.5% (v/v) NP40) with either periodic vortexing or sonication (Sonicator W-375; setting 3; Heat Systems Ultrasonics Inc., Wehnrath, Germany) for 30 min on ice. Subsequently, lysates were centrifuged at 10,000 × g at 4°C for 10 min (IEC, Micromax/Micromax RF; Thermo Fisher Scientific, Inc.) and the pellet discarded. The protein concentration of the resulting supernatant was determined by the Bradford method. BSA was used as the protein standard. Cell lysates were stored at −80°C.
Preparation of nuclear lysates
Cells were lysed on the plate with cold homogenization buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 2 mM DTT, 2 mM MG132, 200 µM DFO), with subsequent centrifugation at 450 g for 5 min. After centrifugation, the cell pellet was resuspended in 200 µl homogenization buffer and disrupted with the aid of a pre-chilled glass pestle and tube. The homogenate was then centrifuged for 10 min at 10,000 × g at 4°C and the pellet was gently re-suspended in 150 µl of extraction buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 1.4 M KCl, 0.2 mM EDTA, 25% glycerol, 2 mM dithiothreithol, 2 mM MG132, 200 µM DFO), followed by a 30 min incubation on ice in an orbital shaker (Madell Corporation, USA). After incubation, cell lysates were centrifuged at 15,000 × g (4°C) for 30 min (IEC Micromax/Micromax RF; Thermo Fisher Scientific, Inc.). The supernatant was then transferred to a fresh centrifuge tube and stored at −80°C.
Lactate Dehydrogenase (LDH) activity
LDH activity was measured in a solution containing 20 mM HEPES buffer (pH 7.3), 0.2 mM NADH, 10 mM pyruvate and 5 µg of sample protein. The conversion of NADH to NAD+ after pyruvate addition was followed spectrophotometrically (340 nm) using a Varian Cary 100 UV-visible spectrophotometer (Agilent Technologies, Inc., Santa Clara, CA, USA). Assays were performed at 30°C.
Western blots
Nuclear lysates were electrophoresed in a 6% SDS-PAGE gel and electroblotted onto a polyvinylidene fluoride membrane using a Bio-Rad Trans-Blot semi-dry transfer apparatus. Membranes were blocked for an hour at room temperature and incubated overnight at 4°C independently with primary antibodies to HIF-1α (1:100; w/v) or the nuclear loading control Lamin B1 (1:500; w/v). Incubations with either an Alexa Fluor 647-conjugated anti-mouse antibody (1:500; w/v) or an infrared fluorophore-conjugated anti-rabbit antibody (1:1,000; w/v) were performed at room temperature (2 h). Membranes were visualized using a VersaDoc MP 4000 Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Glucose oxidase activity
Cellular glucose uptake was assessed by observing the generation of Amplex Red reagent red-fluorescent oxidation product, resorufin, in a Varian Cary Eclipse fluorescence spectrophotometer equipped with a microplate reader (Agilent Technologies, Inc.). Briefly, 5×105 cells were seeded in high glucose DMEM and then treated with either RES or DMSO for 48 h at 0.4% O2 (hypoxia). After 48 h the cells were incubated under the same conditions for 5 h in a 5 mM glucose media. A sample of the media was then mixed with a solution containing HEPES buffer (20 mM, pH 7.3), Glucose oxidase from Aspergillus niger (0.1 U/ml), Horseradish peroxidase (0.2 U/ml) and Amplex Red (50 µM). The formation rate of resorufin was continuously measured for a period of 3 min by using excitation and emission wavelengths of 535 nm and 595 nm, respectively. Maximal reaction rates [Arbitrary Fluorescence Units (AFU) per minute] were calculated during the reaction linear range by using the Cary Eclipse Kinetics software (Walnut Creek, CA, USA). For each experiment a standard curve was used to quantify glucose concentration in media. At the end of each experiment, cells were counted in order to assess cell density and glucose uptake was converted into nmol glucose·106 cells−1·min−1.
Plasmid DNA transfections
HA-HIF1α P402A/P564A-pcDNA3 (Addgene plasmid no. 18955) and HA-HIF1α-pcDNA3 (Addgene plasmid no. 18949) were gifts from William Kaelin. pC1-Hyper-3 (Addgene plasmid no. 42131) was a gift from Vsevolov Belousov. Plasmid DNA was initially isolated and purified from bacterial cultures via a plasmid DNA Miniprep kit (Norgen Biotek Corp., Thorold, ON, Canada). Plasmid DNA purity (260 nm/280 nm absorbance ratio) and concentration were assessed by using a NanoPhotometer (Montreal Biotech Inc., Dorval, QC, Canada) Transfections were performed with Lipofectamine 2000 reagent in accordance with manufacturer's instructions. PC3 cells stably expressing HA-HIF1α P402A/P564A-pcDNA3 or HA-HIF1α-pcDNA3 were selected and maintained with G418.
Fluorescence microscopy and image analysis
Fluorescence images using structured illumination of live PC3 cells were obtained using a Zeiss Axio Observer. Z1 inverted light/epifluorescence microscope equipped with ApoTome.2 optical sectioning and a Hamamatsu ORCA-Flash4.0 V2 digital camera. Cells were cultured on MatTek 35 mm poly-D-lysine-coated glass bottom culture dishes with phenol red-free culture media and were viewed with a Plan-Apochromat 63×/1.40 oil objective. The microscope stage was maintained at 37°C and 5% CO2 (O2 levels were not regulated). MitoTracker Red CMXRos signal was imaged using excitation and emission wavelengths of 587 nm and 610 nm, respectively. Mitochondrial network morphology in MitoTracker Red-labelled PC3 cells were quantitatively analyzed using the ImageJ tool ‘MiNA’ (16).
Cell cycle analysis
Approximately 5×105 cells were harvested via trypsinization and centrifugation (5 min at 240 × g). The resulting pellets were washed once with PBS and then fixed via drop-wise addition of ice-cold ethanol (75% v/v) with routine vortexing. After an overnight incubation at −20°C, the suspension was centrifuged (5 min at 240 × g) and washed twice with ice-cold PBS. The fixed cells were then incubated with 0.5 ml Propidium Iodide (PI)/RNase Staining Buffer (BD Pharmingen, San Diego, CA, USA) in darkness at room temperature for 15 min. The DNA content was immediately measured using a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA). The PI signal was detected using a 562–588 nm band pass filter. For each sample, 100,000 events were recorded using a medium data acquisition rate, as per the manufacturer guidelines. The percentages of cells in G0-G1, S, and G2-M phases were determined using the CFlow Plus software (BD Biosciences).
Cellular respiration measurements
Oxygen consumption rates of intact cultured cells were measured using a Clark-type oxygen electrode (Rank Brothers Dual Digital Model 20 Respirometer; Bottisham, UK) within a chamber maintained at 37°C. Cells were harvested via trypsinization and centrifugation before being re-suspended in 1 ml of complete culture media. The chamber was capped and the rate of oxygen consumption was recorded via an attached polygraph unit. All respiration rates were converted to nanomoles O2 consumed·minute−1·106 cells−1.
Statistics
Where T-tests were performed, they were done using Microsoft Excel (Redmond, USA). t-tests (unpaired, two-tailed) were used for data sets containing two experimental groups. P<0.05 was considered to indicate a statistically significant difference. For data sets including more than two experimental groups, ANOVA followed by Bonferroni comparisons of means were performed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
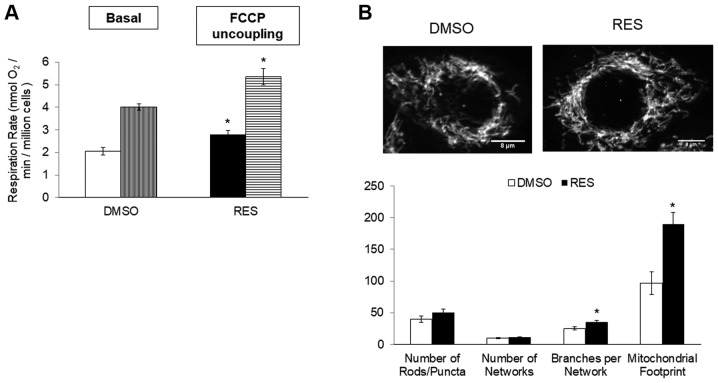
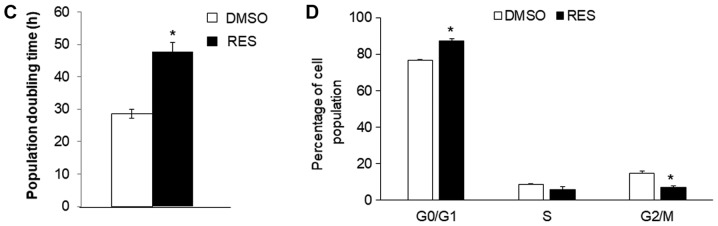
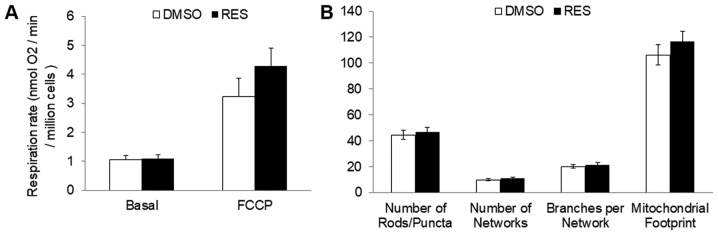
In PC3 cells growing in high glucose media, 48 h treatment with 10 µM RES stimulated cellular respiration rates (Fig. 1A), mitochondrial biogenesis (measured here as ‘mitochondrial footprint’, i.e., area of cell image occupied by mitochondria, Fig. 1B) and increased the mean mitochondrial network size (number of branches per network, Fig. 1B). Coincidentally, growth rates were reduced (Fig. 1C) and cells accumulated in G0/G1 phase (Fig. 1D).
Figure 1.
Resveratrol stimulates PC3 cell respiration and mitochondrial fusion while slowing cell growth. (A) RES increased basal (open bar; solid bar) and FCCP uncoupled (vertical striped bar; horizontal stripe bar) cellular respiration rates. (B) In the same experiment, RES stimulated increased mitochondrial fusion (mean number of branches per network) and mitochondrial footprint (area of cell image occupied by mitochondria). (C) Population doubling time increased and the (D) distribution of cells in different cell cycle phases was altered such that cells accumulated in G0/G1. In all experiments, PC3 cells were treated for 48 h with 10 µM RES (filled bars) or an equal volume of DMSO (open bars). Data shown are means ± SEM of (A) 6; (B) 35; (C) 17; and (D) 6 independent measurements. *P<0.05 RES-treated vs. DMSO vehicle control cells.
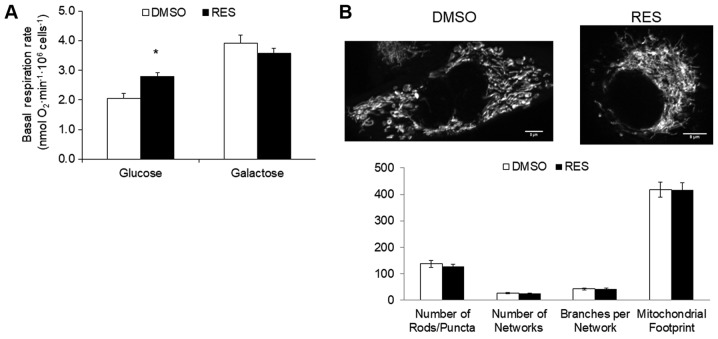
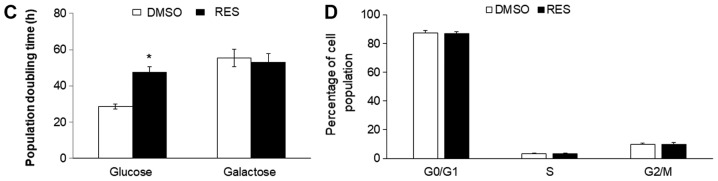
To investigate the potential role of the metabolic shift toward oxidative phosphorylation in RES's growth effects, the experiment was repeated using PC3 cells cultured in the same medium but with galactose replacing glucose as carbon fuel. Cells growing in galactose medium cannot meet ATP demand by glucose fermentation and become reliant on oxidative phosphorylation (15). Growth in galactose medium increased respiration rates relative to high glucose medium (Fig. 2A), but abolished the RES effect on these rates. Similarly, while the mitochondrial footprint (Fig. 2B) was greater in galactose medium compared to glucose medium, the effect of RES on this parameter was absent. Other mitochondrial network features were also not affected by RES treatment in cells growing in galactose (Fig. 2B). Coincidentally with the absence of metabolic effects, RES did not inhibit PC3 cell growth (Fig. 2C) or cell cycle distribution (Fig. 2D) in galactose medium. Thus, in the absence of a switch between glycolytic and oxidative metabolism RES had no metabolic or growth effects.
Figure 2.
Growth in galactose medium abolishes effects of resveratrol. No effect of resveratrol on (A) respiration or (B) mitochondrial network characteristics when PC3 cells were grown in galactose medium. Similarly, RES had no effect on (C) cell growth or (D) cell cycle distribution. Experiments were done as in Fig. 1, but cells were cultured in galactose medium (without glucose) except where indicated. In all experiments, cells were treated for 48 h with 10 µM RES (filled bars) or an equal volume of DMSO (open bars). Data shown are means ± SEM of (A) 6; (B) 35; (C) 12–17; and (D) 6 independent measurements. *P<0.05 RES-treated vs. DMSO-treated cells.
Given the role of HIF-1 as a metabolic regulator, and previous reports of RES effects on HIF-1 activity (17,18), we hypothesized that the metabolic switch observed with RES involves HIF-1 regulation. HIF-1 heterodimer activity is regulated by HIF-1α levels, which are in turn regulated by the hydroxylation by prolyl hydroxylases (PHDs) of key prolines 402 and 564. We used two strategies to prevent this proline hydroxylation and thus stabilize HIF-1α in PC3 cells growing in high glucose medium: PC3 cells were either stably transfected with a mutant HIF-1α lacking prolines 402 and 564 (19), or were treated throughout the experiment with IOX2. IOX2 is a selective PHD inhibitor that stabilizes HIF-1α in normoxia (20).
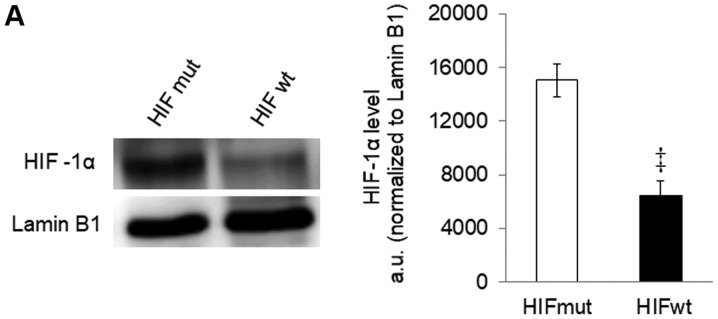
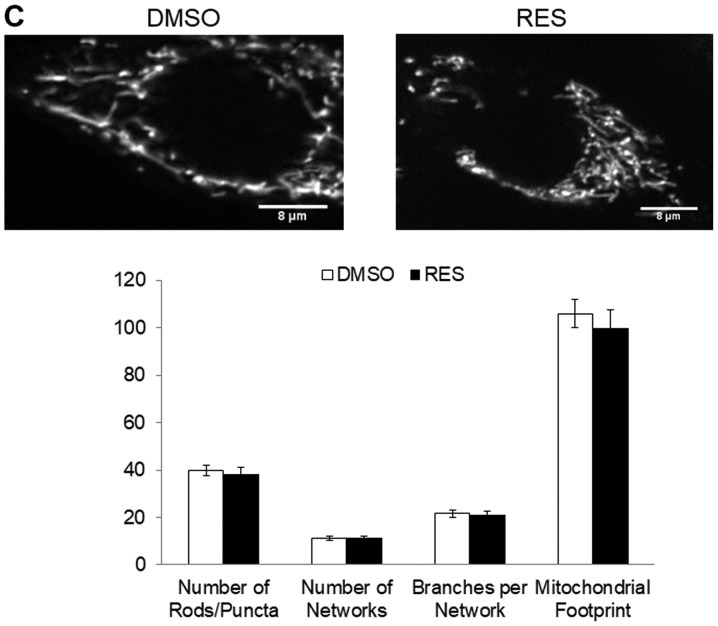
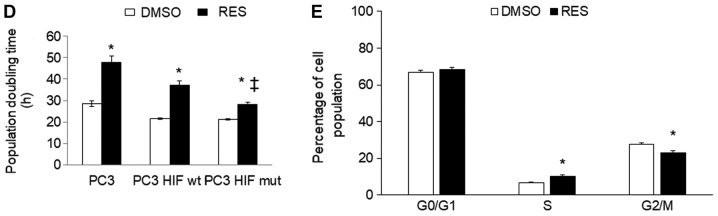
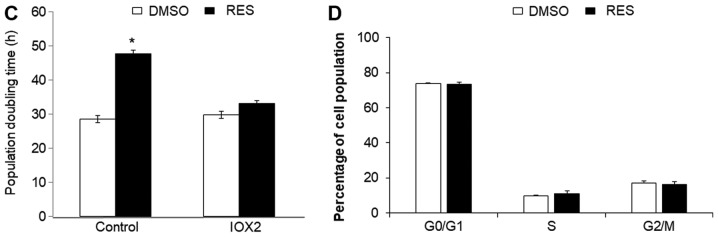
HIF-1α was detectable in cells stably expressing the mutant protein (Fig. 3A). In these cells, the effects of RES on respiration (Fig. 3B) and mitochondrial network characteristics (Fig. 3C) were absent. The effects of RES on cell growth (Fig. 3D) and cell cycle distribution (Fig. 3E) were reduced in PC3 cells expressing mutant HIF-1α, though they were not absent entirely. Interestingly, rather than accumulating in G0/G1 phase cells expressing the RES treated HIF-1α mutants showed a slight accumulation in S-phase. Similar results were observed using IOX2, with the exception that this treatment more fully abolished RES's effects on respiration (Fig. 4A) and mitochondrial network characteristics (Fig. 4B). The effects of RES on cell growth (Fig. 4C) were reduced in PC3 cells treated with IOX2, while RES effects on cell cycle distribution appear to be absent (Fig. 4D). To determine whether the ability of IOX2 to modulate RES's growth effects were specific to PC3 cells, this experiment was repeated with another prostate cancer cell line (LNCaP), and two non-prostate cancer cell lines: C2C12 mouse myoblasts and SHSY5Y human neuroblastoma. Interestingly, although IOX2 similarly abolished RES's effect on growth in LNCaP cells, it was either partially effective or ineffective in the two non-prostate cancer cell lines (Fig. 5).
Figure 3.
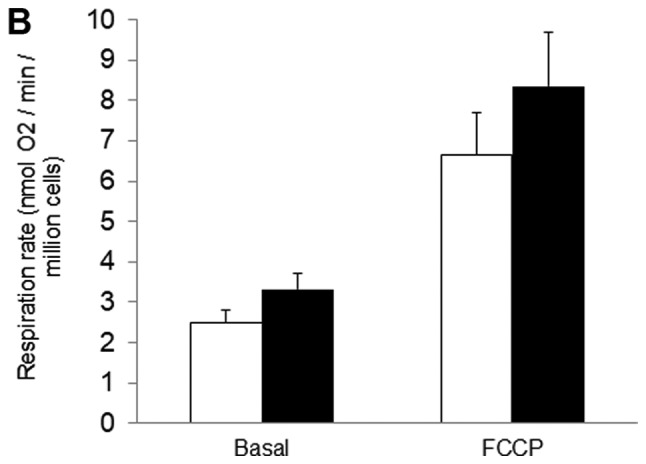
Effects of resveratrol were diminished in PC3 cells expressing a stable HIF-1α mutant. (A) HIF-1α in cells expressing wt or mutant HIF-1α in normoxic conditions. (B) Cellular respiration rates and (C) mitochondrial network characteristics in cells expressing mutant HIF-1α and treated with either 10 µM RES or vehicle control (DMSO) for 48 h. (D) Population doubling time and (E) cell cycle distribution for cells expressing mutant HIF-1α. In (B-E) open bars are DMSO vehicle control and filled bars are 10 µM RES. Data shown are means ± SEM of (A) 4; (B) 5–6; (C) 44; (D) 9 and (E) 6 independent measurements. *P<0.05 RES-treated vs. DMSO-treated cells; ‡P<0.05 wtHIF-1α vs. mutHIF-1α.
Figure 4.
Inhibition of prolyl hydroxylase activity with IOX2 abolished resveratrol effects on (A) cellular respiration, (B) mitochondrial network features, (C) cell growth, (D) cell cycle distribution. In all experiments, PC3 cells were treated for 48 h with 10 µM RES (filled bars) or an equal volume of DMSO (open bars). In both DMSO and RES groups, cells were simultaneously treated with IOX2 as in Material and Methods, except where indicated in (C). Data shown are means ± SEM of (A) 6; (B) 32; (C) 10–12; and (D) 5 independent measurements. *P<0.05 RES-treated vs. DMSO-treated cells.
Figure 5.
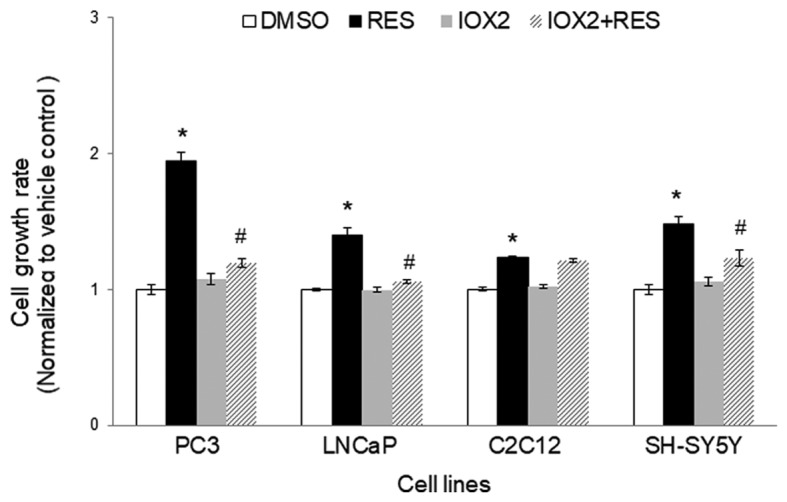
IOX2 inhibition of prolyl hydroxylase activity abolishes resveratrol's effects on growth in PC3 and LNCaP, but not C2C12 or SHSY5Y cells. Cells were treated with the PHD inhibitor IOX2 (25 µM) or vehicle control (DMSO) overnight prior to commencing experiments. RES treatments were started on the following day and continued for 48 h. All treatments (IOX2 and RES) were refreshed every 24 h. Data shown are means ± SEM of 6 independent measurements. *P<0.05 RES-treated vs. DMSO-treated cells; #P<0.05 RES-treated vs. IOX2+ RES treated.
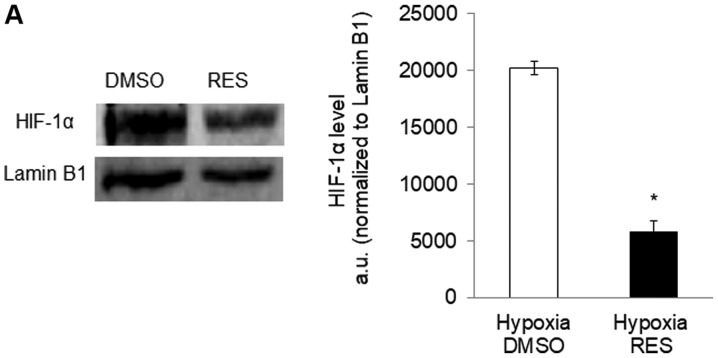
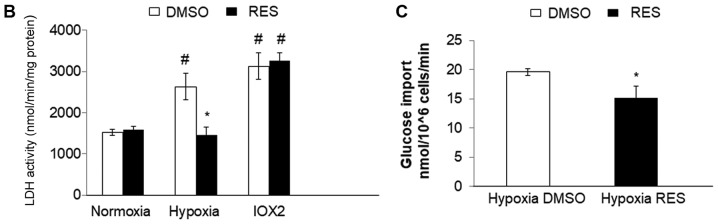
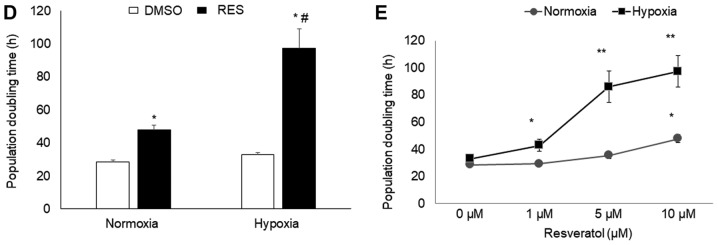
Given the implication of HIF-1 in mediating RES's effects on PC3 cells, we hypothesized that RES might be a particularly effective inhibitor of PC3 cell growth under hypoxic conditions, as would be prevalent in vivo (21). Indeed, RES treatment prevented the stabilization of HIF-1α under hypoxia (Fig. 6A and B), prevented the increase in LDH activity (Fig. 6C), associated with hypoxia and reduced the rate of glucose import (Fig. 6D). RES strongly inhibited PC3 cell growth under hypoxic conditions (Fig. 6E). Importantly, PC3 cell growth under hypoxic conditions was inhibited at lower concentrations of RES (Fig. 6F).
Figure 6.
Resveratrol strongly inhibits PC3 cell growth under hypoxic conditions. RES prevents HIF-1α stabilization under hypoxia. (A) Representative western blot and normalized HIF-1α levels in DMSO and RES treated cells. RES reduces (B) lactate dehydrogenase activity and (C) glucose import in hypoxia. (D) RES more strongly inhibits cell growth in hypoxia vs. normoxia. (E) Low concentrations of RES are effective at inhibiting cell growth under hypoxic, but not normoxic conditions. In all experiments, PC3 cells were treated for 48 h with 10 µM RES (filled bars) or an equal volume of DMSO (open bars). Data shown are means ± SEM of (A) 4; (B) 7–9; (C) 5; (D) 9–12; (E) 9–12 independent measurements. *P<0.05 RES-treated vs. DMSO-treated cells; **P<0.01 RES-treated vs. DMSO-treated cells, #P<0.05 hypoxia vs. normoxia.
Discussion
Cancer cells are generally typified by a high flux of glucose through glycolysis and the pentose phosphate pathway and this metabolic predisposition has emerged as a pharmacological target (22,23). Strategies aimed at inhibiting these pathways and/or shifting glucose catabolism toward complete mitochondrial oxidation have been effective in slowing cancer cell and tumour growth (5,6,24).
In our experiments, RES treatment stimulated a shift toward oxidative metabolism in PC3 cells concomitantly with the inhibition of growth, and when this metabolic shift was absent growth effects were also absent. The RES effects on PC3 cell mitochondrial and growth characteristics were completely abolished when the cells were grown in galactose medium containing no glucose. Growth of mammalian cells in galactose medium increases the contribution of oxidative phosphorylation to glucose oxidation (15), apparently related to the relatively slow multi-step Leloir pathway converting galactose to glucose-1-phosphate (25) that limits rates of ATP synthesis from glycolysis. PC3 cells grown in galactose had higher rates of respiration and more mitochondria (mitochondrial footprint, Fig. 2), suggesting the metabolic shift toward oxidative metabolism observed in glucose-grown PC3 cells treated with RES was already present. The inability of RES to affect cell growth under these conditions suggests that the metabolic shift from glucose fermentation to oxidation is an essential component of RES's growth inhibition effect. Interestingly, we have observed similar results previously (26); rho° PC3 cells that are unable to respire also show no growth inhibition by RES. Similarly, mouse embryonic fibroblast cells lacking the mitofusin-2 have compromised respiration and growth that is insensitive to RES (11). Taken together, these observations suggest that RES's growth inhibitory effect in PC3 cells (and perhaps also other cell types) is related to its ability to shift metabolism toward oxidative phosphorylation.
HIF-1 is a key mediator of the Warburg effect in cancer cells (22,27), and thus a target of strategies to promote oxidative metabolism and slow growth. Our results strongly suggest that HIF-1 is involved in the metabolic switch stimulated by RES treatment, since changes in HIF-1α stabilization were required for RES's effects on cellular respiration, mitochondrial network characteristics, and cell growth to manifest. The most complete abolishment of RES's effects was observed when PHD was inhibited by IOX2. PHD post-translationally modifies multiple proteins to exert broad effects on cellular metabolism (28–30). Though we focused here on HIF-1α, other PHD targets including HIF-2α may also be involved. This could explain why HIF-1α stabilization only partly abrogated RES's effects.
Given the evidence above for a role of PHD and HIF-1, it follows that RES might be a particularly strong inhibitor of growth in hypoxic PC3 cells where the importance of the HIF-1 pathway is enhanced. We found this to be the case, as PC3 cell population doubling time (PDT) was increased by more than two-fold in hypoxia, compared to an approximately 50% increase in PDT under atmospheric oxygen levels. Perhaps even more importantly, much lower concentrations of RES could elicit increases in PDT in hypoxia (1 µM in hypoxia vs. 10 µM in atmospheric oxygen). This observation has practical significance. One of the contradictions apparent in the literature on RES and cancer is that, while 10–100 µM RES is typically required to slow cell growth in vitro, the RES concentrations reached in mammalian blood plasma and tissues in vivo are much lower (10). And yet, RES treatment by various means has nonetheless been shown to slow tumour growth in many instances, including prostate cancer (31). The observation that RES interferes with HIF-1 activity may explain this apparent disjunction, given HIF-1's role in the growth of some cancers. In this respect, it is interesting that LNCaP cells responded similarly to the PHD inhibitor IOX2 in terms of the RES effect on growth, while this was largely absent in C2C12 or SHSY5Y cells. It will be interesting to investigate the connections between RES, HIF-1, mitochondrial respiration, and growth in other cancer cell types in which HIF-1 is known to play a particularly critical role (32).
Acknowledgements
Not applicable.
Funding
This study was supported by a Natural Sciences and Engineering Research Council (NSERC) Discovery Grant to JAS (RGPIN-2015-05645), an NSERC Undergraduate Summer Research Award to SMS, and an Ontario Graduate Scholarship to LAM.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JS and JF conceived and supervised experiments. JF, FM, LAM, BFT, and SMS performed experiments. JF performed all data analyses. JS and JF wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Conflict of interest statement
The authors declare that they have no competing interests.
References
- 1.Diaz-Ruiz R, Rigoulet M, Devin A. The warburg and crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim Biophys Acta. 2011;1807:568–576. doi: 10.1016/j.bbabio.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3108. [DOI] [PubMed] [Google Scholar]
- 5.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masoud GN, Li W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han G, Xia J, Gao J, Inagaki Y, Tang W, Kokudo N. Anti-tumor effects and cellular mechanisms of resveratrol. Drug Discov Ther. 2015;9:1–12. doi: 10.5582/ddt.2015.01007. [DOI] [PubMed] [Google Scholar]
- 10.Stuart JA, Robb EL. Springer Press; New York, NY: 2013. Bioactive polyphenols from wine grapes; p. 77. [Google Scholar]
- 11.Robb EL, Moradi F, Maddalena LA, Valente AJF, Fonseca J, Stuart JA. Resveratrol stimulates mitochondrial fusion by a mechanism requiring mitofusin-2. Biochem Biophys Res Commun. 2017;485:249–254. doi: 10.1016/j.bbrc.2017.02.102. [DOI] [PubMed] [Google Scholar]
- 12.de Oliveira MR, Nabavi SF, Manaya A, Daglia M, Hajheydari Z, Nabavi SM. Resveratrol and the mitochondria: From triggering the intrinsic apoptotic pathway to inducing biogenesis, a mechanistic view. Biochim Biophys Acta. 2016;1860:727–745. doi: 10.1016/j.bbagen.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Tang X, Lu QY, Zhang ZF, Brown J, Le AD. Resveratrol inhibits hypoxia-induced accumulation of hypoxia-inducible factor-1alpha and VEGF expression in human tongue squamous cell carcinoma and hepatoma cells. Mol Cancer Ther. 2005;4:1465–1474. doi: 10.1158/1535-7163.MCT-05-0198. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Li W, Yu L, Wu S. The suppressive effect of resveratrol on HIF-1α and VEGF expression after warm ischemia and reperfusion in rat liver. PLoS One. 2014;9:e109589. doi: 10.1371/journal.pone.0109589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossignol R, Gilerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.CAN-03-1101. [DOI] [PubMed] [Google Scholar]
- 16.Valente AJ, Maddalena LA, Robb EL, Moradi F, Stuart JA. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Zhou J, Liu J, Tang B, Zhao F, Qu Y. Biological characteristics of prostate cancer cells are regulated by hypoxia-inducible factor 1α. Oncol Lett. 2014;8:1217–1221. doi: 10.3892/ol.2014.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Benzonana LL, Zhao H, Watts HR, Perry NJ, Bevan C, Brown R, Ma D. Prostate cancer cell malignancy via modulation of HIF-1α pathway with isoflurane and propofol alone and in combination. Br J Cancer. 2014;111:1338–1349. doi: 10.1038/bjc.2014.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Q, Bartz S, Mao M, Li L, Kaelin WG., Jr The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol. 2007;27:2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen A, Ren S, Lerchenmüller C, Sun J, Weiss N, Most P, Peppel K. MicroRNA-138 regulates hypoxia-induced endothelial cell dysfunction by targeting S100A1. PLoS One. 2013;8:e78684. doi: 10.1371/annotation/53080a85-89cc-4a84-8fd9-0eb0c19cc05d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirschey MD, DeBerardinis RJ, Diehl AME, Drew JE, Frezza C, Green MF, Jones LW, Ko YH, Le A, Lea MA, et al. Dysregulated metabolism contributes to oncogenesis. Semin Cancer Biol. 2015;35(Suppl):S129–S150. doi: 10.1016/j.semcancer.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amoedo ND, Obre E, Rossignol R. Drug discovery strategies in the field of tumor energy metabolism: Limitations by metabolic flexibility and metabolic resistance to chemotherapy. Biochim Biophys Acta Bioenerg. 2017;1858:674–685. doi: 10.1016/j.bbabio.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Kankotia S, Stacpoole PW. Dichloroacetate and cancer: New home for an orphan drug? Biochim Biophys Acta. 2014;1846:617–629. doi: 10.1016/j.bbcan.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 26.Robb EL, Stuart JA. The stilbenes resveratrol, pterostilbene and piceid affect growth and stress resistance in mammalian cells via a mechanism requiring estrogen receptor beta and the induction of Mn-superoxide dismutase. Phytochemistry. 2014;98:164–173. doi: 10.1016/j.phytochem.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Courtnay R, Ngo DC, Malik N, Ververis K, Tortorell SM, Kargiannis TC. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol Biol Rep. 2015;42:841–851. doi: 10.1007/s11033-015-3858-x. [DOI] [PubMed] [Google Scholar]
- 28.Boulahbel H, Durán RV, Gottlieb E. Prolyl hydroxylases as regulators of cell metabolism. Biochem Soc Trans. 2009;37:291–294. doi: 10.1042/BST0370291. [DOI] [PubMed] [Google Scholar]
- 29.Jokilehto T, Jaakola PM. The role of HIF prolyl hydroxylases in tumour growth. J Cell Mol Med. 2010;14:758–770. doi: 10.1111/j.1582-4934.2010.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen TL, Durán RV. Prolyl hydroxylase domain enzymes and their role in cell signaling and cancer metabolism. Int J Biochem Cell Biol. 2016;80:71–80. doi: 10.1016/j.biocel.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 31.Li G, Rivas P, Bedolla R, Thapa D, Reddick RL, Ghosh R, Kumar AP. Dietary resveratrol prevents development of high-grade prostatic intraepithelial neoplastic lesions: Involvement of SIRT1-S6K axis. Cancer Prev Res (Phila) 2013;6:27–39. doi: 10.1158/1940-6207.CAPR-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Shi Y, Yuan J, Han Y, Qin R, Wu Q, Jia B, Wei B, Wei L, Dai G, Jiao S. HIF-1 alpha overexpression correlates with poor overall survival and disease-free survival in gastric cancer patients post-gastrectomy. PLoS One. 2014;9:e90678. doi: 10.1371/journal.pone.0090678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.