Abstract
Taxol-based chemotherapy is a conventional therapeutic approach for the treatment of triple-negative breast cancer (TNBC). However, the insensitivity of TNBC cells to Taxol greatly limits the anticancer effect of the drug and leads to patient mortality. The present study first evaluated the expression levels of microRNA (miR)-1207-5p in human normal breast epithelial MCF-10A cells and TNBC cell lines (MDA-MB-231, MDA-MB-436 and MDA-MB-453). The results revealed that the highest miR-1207-5p level was in MDA-MB-231, which suggested an oncogenic role of miR-1207-5p in TNBC. Therefore, MDA-MB-231 served as the present study's research model in subsequent experiments. The mRNAs that functioned as tumor suppressor factors for miR-1207-5p were then determined. Leucine zipper tumor suppressor gene 1 (LZTS1), which was predicted by TargetScan 6.2 and was supported by the results of a dual luciferase assay, was identified as a target of miR-1207-5p. AntagomiR-1207-5p increased LZTS1 mRNA and protein expressions, enhanced cell growth arrest and cell apoptosis induced by Taxol in MDA-MB-231 cells. Additionally, it was observed that, when compared with Taxol treatment, the combination of Taxol and antagomiR-1207-5p induced a sharp decrease in B-cell lymphoma 2 (Bcl-2) and phosphorylated-protein kinase B expression accompanied by an increase in the Bcl-2-associated X protein expression. Finally, miR-1207-5p expression was significantly increased, while LZTS1 expression was significantly decreased, in TNBC tissues when compared with normal adjacent tissues, and there was a negative correlation between miR-1207-5p and LZTS1 expression. In addition, there was a notable elevation in the expression of miR-1207-5p and a reduction in the expression of LZTS1 in the Taxol non-responsive TNBC tissues when compared with the Taxol-responsive TNBC tissues. The results of the present study suggested that miR-1207-5p may be a promising predictor of sensitivity towards Taxol in TNBC.
Keywords: microRNA-1207-5p, triple negative breast cancer, taxol, leucine zipper tumor suppressor gene 1
Introduction
Despite improvements in diagnostic technologies and breakthroughs in effective treatment in recent years, breast cancer remains the leading cause of cancer-related deaths in females worldwide, and the incidence and mortality rate generally increase with age (1). Triple-negative breast cancer (TNBC) is a sub-group of breast cancer that is characterized by the lack of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) expressions. TNBC accounts for approximately 15–20% of breast cancer cases and is associated with a poorer overall survival compared with other types of breast cancer (2). TNBC is the most aggressive breast cancer sub-type with a high possibility of metastasis (3). Moreover, due to the lack of ER, PR and HER2, TNBC is unresponsive to any hormonal treatment and there is currently no available targeted therapy, thereby increasing the chance of relapse, worsening prognosis and making TNBC difficult to treat (4,5). Therefore, there is an urgent need to identify novel therapeutic targets for TNBC, and our study focused only on TNBC.
TNBC patients are commonly treated with chemotherapy drugs, such as Taxol. Taxol is able to polymerize tubulin, promote the assembly/stabilization of microtubules, disrupt normal microtubule dynamics and arrest cells in mitosis. Cancer cells in TNBC should be killed by Taxol, but they maintain their viability via cellular responses, thereby promoting malignance (6). TNBC frequently recurs and metastasizes due to the acquirement of resistance to Taxol (7), which is one of the major obstacles to effective treatment.
MiRNAs are small, non-coding, endogenous RNAs, 22–25 nucleotides in length, which regulate gene expression (8). Mounting evidence has indicated that miRNAs act as oncogenes or tumor suppressor genes in different types of cancer, including TNBC (9), indicating their potential as therapeutic targets (10). MiR-1207 was identified to be upregulated in younger breast cancer patients, and related to cell motility, invasion and proliferation (11). MiR-1207 overexpression was also found to promote cancer stem cell-like traits in ovarian cancer (12). In colorectal cancer, significant overexpression of miR-1207 was detected (13). These findings demonstrate the participation of miR-1207 in cancer progression. Consequently, the function of miR-1207 in the sensitivity of TNBC to Taxol was investigated in the present study.
In summary, we observed a high level of miR-1207-5p in MDA-MB-231 cells. Treatment with antagomiR-1207-5p enhanced the cell growth arrest and cell apoptosis induced by Taxol in MDA-MB-231 cells, by regulating and increasing the protein level of LZTS1. Combined treatment with Taxol and antagomiR-1207-5p induced a sharp decrease in Bcl-2 and p-Akt expression, and an increase in the Bax protein expression level. A notable elevation in the expression of miR-1207-5p and a reduction in the expression of LZTS1 were identified in TNBC tissues but not adjacent tissues and Taxol non-responsive TNBC tissues but not responsive TNBC tissues. Our study showed the novelty of the interaction of miR-1207-5p regulating the LZTS1 gene expression and therefore, its role in taxol sensitivity. Our data suggest that miR-1207-5p may be a promising predictor of sensitivity towards Taxol in TNBC.
Materials and methods
Cell culture
Human normal breast epithelial MCF-10A cells, TNBC cell lines (MDA-MB-231, MDA-MB-436 and MDA-MB-453) and 293 cells were purchased from ATCC, authenticated by cytogenetic analysis and were used within 6 months or stored in liquid nitrogen. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) in an incubator at 37°C with 5% CO2.
Transient transfection
For cell transfection, MDA-MB-231 cells were seeded onto 6-well plates at a density of 2×105 cells/well. In brief, antagomiR-1207-5p (5′-CCCCUCCCAGCCUCCCUGCCA-3′, 100 nM; Guangzhou RiboBio Co., Ltd., Guangzhou, China), an antisense-based specific inhibitor against miR-1207-5p which was applied for specific silencing of endogenous miR-1207-5p, or antagomiR-NC (5′-UUCUCCGAACGUGUCACGU-3′, 100 nM; Guangzhou RiboBio Co., Ltd.) was diluted in DMEM with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The mixture was added into the 6-well plates to obtain a final concentration of 20 nmol/L and incubated for 48 h before the subsequent experiments.
Taxol treatment
Taxol (Selleck Chemicals, Shanghai, China) was stored in DMSO as a 10 µmol/L stock solution. In brief, prior to treatment with 10 nmol/L Taxol, 1×106 MDA-MB-231 cells were first seeded onto 60-mm culture dishes and were cultured for 24 h. After incubation with Taxol for 3 days, fresh 10 nmol/L Taxol-containing DMEM was added and incubated for another 2 days. Finally, MDA-MB-231 cells were washed with PBS and were cultured in drug-free DMEM. The medium was replaced every 2 days until the commencement of the subsequent experiments.
Bioinformatics analysis
Bioinformatics analysis was performed using the online software program TargetScan 6.2 (www.targetscan.org/).
Tissue specimens
Tissue specimens (tumor tissues and non-cancerous tissues) were obtained from 30 TNBC patients who had undergone surgery at Linfen People's Hospital. Non-cancerous tissues were at least 2-cm distal to tumor margins. The definition of response to Taxol-based chemotherapy in TNBC patients was based on the RECIST criteria (14). Patients were categorized into two groups: The Taxol responsive group and the Taxol non-responsive group. Patients in the Taxol responsive group achieved a complete response (CR), while patients in the Taxol non-responsive group exhibited persistent disease (PR, SD and PD). There were 18 TNBC patients in the Taxol responsive group and 12 patients in the Taxol non-responsive group. Written informed consent was obtained from each patient. The study was conducted in accordance with Declaration of Helsinki and was approved by the Institutional Review Board of Linfen People's Hospital, China.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from TNBC tissues and TNBC cells using TRIzol (Tiangen Biotech Co., Ltd., Beijing, China). Briefly, RNA was reverse transcribed into cDNA using a Reverse Transcription kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR analyses were performed using Power SYBR Green (Takara Biotechnology Co., Ltd.) in a 7500HT Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). RT-qPCR data were analyzed using the 2−ΔΔCq method. The thermocycling conditions were as follows: 95°C for 5 min (pre-incubation), 30 cycles at 95°C for 30 sec (denaturation), 60°C for 1 min (annealing), and 72°C for 30 sec (elongation). The expression of GAPDH was used to standardize the amount of mRNA in each PCR tube. U6 was used to standardize the amount of miRNA in each PCR tube. Primers were as followed: LZTS1 forward, 5′-ACCTCTAGAAACCCAGAACTCA-3′ and reverse, 5′-TCCAGAAGAGCCCATATCACTA-3′; GAPDH forward, 5′-GCGCCCAATACGACCAA-3′ and reverse, 5′-CTCTCTGCTCCTCCTGTTC-3′; miR-1207-5p forward, 5′-GCCAGATCTTGATTGACTTACAGCCCAGTT-3′ and reverse, 5′-GCCGAATTCCACCTGTCTTTATTCCACCC-3′; U6 forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse 5′-CGCTTCACGAATTTGCGT-3′.
Western blot analysis
Cell lysates were prepared using RIPA buffer (Roche, Shanghai, China). Proteins were resolved by sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA) for the detection of proteins with primary antibodies against AKT1/2/3 (1:500, sc-8312; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), p-AKT1/2/3 (Ser 473)-R (1:500, sc-7985-R; Santa Cruz Biotechnology, Inc.), LZTS1 (1:1,000, LBP62153; Novus Biologicals, LLC, Littleton, CO, USA), Bax (1:1,000, 2772; Cell Signaling Technology, Inc., Danvers, MA, USA), Bcl-2 (1:1,000, 3498 Cell Signaling Technology, Inc.) and GAPDH (1:5,000, A5060; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C overnight. Membranes were then incubated with goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000, ab97080; Abcam, Shanghai, China) at room temperature for 1 h. Protein bands were treated with an enhanced chemiluminescence detection system (ECL; Bio-Rad Laboratories, Inc.), visualized with the ChemiDoc XRS system (Bio-Rad Laboratories, Inc.) and analyzed using Quality One 4.5.2 (Bio-Rad Laboratories, Inc.). Protein expression levels were normalized to GAPDH.
Luciferase reporter assay
3′UTR of LZTS1 was amplified from cDNA of 293 cells. Oligonucleotides that contained LZTS1 cDNA fragments, including miR-1207-5p binding sites were amplified and cloned into pmirGLO plasmids (Promega Corporation, Madison, WI, USA) to obtain luciferase reporter plasmids, pmirGLO-LZTS1-WT. Mutant LZTS1 (pmirGLO-LZTS1-MUT) acted as a negative control and was obtained by site-directed mutagenesis PCR with platinum pfx DNA polymerase, according to the manufacturer's protocol. Cells (3×104) were seeded onto 24-well plates and cultured for 24 h. Then, pmirGLO-LZTS1-WT or pmirGLO-LZTS1-MUT and miR-1207-5p mimics (5′-UGGCAGGGAGGCUGGGAGGGG-3′) or miR-NC mimics (5′-UUCUCCGAACGUGUCACGU-3′) and 3 ng pRL-TK Renilla plasmid were transfected into cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h after transfection, a Dual Luciferase Reporter Assay (Promega Corporation) was performed in a luminometer, according to the manufacturer's protocol, to examine the relative luciferase activity.
Flow cytometric analysis
Cell apoptosis was detected using an Annexin-V/Dead Cell Apoptosis kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. In brief, MDA-MB-231 cells were trypsinized and suspended in 1 × annexin binding buffer. Subsequently, propidium iodide (PI) and Annexin V-FITC were added to the cell suspension and cultured for 15 min. Stained cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
MTT assay
An MTT assay (Invitrogen; Thermo Fisher Scientific, Inc.) was performed to evaluate the cell proliferation rate in each group. In brief, MDA-MB-231 cells were washed with PBS, trypsinized, and seeded onto 96-well plates. Then, MTT reagent (10 µl) was added and plates were incubated in an incubator at 37°C with 5% CO2 until purple precipitate was observed. Thereafter, 100 µl dimethylsulfoxide was added to dissolve the formazan crystals and the plates were incubated at 37°C for 2 h in the dark. Absorbance was read at 570 nm by a microplate reader.
Statistical analysis
Data were analyzed using SPSS version v.13 (SPPS, Inc., Chicago, IL, USA) and are expressed as the mean ± standard deviation. Comparisons between two groups were analyzed using Student's t-test, and comparisons among multiple groups were analyzed by one-way analysis of variance followed by Student-Newman-Keuls test. The correlation between LZTS1 and miR-1207-5pb was analyzed by Spearman's correlation analysis. P<0.05 was considered to indicate a statistically significant difference. Each experiment was repeated three times.
Results
miR-1207-5p expression was significantly elevated in MDA-MB-231 cells
TNBC is the most aggressive breast cancer sub-type with a high possibility of metastasis (3). Moreover, TNBC is unresponsive to any hormonal treatment, making TNBC difficult to treat (4,5). Therefore, there is an urgent need to identify novel therapeutic targets for TNBC, our study focused only on TNBC, and we chose TNBC MDA-MB-231, MDA-MB-436 and MDA-MB-453 cell lines for our study instead of ER+, PR+/-, HER2- T47D or MCF7 cells despite of their response to chemotherapy (15,16).
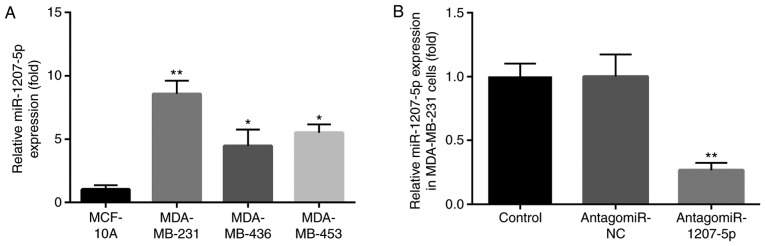
We evaluated the expression of miR-1207-5p in TNBC MDA-MB-231, MDA-MB-436 and MDA-MB-453 cell line. Compared with normal MCF-10A cells, miR-1207-5p expression was increased in MDA-MB-436 and MDA-MB-453 cells (P<0.05). However, the greatest increase in the miR-1207-5p expression level was in MDA-MB-231 cells (P<0.01; Fig. 1A). Therefore, the MDA-MB-231 cells were selected for our research model.
Figure 1.
miR-1207-5p expression is significantly elevated in MDA-MB-231 cells. (A) When compared with the normal MCF-10A cells, miR-1207-5p expression was increased in MDA-MB-436 and MDA-MB-453 cells; however, the greatest increase in miR-1207-5p expression level was observed in MDA-MB-231 cells. *P<0.05 and **P<0.01 vs. MCF-10A. (B) There was no significant difference in miR-1207-5p expression between the control and the antagomiR-NC groups; however, the expression of miR-1207-5p was significantly decreased following treatment with antagomiR-1207-5p when compared with expression in the antagomiR-NC group. **P<0.01 vs. antagomiR-NC group. miR, microRNA; NC, negative control.
The effect of antagomiR-1207-5p on miR-1207-5p expression was evaluated in MDA-MB-231 cells by RT-qPCR. There was no significant difference in miR-1207-5p expression between the control group and the antagomiR-NC group. However, the expression level of miR-1207-5p was significantly decreased after treatment with antagomiR-1207-5p, compared with expression in the antagomiR-NC group (P<0.01; Fig. 1B).
Since miR-1207-5p was found to be up-regulated in the MDA-MB-231 cells compared with expression in the MCF-10A cells, this suggested an oncogenic role of miR-1207-5p in TNBC. Therefore, using the online software programs TargetScan 6.2, we aimed to identify target mRNAs of miR-1207-5p that functioned as tumor suppressor factors during tumor progression.
LZTS1 was a direct target for miR-1207-5p
LZTS1 was previously found to be decreased in cutaneous squamous cell carcinoma (17) and osteosarcoma (18), and was identified to suppress colorectal cancer proliferation (19). Moreover, LZTS1 reduction conferred Taxol resistance and was associated with a poor prognosis in patients with breast cancer (20). Additionally, loss of LZTS1 contributed to the lymph node metastasis of breast invasive micropapillary carcinoma (21). Notably, LZTS1 was predicted to be a potential target for miR-1207-5p in our study, and the predicted binding sites between LZTS1 and miR-1207-5p are presented in Fig. 2A.
Figure 2.
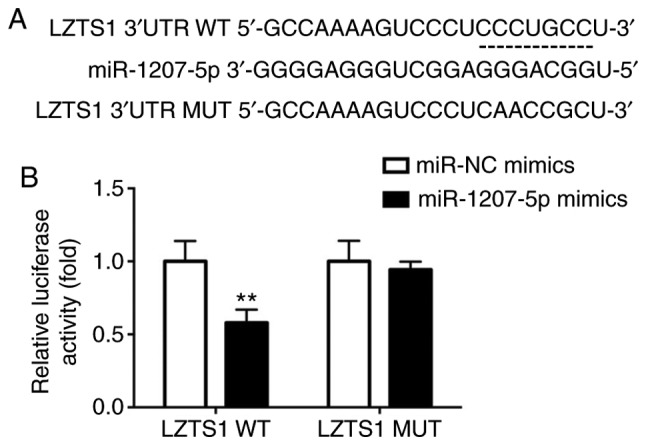
LZTS1 is a direct target for miR-1207-5p. (A) The putative binding sites between LZTS1 and miR-1207-5p are presented. (B) Compared with the activity in cells transfected with pmirGLO-LZTS1-WT and miR-NC mimics, miR-1207-5p mimics significantly inhibited the luciferase in cells transfected with pmirGLO-LZTS1-WT and miR-1207-5p mimics. **P<0.01 vs. miR-NC mimics (n=3). LZTS1, leucine zipper tumor suppressor gene 1; miR, microRNA; NC, negative control; WT, wild-type.
The interaction between LZTS1 and miR-1207-5p was verified by luciferase reporter assay. There was a significant reduction in luciferase activity in cells transfected with pmirGLO-LZTS1-WT and miR-1207-5p mimics compared with activity in cells transfected with pmirGLO-LZTS1-WT and miR-NC mimics (P<0.01). Meanwhile, no significant difference was found between cells transfected with pmirGLO-LZTS1-MUT and miR-NC mimics or pmirGLO-LZTS1-MUT and miR-1207-5p mimics, as presented in Fig. 2B.
Based on these results, we inferred that miR-1207-5p regulated LZTS1. However, antagomiR-1207-5p could not bind to LZTS1, this luciferase reporter assay cannot demonstrate ‘direct interaction’ of molecules, which was a limitation. The function of miR-1207-5p and LZTS1 in the sensitivity of TNBC to Taxol has not been studied previously. Therefore, the present study carried out experiments to investigate this.
LZTS1 expression was repressed by miR-1207-5p
The influence of miR-1207-5p on the mRNA and protein expression level of LZTS1 was evaluated by RT-qPCR and western blotting, respectively. The results demonstrated that, compared with cells in the antagomiR-NC group, there was a significantly higher protein expression level (Fig. 3A) as well as mRNA level (P<0.01; Fig. 3B) of LZTS1 in cells transfected with antagomiR-1207-5p. These results suggested that miR-1207-5p inhibited the expression of LZTS1.
Figure 3.
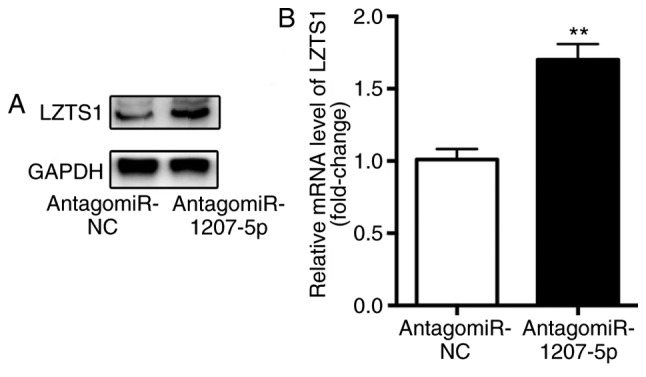
Inhibition of miR-1207-5p increases LZTS1 expression levels. When compared with cells in the antagomiR-NC group, there were (A) markedly higher protein expression levels and (B) significantly greater mRNA levels of LZTS1 in cells transfected with antagomiR-1207-5p. **P<0.01 vs. antagomiR-NC (n=3). LZTS1, leucine zipper tumor suppressor gene 1; miR, microRNA; NC, negative control.
AntagomiR-1207-5p enhanced the Taxol-induced reduction in cell proliferation and increase in cell apoptosis
As presented in Fig. 4, compared with the control group, Taxol significantly reduced the cell proliferation rate (P<0.01), which was further reduced by the co-administration of Taxol and antagomiR-1207-5p (P<0.01).
Figure 4.
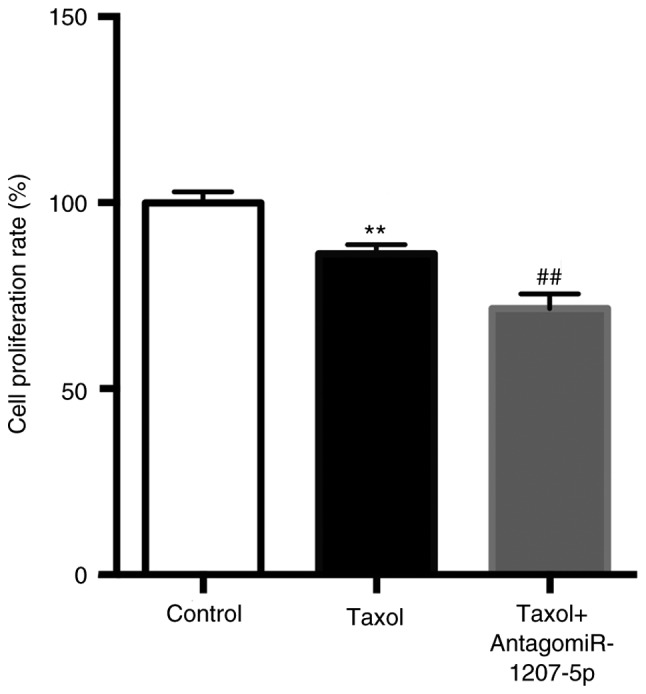
Inhibition of miR-1207-5p enhances the Taxol induced cell proliferation arrest. When compared with the control group, Taxol significantly reduced the cell proliferation rate, which was further reduced by the co-administration of Taxol and antagomiR-1207-5p. **P<0.01 vs. control; ##P<0.01 vs. Taxol (n=3). miR, microRNA.
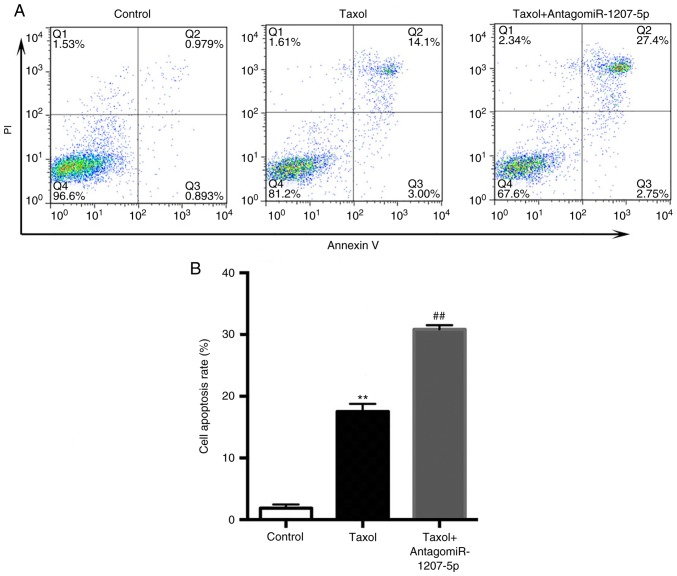
As presented in Fig. 5A and B, compared with the control group, Taxol significantly increased the cell apoptosis rate (P<0.01), which was further increased by the co-administration of Taxol and antagomiR-1207-5p (P<0.01).
Figure 5.
(A) Inhibition of miR-1207-5p enhances the Taxol induced increase in cell apoptosis. (B) When compared with the control group, Taxol significantly increased the cell apoptosis rate, which was further increased by the co-administration of Taxol and antagomiR-1207-5p. **P<0.01 vs. control; ##P<0.01 vs. Taxol (n=3). miR, microRNA; PI, propidium iodide.
These results regarding the effects of miR-1207-5p on cancer cell proliferation and apoptosis were consistent with those of a previous report (12).
AntagomiR-1207-5p enhanced Taxol-induced Bax upregulation, and Bcl-2 and p-Akt downregulation
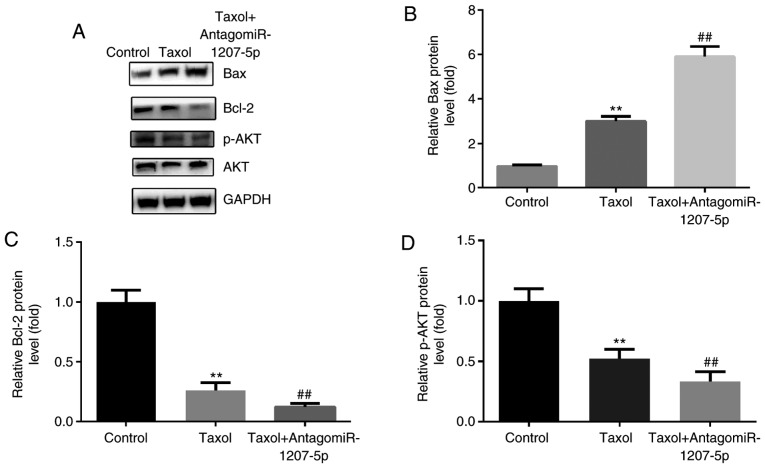
According to the aforementioned findings, we inferred that miR-1207-5p affected cell proliferation and apoptosis after Taxol treatment. However, the molecules that were regulated by miR-1207-5p were unknown. Therefore, we investigated pathways or molecules that were associated with cell proliferation and apoptosis. We found that the PI3K/Akt signaling pathway is important in regulating cell proliferation, migration, apoptosis, and angiogenesis (22,23); meanwhile, the Bcl-2 protein family including Bax (pro-apoptosis) and Bcl-2 (anti-apoptosis), takes part in the apoptotic process (24). Therefore, we wanted to investigate whether expression of Bax, Bcl-2 and p-Akt could be influenced by miR-1207-5p after administration of Taxol. We detected the protein expression of p-Akt, Bax and Bcl-2 after antagomiR-1207-5p treatment in MDA-MB-231 cells. The results indicated that, compared with the control group, Taxol induced notable up-regulation of Bax, and downregulation of Bcl-2 and p-Akt (P<0.01), which was further induced by the co-administration of antagomiR-1207-5p (P<0.01; Fig. 6A-D).
Figure 6.
Inhibition of miR-1207-5p enhances Taxol-induced Bax upregulation, and Bcl-2 and p-Akt downregulation. (A) Western blot analysis. There was notable upregulation of (B) Bax, and downregulation of (C) Bcl-2 and (D) p-Akt following Taxol treatment, which was further induced by the co-administration of antagomiR-1207-5p. **P<0.01 vs. control; ##P<0.01 vs. Taxol (n=3). miR, microRNA; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein; p-Akt, phosphorylated protein kinase B.
These results suggested that miR-1207-5p functioned through regulation of apoptosis-related pathways and molecules via regulating the expression of LZTS1. The role of miR-1207-5p in regulating the expression of LZTS1 was evidenced by the results as follows: 1. Luciferase assay; 2. the elevated mRNA/protein levels of LZTS1 after AntagomiR-1207-5p treatment.
miR-1207-5p was negatively correlated with LZTS1 in TNBC patients
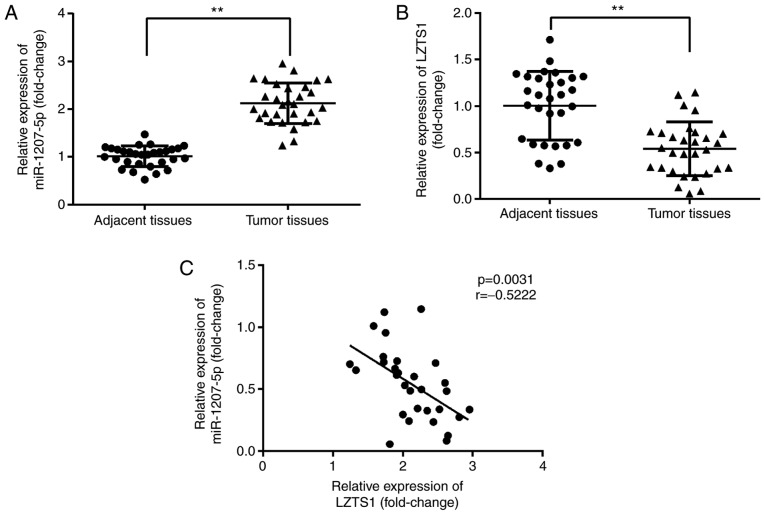
We investigated whether there were differences in miR-1207-5p and LZTS1 expression levels between the adjacent normal tissues and tumor tissues from TNBC patients using RT-qPCR. As shown in Fig. 7A and B, miR-1207-5p expression was significantly increased (P<0.01), while LZTS1 expression was significantly decreased (P<0.01) in tumor tissues compared with normal adjacent tissues. Moreover, there was a negative correlation between miR-1207-5p and LZTS1 expression (P=0.0031, r=−0.5222; Fig. 7C).
Figure 7.
miR-1207-5p is negatively associated with LZTS1 in TNBC patients. (A) miR-1207-5p expression was significantly increased and (B) LZTS1 expression was significantly decreased in the 30 tumor tissues when compared with the 30 normal adjacent tissues. (C) In addition, there was a negative correlation between miR-1207-5p and LZTS1 expression. **P<0.01, as indicated (n=30). LZTS1, leucine zipper tumor suppressor gene 1; miR, microRNA; TNBC, triple negative breast cancer.
Elevated miR-1207-5p and reduced LZTS1 in Taxol non-responsive TNBC tissues
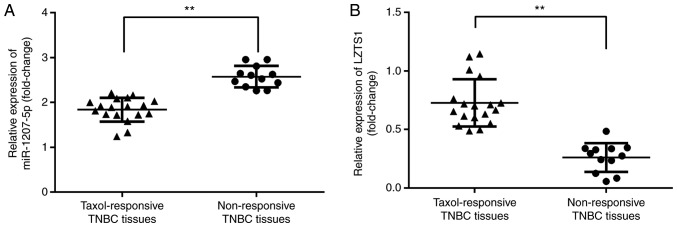
Among the 30 TNBC patients, we found that there were 18 TNBC patients who responded to Taxol and 12 patients who were non-responsive to Taxol. Furthermore, we evaluated the expression of miR-1207-5p and LZTS1 in Taxol-responsive and Taxol non-responsive TNBC tissues. The results demonstrated that, compared with Taxol-responsive TNBC tissues, there was a significant elevation in the expression of miR-1207-5p (P<0.01; Fig. 8A) and a significant reduction in the expression of LZTS1 (P<0.01; Fig. 8B) in the Taxol non-responsive TNBC tissues.
Figure 8.
Elevated miR-1207-5p and reduced LZTS1 in Taxol non-responsive TNBC tissues. (A) miR-1207-5p expression was significantly increased and (B) LZTS1 expression was significantly decreased in the 12 Taxol non-responsive TNBC tissues when compared with the 18 Taxol-responsive TNBC tissues. **P<0.01, as indicated. LZTS1, leucine zipper tumor suppressor gene 1; miR, microRNA; TNBC, triple negative breast cancer.
Discussion
Due to the lack of ER, PR and HER2 expression, TNBC is unresponsive to any hormonal treatment (2,5). Although there have been multiple effective treatment strategies developed for breast cancer, TNBC patients are commonly treated with chemotherapy agents, such as Taxol (6), resistance to which makes TNBC patients more susceptible to relapse (7). Therefore, there is an urgent requirement to identify novel therapeutic targets.
Decreases in tumor suppressor miRNAs or increases in onco-miRNAs are known to be involved in the pathogenesis of human cancer (25). MiRNAs also have the potential to be therapeutic targets (10).
MiR-1207 was identified to be up-regulated in younger breast cancer patients (11) and colorectal cancer patients (13), and overexpression of miR-1207 also promoted the cancer stem cell-like traits of ovarian cancer (12). These reports suggest a potential oncogenic role of miR-1207 in cancer. Moreover, a previous report about the effects of miR-1207-5p expression in peripheral blood on cisplatin-based chemosensitivity in primary gallbladder carcinoma patients, indicating the lower miR-1207-5p expression in effective group than in the ineffective group after chemotherapy (26), suggesting its role in chemosensitivity. Therefore, the present study first detected the expression level of miR-1207-5p in normal MCF-10A cells and TNBC MDA-MB-436, MDA-MB-453 and MDA-MB-231 cell lines, and found that, compared with normal MCF-10A cells, there was a higher miR-1207-5p expression level in the TNBC cell lines, with the highest level being observed in the MDA-MB-231 cells. This suggested an oncogenic role for miR-1207-5p in TNBC. Therefore, we aimed to identify the target mRNAs that functioned as tumor suppressor factors during tumor progression for miR-1207-5p using the online software programs TargetScan 6.2.
LZTS1 was found to be decreased in cutaneous squamous cell carcinoma (17) and osteosarcoma (18), suggesting a tumor suppressor role for LZTS1 in cancer. Interestingly, the tumor-suppressor gene LZTS1 suppressed colorectal cancer cell proliferation through inhibition of the Akt/mTOR signaling pathway (19), and inhibited hepatocellular carcinoma cell proliferation through inhibition of the PI3K/Akt signaling pathway (27). LZST1 reduction was correlated with a poor prognosis, increased cell motility/invasion and epithelial-to-mesenchymal transition of breast carcinoma (28). Meanwhile, LZST1 reduction conferred Taxol resistance and was associated with a poor prognosis in breast cancer (20). However, there was no report about the relationship between miR-1207-5p and LZTS1. In the present study, LZTS1 was first predicted and verified to be a potential target for miR-1207-5p. The function of miR-1207-5p and LZTS1 in the sensitivity of TNBC cells to Taxol treatment has not yet been elucidated. Therefore, we carried out experiments to investigate this.
Compared with the control group, Taxol dramatically reduced the cell proliferation rate and increased the cell apoptosis rate in MDA-MB-231 cells. These effects were further promoted by the co-administration of Taxol and antagomiR-1207-5p. Accordingly, we inferred that miR-1207-5p affected cell proliferation and apoptosis after Taxol treatment. However, it was unknown which molecules could be regulated by miR-1207-5p or LZTS1.
We investigated signaling pathways or molecules that were correlated with cell proliferation and apoptosis. LZTS1 suppressed the proliferation of colorectal cancer and hepatocellular carcinoma cells by inhibiting the Akt/mTOR signaling pathway (19) and the PI3K/Akt signaling pathway (27), respectively. Bax and Bcl-2 are involved in the apoptotic process (24). Therefore, we tested the protein levels of Bax and Bcl-2, as well as the activation level of p-Akt by western blotting. Up-regulation of Bax, and downregulation of Bcl-2 and p-Akt, was observed after transfection of antagomiR-1207-5p compared with the control and Taxol groups. This suggested that miR-1207-5p functioned through regulating apoptotic-related pathways and molecules via regulating the expression of LZTS1.
A significant increase in miR-1207-5p expression and a decrease in LZTS1 expression were observed in TNBC tissues compared with normal adjacent tissues. Additionally, there was a negative correlation between miR-1207-5p and LZTS1 expression. Moreover, in comparison with Taxol-responsive TNBC tissues, there was a notable elevation in miR-1207-5p expression and a reduction in LZTS1 expression in non-responsive TNBC tissues. Compare with the literature that previously reported LZTS1 gene as tumor suppressor gene related to paclitaxel resistant in cancer therapy in, our results was consistent with it (20).
In conclusion, miR-1207-5p may be a promising predictor of sensitivity to Taxol in TNBC. Our study showed the novelty of the interaction of miR-1207-5p regulating the LZTS1 gene expression and therefore, its role in taxol sensitivity. Unfortunately, there are limitations of our study: 1. The absence of data on breast cancer cell lines that are not of the triple negative subtype; 2. the use of a single TNBC cell line; 3. miR-1207-5p may target not only LZTS1 but also other molecules critical for MDA-MB-231 cell proliferation, apoptosis and TNBC development. Comprehensive gene expression analysis of these cells with or without miR-1207-5p inhibitor should be performed in future studies to understand its real effects.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XH, ZN and LL performed the experiments and analyzed the data. QG, HL and XY conducted the experiments. XZ conceived the study, analyzed the data and prepared the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from each patient. The present study was conducted in accordance with Declaration of Helsinki and was approved by the Institutional Review Board of Linfen People's Hospital (Shanxi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there were no competing interests.
References
- 1.American Cancer Society: Breast Cancer Facts & Figures 2013–2014. American Cancer Society; Atlanta, GA: 2013. [Google Scholar]
- 2.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 4.von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 5.Heitz F, Harter P, Lueck HJ, Fissler-Eckhoff A, Lorenz-Salehi F, Scheil-Bertram S, Traut A, du Bois A. Triple negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer. 2009;45:2792–2798. doi: 10.1016/j.ejca.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Zasadil LM, Andersen KA, Yeum D, Rocque GB, Wilke LG, Tevaarwerk AJ, Raines RT, Burkard ME, Weaver BA. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci Transl Med. 2014;6:229ra43. doi: 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamdade VS, Sethi N, Mundhe NA, Kumar P, Lahkar M, Sinha N. Therapeutic targets of triple-negative breast cancer: A review. Br J Pharmacol. 2015;172:4228–4237. doi: 10.1111/bph.13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Yang F, Zhang W, Shen Y, Guan X. Identification of dysregulated microRNAs in triple-negative breast cancer (review) Int J Oncol. 2015;46:927–932. doi: 10.3892/ijo.2015.2821. [DOI] [PubMed] [Google Scholar]
- 10.Ghelani HS, Rachchh MA, Gokani RH. MicroRNAs as newer therapeutic targets: A big hope from a tiny player. J Pharmacol Pharmacother. 2012;3:217–227. doi: 10.4103/0976-500X.99416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peña-Chilet M, Martínez MT, Pérez-Fidalgo JA, Peiró-Chova L, Oltra SS, Tormo E, Alonso-Yuste E, Martinez-Delgado B, Eroles P, Climent J, et al. MicroRNA profile in very young women with breast cancer. BMC Cancer. 2014;14:529. doi: 10.1186/1471-2407-14-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu G, Liu A, Zhu J, Lei F, Wu S, Zhang X, Ye L, Cao L, He S. MiR-1207 overexpression promotes cancer stem cell-like traits in ovarian cancer by activating the Wnt/β-catenin signaling pathway. Oncotarget. 2015;6:28882–28894. doi: 10.18632/oncotarget.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagy ZB, Barták BK, Kalmár A, Galamb O, Wichmann B, Dank M, Igaz P, Tulassay Z, Molnár B. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol Oncol Res. 2017 Oct 4; doi: 10.1007/s12253-017-0308-1. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 14.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European organization for research and treatment of cancer, National cancer institute of the United States, National cancer institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Neut M, Rao VR, Gentile S. hERG1/Kv11.1 activation stimulates transcription of p21waf/cip in breast cancer cells via a calcineurin-dependent mechanism. Oncotarget. 2016;7:58893–58902. doi: 10.18632/oncotarget.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singel SM, Cornelius C, Batten K, Fasciani G, Wright WE, Lum L, Shay JW. A targeted RNAi screen of the breast cancer genome identifies KIF14 and TLN1 as genes that modulate docetaxel chemosensitivity in triple-negative breast cancer. Clin Cancer Res. 2013;19:2061–2070. doi: 10.1158/1078-0432.CCR-13-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olasz EB, Seline LN, Schock AM, Duncan NE, Lopez A, Lazar J, Flister MJ, Lu Y, Liu P, Sokumbi O, et al. MicroRNA-135b regulates leucine zipper tumor suppressor 1 in cutaneous squamous cell carcinoma. PLoS One. 2015;10:e0125412. doi: 10.1371/journal.pone.0125412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Z, Wang T. miR-214 promotes the proliferation and invasion of osteosarcoma cells through direct suppression of LZTS1. Biochem Biophys Res Commun. 2014;449:190–195. doi: 10.1016/j.bbrc.2014.04.140. [DOI] [PubMed] [Google Scholar]
- 19.Zhou W, He MR, Jiao HL, He LQ, Deng DL, Cai JJ, Xiao ZY, Ye YP, Ding YQ, Liao WT, Liu SD. The tumor-suppressor gene LZTS1 suppresses colorectal cancer proliferation through inhibition of the AKT-mTOR signaling pathway. Cancer Lett. 2015;360:68–75. doi: 10.1016/j.canlet.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Lovat F, Ishii H, Schiappacassi M, Fassan M, Barbareschi M, Galligioni E, Gasparini P, Baldassarre G, Croce CM, Vecchione A. LZST1 downregulation confers paclitaxel resistance and is associated with worse prognosis in breast cancer. Oncotarget. 2014;5:970–977. doi: 10.18632/oncotarget.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XX, Liu BB, Wu X, Su D, Zhu Z, Fu L. Loss of leucine zipper putative tumor suppressor 1 (LZTS1) expression contributes to lymph node metastasis of breast invasive micropapillary carcinoma. Pathol Oncol Res. 2015;21:1021–1026. doi: 10.1007/s12253-015-9923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;1(16 Suppl):S12–S19. doi: 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- 23.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: Of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 24.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/S0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen ED, Liu B, Yu XS, Xiang ZF, Huang HY. The effects of miR-1207-5p expression in peripheral blood on cisplatin-bsed chemosensitivity of primary gallbladder carcinoma. Onco Targets Ther. 2016;9:3633–3642. doi: 10.2147/OTT.S101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y, Liu X. The tumor-suppressor gene LZTS1 suppresses hepatocellular carcinoma proliferation by impairing PI3K/Akt pathway. Biomed Pharmacother. 2015;76:141–146. doi: 10.1016/j.biopha.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Wang XX, Zhu Z, Su D, Lei T, Wu X, Fan Y, Li X, Zhao J, Fu L, Dong JT, Fu L. Down-regulation of leucine zipper putative tumor suppressor 1 is associated with poor prognosis, increased cell motility and invasion, and epithelial-to-mesenchymal transition characteristics in human breast carcinoma. Hum Pathol. 2011;42:1410–1419. doi: 10.1016/j.humpath.2010.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.