Abstract
The PRL-3 gene is involved in the liver metastasis of colorectal cancer (CRC) and oncogene addiction to anticancer therapy. In the present study genomic gains in PRL-3 and its pathway genes, c-myc and EGFR, were investigated in order to determine their clinical relevance during metastatic formation in primary CRC and corresponding liver metastases. The genomic gain statuses of PRL-3, EGFR, and c-myc were investigated using quantitative polymerase chain reaction (qPCR) analysis in 35 samples of CRC and corresponding liver metastases. In the primary CRC specimens, genomic gains in PRL-3, c-myc, and EGFR were observed in 4, 4, and 13 cases, respectively. A genomic gain in one gene was observed in 18 cases, and these genomic gains were mutually exclusive. In the liver metastasis specimens, genomic gains were observed in 14, 8, and 13 cases, respectively. The copy numbers of PRL-3 and c-myc were significantly higher in the liver metastases than in the primary CRC specimens (P=0.03, P=0.009, respectively). A genomic gain in PRL-3 was the most frequent gain in the liver metastases (P=0.004) and was partially redundant with a c-myc genomic gain. EGFR genomic gains were consistent between the primary CRC and the liver metastases (P=0.0000008). In addition, a genomic gain in any of the 3 genes was observed in 23 cases (66%). Among the clinicopathological factors that were assessed, an EGFR genomic gain was significantly associated with tumour size in the primary CRC and the liver metastases (P=0.04). A c-myc genomic gain was also significantly associated with the v factor of the primary tumours in the liver metastases (P<0.01). In conclusion, the genomic copy numbers of PRL-3, c-myc and EGFR were frequently characterised by aberrations in genomic gain in liver metastases from CRC; thus, these gene statuses exhibit potential for the identification of patients who are likely to respond positively to anticancer therapies.
Keywords: PRL-3, c-myc, EGFR, colorectal cancer, genomic gain
Introduction
The prevalence of colorectal cancer (CRC) is increasing, and CRC has one of the highest cancer morbidities reported worldwide. Over 47,000 CRC-related deaths occurred in Japan in 2012; in terms of site-specific cancer mortality, these figures corresponded to the third highest in men and the highest in women (1). CRC without lymph node metastasis can be largely cured by surgical resection alone (2), but the prognosis becomes poor once CRC has progressed to lymph node metastasis or distant organ metastasis. With lymph node metastasis, CRC is more likely to recur than CRC without lymph node metastasis (3), and distant metastases greatly reduce the 5-year survival rate. The liver is the most common metastatic organ (4,5), regardless of whether metastasis occurs in a synchronous or metachronous manner, and liver metastasis is a major factor affecting the prognosis of CRC. For CRC treatment strategies, the prevention of liver metastasis is one of the most important issues. Thus, multimodality therapies have been rigorously established, including surgical resection (6), radiation (7,8), chemotherapy (EORTC trial) (9), and molecular target therapies (PRIME trial, FIRE-3 trial) (10,11).
Various genetic changes have been reported as factors involved in the therapeutic effect of anti-epidermal growth factor receptor (EGFR) monoclonal antibody, including K-ras genomic mutation (12,13) and EGFR genomic amplification (14,15). These factors have already been applied in clinical practice, but treatment failure continues to occur frequently. Recently, the phosphatase of regenerating liver-3 (PRL-3)-induced activation of EGFR was reported to be primarily attributable to the transcriptional downregulation of protein tyrosine phosphatase 1B (PTP1B), an inhibitory phosphatase for EGFR, and oncogene addiction to EGFR, which enhances tumour sensitivity in anti-EGFR cancer therapy in patients with CRC (16). Accordingly, the PRL-3 gene status has attracted attention as a new therapeutic biomarker for CRC.
A number of genetic and epigenetic abnormalities are related to colorectal tumour progression (17). Normal colorectal mucosa changes into adenoma, accompanied by APC gene mutations (18), and subsequent progression to atypical adenoma is related to K-ras gene mutations (19). p53 gene mutations finally cause malignant transformation (20). Moreover, there have been critical reports that 8q chromosome amplification is associated with the acquisition of metastatic potential (21). This locus includes many oncogene candidates, such as PRL-3 and c-myc. PRL-3 and c-myc genes exhibit gene amplification accompanied by gene overexpression in CRC (21), and an association with the progression of CRC or vascular invasion has been reported (22). PRL-3 is a member of the protein tyrosine phosphatase family, the members of which play important roles in signalling pathways. We recently reported that PRL-3 genomic amplification and overexpression were confirmed in gastric cancer, oesophageal squamous cell cancer, and CRC (23–26), and we revealed a relationship between these changes and cancer invasion and a poor prognosis. Especially in terms of CRC, PRL-3 genomic gains (defined larger as over two-fold) were significantly increased in liver metastases (26).
In the current study, the genomic copy numbers of PRL-3, c-myc and EGFR were simultaneously investigated in both primary CRC and corresponding liver metastases to clarify the clinical relevance of these oncogenes during metastatic formation.
Patients and methods
Patients
A total of 35 patients with histologically confirmed liver metastases (synchronous, n=11 and metachronous, n=24) of CRC underwent surgical resection of the liver and primary tumours at the Department of Surgery, Kitasato University School of Medicine (Sagamihara, Japan), between 1993 and 2007. Nakayama et al previously reported 44 cases in this series (26), but 9 of those cases were excluded because additional exploration was impossible due to a lack of available DNA from the tumour tissues. The 35 cases in the present series were composed of stage I (n=3), stage IIA (n=7), stage IIB (n=1), stage IIIB (n=9), stage IIIC (n=2), stage IVA (n=9), and stage IVB (n=4) disease at the time of the diagnosis of the primary CRC.
Clinicopathological factors
The included clinicopathological factors were age, sex, tumour portion, tumour size, 7th UICC pT factor, 7th UICC pN factor, ly factor (lymphatic permeation), v factor (vascular permeation), synchronous or metachronous liver metastasis, and 7th UICC stage, in addition to the genomic copy statuses of the PRL-3, c-myc and EGFR genes.
DNA extraction from the tissues
Tissue sections from the tumours (primary CRC or the corresponding liver metastases from CRC) and corresponding normal mucosa obtained at least 5 cm from the tumour edge were precisely dissected on haematoxylin and eosin-stained slides; genomic DNA was subsequently extracted using a QIAamp® DNA FFPE kit (Qiagen GmbH, Hilden, Germany).
PRL-3, c-myc, and EGFR genomic gain statuses in primary CRC and liver metastases
Quantitative polymerase chain reaction (qPCR) was performed to quantify each genomic amplification in triplicate samples using iQ™ Supermix and the iCycler iQ™ Real-Time PCR Detection system (both Bio-Rad Laboratories, Hercules, CA, USA). To normalise each gene copy number per cell, β-actin was used as an endogenous reference, as previously described (23,24). The ΔCt values were calculated as Ct (PRL-3)-Ct (β-actin) for each sample. The copy number relative to the corresponding normal tissue was determined as 2−ΔΔCt, where ΔΔCt=ΔCt (tumour)-ΔCt (corresponding normal tissues). The PCR conditions and sequences for the β-actin/PRL-3 primers and probes have been previously described (23) (β-actin: Forward, 5′-tggtgtttgtctctctgactaggtg-3′; reverse, 5′-ctaagtgtgctggggtcttgg-3′; probe, 5′-tggctcgtgtgacaaggccatg-3′, PRL-3: 5′-aaagattggcgagaacagca-3′; reverse, 5′-atcccagacacacaccgaac-3′; probe, 5′-tggtgtttgtctctctgactaggtg-3′). The EGFR primers and probe were based on those reported by Moroni et al (15) (forward, 5′-gaattcggatgcagagcttc-3′; reverse, 5′-gacatgctgcggtgttttc-3′; probe, 5′-ctctgtttcagggcatgaactact-3′). The c-myc primers and probe were prepared using Primer 3 software (forward, 5′-agcccactggtcctcaagag-3′; reverse, 5′-cttggcagcaggatagtccttc-3′; probe, 5′-tctccacacatcagcacaactacgcagc-3′). DNA ratios for the tumour (primary CRC or liver metastasis) tissues (T) relative to the corresponding normal tissues (N) (T/N ratio) that were equal to or greater than 2-fold were defined as positive genomic gains.
Immunohistochemical study
Liver metastases from CRC were immunohistochemically stained for PRL-3, c-myc, EGFR, and E-cadherin using the following respective antibodies: anti-human/-mouse/-rat PRL-3 antibody Clone 334407, Human c-Myc Antibody Monoclonal Mouse IgG1 Clone #9E10 cat. no. MAB3696 (both R&D Systems Inc., Minneapolis, MN, USA), Anti EGFR UltraMAB (R) UM570070 (OriGene Technologies Inc., Rockville, MD, USA), and G-10:sc-8426 diluted 1:200 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
FFPE tissue blocks of cases 24 and 26 were cut into thin sections (4 µm thick) that were then deparaffinized with xylene and dehydrated through a stepwise series of ethanol. For antigen activation, samples were immersed in pH 6 citrate buffer and boiled in a microwave for 15 min. The sections were then incubated in 3% aqueous hydrogen peroxide for 15 min to inactivate endogenous peroxidases. The sections were incubated with each of the above primary antibodies overnight at 4°C. The secondary antibody reaction was performed using the Histofine Simple Stain MAX-PO (MULTI) kit (Nichirei, Tokyo, Japan) according to the manufacturer's protocol. Color was developed by incubating with ImmPACT DAB (Vector Laboratories, Inc., Burlingame, CA, USA) for 5 min. Mayer's Hematoxylin Solution was used to stain nuclei. The slides were observed with an optical microscope at magnification, 400.
Statistical analysis
Continuous variables were evaluated using a paired Student t-test, and categorical variables were evaluated using the Fisher exact test or the Chi square test, as appropriate. P<0.05 was considered to indicate a statistically significant difference. All the calculations were performed using JMP® 10 software (SAS Institute Inc., Cary, NC, USA).
Results
Genomic gains in PRL-3, c-myc and EGFR in primary CRC and corresponding liver metastases as evaluated using Q-PCR
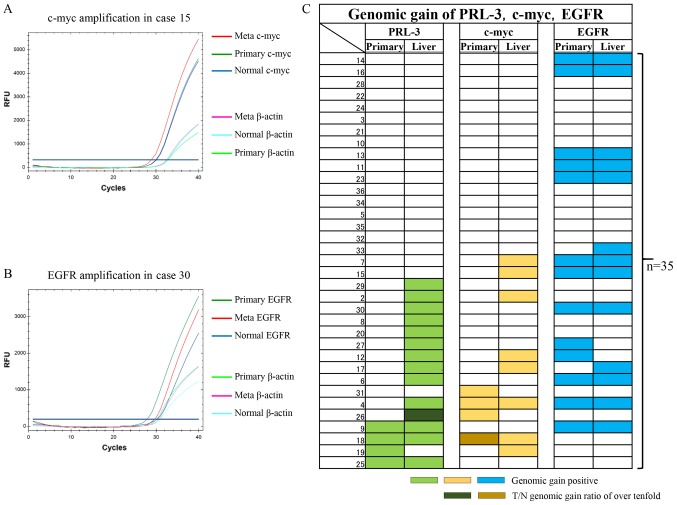
In this study, Q-PCR protocols for the c-myc gene and the EGFR gene were initially developed to assess the genomic copy number, and these protocols were used in addition to the previously reported protocol for the PRL-3 gene (26). Representative genomic quantifications for the c-myc gene and the EGFR gene are shown in Fig. 1A and B, respectively. In Fig. 1A, c-myc genomic gains (2.1-fold for both) were recognised only in the liver metastases, compared with the corresponding primary tumours (Primary) and the corresponding normal tissues (Normal). In Fig. 1B, EGFR genomic gains (5.7-fold and 9.6-fold, respectively) were recognised in both the liver metastases and the corresponding primary tumours (Primary), compared with the corresponding normal tissues (Normal).
Figure 1.
qPCR analysis examining genomic gains in c-myc and EGFR genes as well as gains in the PRL-3 gene in 35 primary CRC and corresponding liver metastases. (A) Representative case with a c-myc gain. The β-actin gene was used as an internal control for the DNA. Note that the β-actin levels of the Meta, Primary and Normal tissues were almost the same, unlike the levels of the c-myc gene. (B) Representative case with an EGFR gain. The β-actin gene was used as an internal control for the DNA. Note that the β-actin levels of the Meta, Primary and Normal tissue were almost the same, unlike the levels of the EGFR gene. (C) Panel showing the genomic gain statuses of the PRL-3 (light green), c-myc (yellow) and EGFR (blue) genes using coloured bars. The two dark-coloured bars show T/N genomic gain ratios of over 10-fold. qPCR, quantitative polymerase chain reaction analysis; CRC, colorectal cancer; EGFR, epidermal growth factor receptor; PRL-3, phosphatase of regenerating liver-3; RFU, relative fluorescence unit.
The genomic gain statuses of c-myc and EGFR were then compared with the PRL-3 genomic gain status in the 35 previously reported primary CRC and corresponding liver metastases (Fig. 1C) (26). In the primary CRC tumours, genomic gains in PRL-3, c-myc, and EGFR were seen in 4 (11%), 4 (11%), and 13 (37%) cases, respectively. Among the 3 genes, EGFR genomic gains were the most frequent, followed by PCR-3 and c-myc, in the primary CRC tissues. Genomic gains in the 3 genes were mutually exclusive in the primary CRC tissues. As a result, a genomic gain in any of the 3 genes was observed in 18 cases (51%).
In the corresponding liver metastases, genomic gains in PRL-3, c-myc, and EGFR were seen in 14 (40%), 8 (23%), and 13 (37%) cases, respectively. A genomic gain in any of the 3 genes was observed in 23 cases (66%). Among the 3 genes, a genomic gain in PRL-3 was the most frequent, followed by EGFR and c-myc. A genomic gain in c-myc was partially redundant with PRL-3 in the liver metastases. Interestingly, genomic gains in the EGFR gene were consistent between both primary CRC and liver metastases (P=0.0000008).
T/N ratios of PRL-3, c-myc and EGFR genetic copies between primary CRC and corresponding liver metastases
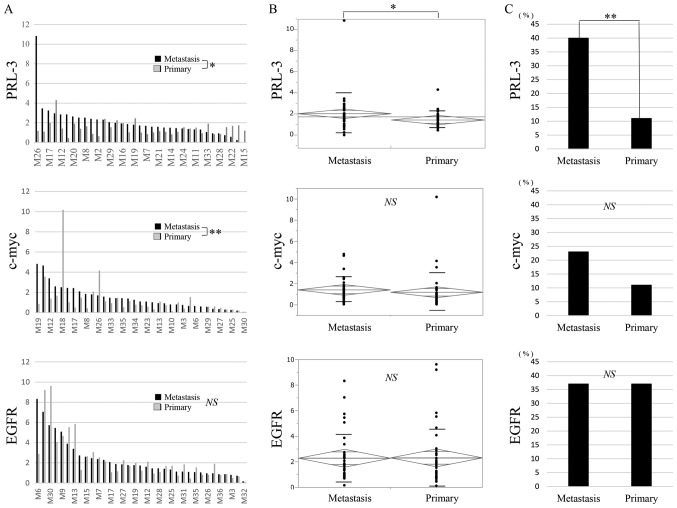
The T/N ratios of the PRL-3 gene are shown for primary CRC and the corresponding liver metastases in Fig. 2A. The T/N ratio ranged from 0.4 to 4.3 (mean, 1.4) in the primary CRC and from 0 to 10.8 (mean, 2.0) in the corresponding liver metastases. A significant difference in the T/N ratios of the PRL-3 gene was observed between the primary CRC and the corresponding liver metastases when compared using a t-test (Fig. 2A; P=0.03) and an analysis of variance (ANOVA) (Fig. 2B; P=0.03). A genomic gain in PRL-3 was more frequently seen in the liver metastases than in the corresponding primary CRC (Fig. 2C; P=0.004).
Figure 2.
DNA T/N ratio in primary CRC and corresponding liver metastases. (A) The T/N ratios of the PRL-3 gene and the c-myc gene exhibited significant differences between the primary CRC (grey bar) and the corresponding liver metastases (black bar), respectively. (B) In an ANOVA, the T/N ratio of the PRL-3 gene was the only significant difference between primary CRC and the corresponding liver metastases. Error bars indicate respective standard deviation. (C) Only a PRL-3 genomic gain was observed significantly more frequently in the liver metastases than in the corresponding primary CRC. *P<0.05; **P<0.01. PRL-3, phosphatase of regenerating liver-3; NS, not significant; CRC, colorectal cancer; PRL-3, phosphatase of regenerating liver-3; EGFR, epidermal growth factor receptor; ANOVA, analysis of variance.
The T/N ratios of the c-myc gene are also shown for primary CRC and the corresponding liver metastases in Fig. 2A. The T/N ratio ranged from 0.05 to 10.2 (mean, 1.2) in the primary CRC and from 0.05 to 4.8 (mean, 1.4) in the corresponding liver metastases. A significant difference in the T/N ratios of the c-myc gene was observed between the primary CRC and the corresponding liver metastases when compared using a Wilcoxon signed-rank sum analysis (Fig. 2A; P=0.009), but a significant difference was not seen using an ANOVA (Fig. 2B). A genomic gain in c-myc was more frequently seen in the liver metastases than in the corresponding primary CRC (Fig. 2C).
The T/N ratios of the EGFR gene are also shown for primary CRC and the corresponding liver metastases in Fig. 2A. The T/N ratio ranged from 0.1 to 9.6 (mean, 2.3) in the primary CRC and from 0.2 to 8.3 (mean, 2.3) in the corresponding liver metastases. A significant difference in the T/N ratio of the EGFR gene was not observed between the primary CRC and the corresponding liver metastases when compared using a Wilcoxon analysis (Fig. 2A) or an ANOVA (Fig. 2B). The genomic gain in EGFR was equal between the primary CRC and the corresponding liver metastases (Fig. 2C).
Clinicopathological analysis of PRL-3, c-myc and EGFR genomic gains in primary CRC tissues and corresponding liver metastases
We then investigated the associations between PRL-3, c-myc, and EGFR genomic gains and clinicopathological factors in the 35 primary CRC. In the primary CRC tissues, genomic gains in the PRL-3 and c-myc genes were not correlated with any factors, while that of EGFR was significantly associated with tumour size (Table I, P=0.04). In the liver metastases, the c-myc genomic status was significantly associated with the v factor (P<0.01), and the EGFR genomic status was significantly correlated with tumour size (P=0.04).
Table I.
Univariate relations of clinicopathological factors to the three genomic gain in liver metastasis of CRC.
| PRL-3 genomic gain (%) | c-myc genomic gain (%) | EGFR genomic gain (%) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number (%) | Primary | Liver | Primary | Liver | Primary | Liver | |||||||||||||
| Patient variable | n=35 | (−) n=31 | (+) n=4 | P-value | (−) n=21 | (+) n=14 | P-value | (−) n=31 | (+) n=4 | P-value | (−) n=27 | (+) n=8 | P-value | (−) n=22 | (+) n=13 | P-value | (−) n=22 | (+) n=13 | P-value |
| Age, years median (range) | 62 (37–82) | NS | NS | NS | NS | NS | NS | ||||||||||||
| ≥60 | 19 (54) | 16 (46) | 3 (9) | 13 (37) | 6 (17) | 16 (46) | 3 (9) | 14 (40) | 5 (14) | 14 (40) | 5 (14) | 13 (37) | 6 (17) | ||||||
| >60 | 16 (46) | 15 (43) | 1 (3) | 8 (23) | 8 (23) | 15 (43) | 1 (3) | 13 (37) | 3 (9) | 8 (23) | 8 (23) | 9 (26) | 7 (20) | ||||||
| Sex | NS | NS | NS | NS | NS | ||||||||||||||
| Male | 24 (69) | 22 (63) | 2 (6) | 14 (37) | 10 (29) | 22 (63) | 2 (6) | 18 (51) | 6 (17) | 14 (40) | 10 (29) | 15 (43) | 9 (26) | ||||||
| Female | 11 (34) | 9 (26) | 2 (6) | 7 (20) | 4 (11) | 9 (26) | 2 (6) | 9 (26) | 2 (6) | 8 (23) | 3 (9) | 7 (20) | 4 (11) | ||||||
| Tumour size, cm (mean ± SD) | 4.72 (±1.97) | NS | NS | NS | NS | 0.04 | 0.04 | ||||||||||||
| ≥4.7 | 19 (54) | 17 (49) | 2 (6) | 12 (34) | 7 (20) | 18 (51) | 1 (3) | 14 (40) | 5 (14) | 9 (26) | 10 (28) | 9 (26) | 10 (28) | ||||||
| >4.7 | 16 (46) | 14 (40) | 2 (6) | 9 (26) | 7 (20) | 13 (37) | 3 (9) | 13 (37) | 3 (9) | 13 (37) | 3 (9) | 13 (37) | 3 (9) | ||||||
| Tumour position | NS | NS | NS | NS | NS | NS | |||||||||||||
| Colon | 28 (80) | 25 (71) | 3 (9) | 16 (46) | 12 (34) | 22 (63) | 6 (17) | 24 (69) | 4 (11) | 17 (29) | 11 (31) | 18 (51) | 10 (29) | ||||||
| Rectum | 7 (20) | 6 (17) | 1 (3) | 5 (14) | 2 (6) | 5 (14) | 2 (6) | 7 (20) | 0 | 5 (14) | 2 (6) | 4 (11) | 3 (9) | ||||||
| N factor | NS | NS | NS | NS | NS | NS | |||||||||||||
| N0 | 12 (34) | 11 (31) | 1 (3) | 7 (20) | 5 (14) | 9 (26) | 3 (9) | 8 (23) | 4 (11) | 7 (20) | 5 (14) | 8 (23) | 4 (11) | ||||||
| N1 | 13 (37) | 11 (31) | 2 (6) | 9 (26) | 4 (11) | 12 (34) | 1 (3) | 10 (29) | 3 (9) | 8 (23) | 4 (11) | 9 (26) | 4 (11) | ||||||
| N2 | 10 (29) | 9 (26) | 1 (3) | 5 (14) | 5 (14) | 10 (29) | 0 | 9 (26) | 1 (3) | 6 (17) | 4 (11) | 5 (14) | 5 (14) | ||||||
| ly factor | NS | NS | NS | NS | NS | NS | |||||||||||||
| ly0 | 2 (6) | 2 (6) | 0 | 2 (6) | 0 | 2 (6) | 0 | 2 (6) | 0 | 2 (6) | 0 | 2 (6) | 0 | ||||||
| ly1 | 12 (34) | 12 (34) | 0 | 8 (23) | 4 (11) | 9 (26) | 3 (9) | 9 (26) | 3 (9) | 6 (17) | 6 (17) | 6 (17) | 6 (17) | ||||||
| ly2 | 13 (37) | 8 (23) | 4 (11) | 6 (17) | 6 (17) | 11 (31) | 1 (3) | 9 (26) | 3 (9) | 7 (20) | 5 (14) | 7 (20) | 5 (14) | ||||||
| ly3 | 8 (23) | 8 (23) | 0 | 4 (11) | 4 (11) | 8 (23) | 0 | 6 (17) | 2 (6) | 6 (17) | 2 (6) | 6 (17) | 2 (6) | ||||||
| v factor | NS | NS | NS | <0.01 | NS | NS | |||||||||||||
| v0 | 3 (9) | 3 (9) | 0 | 3 (9) | 0 | 2 (6) | 1 (3) | 3 (9) | 0 | 3 (9) | 0 | 3 (9) | 0 | ||||||
| v1 | 10 (29) | 9 (26) | 1 (3) | 7 (20) | 3 (9) | 10 (29) | 0 | 9 (26) | 1 (3) | 7 (20) | 3 (9) | 9 (26) | 4 (11) | ||||||
| v2 | 13 (37) | 11 (31) | 2 (6) | 7 (20) | 6 (17) | 12 (34) | 1 (3) | 11 (31) | 2 (6) | 6 (17) | 7 (20) | 7 (20) | 6 (17) | ||||||
| v3 | 9 (26) | 8 (23) | 1 (3) | 4 (11) | 5 (14) | 7 (20) | 2 (6) | 4 (11) | 5 (14) | 6 (17) | 3 (9) | 6 (17) | 3 (9) | ||||||
| 7th UICC stage | NS | NS | NS | NS | NS | NS | |||||||||||||
| I | 3 (9) | 3 (9) | 0 | 1 (3) | 2 (6) | 2 (6) | 1 (3) | 2 (6) | 1 (3) | 3 (9) | 0 | 3 (9) | 0 | ||||||
| IIA | 7 (20) | 7 (20) | 0 | 5 (14) | 2 (6) | 7 (20) | 0 | 5 (14) | 2 (6) | 2 (6) | 5 (14) | 3 (9) | 4 (11) | ||||||
| IIB | 1 (3) | 0 | 1 (3) | 0 | 1 (3) | 0 | 1 (3) | 0 | 1 (3) | 1 (3) | 0 | 1 (3) | 0 | ||||||
| IIC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| IIIA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| IIIB | 9 (26) | 6 (17) | 3 (9) | 3 (9) | 6 (17) | 8 (23) | 1 (3) | 6 (17) | 3 (9) | 5 (14) | 4 (11) | 6 (17) | 3 (9) | ||||||
| IIIC | 2 (6) | 2 (6) | 0 | 2 (6) | 0 | 2 (6) | 0 | 2 (6) | 0 | 2 (6) | 0 | 1 (3) | 1 (3) | ||||||
| IVA | 9 (26) | 9 (26) | 0 | 7 (20) | 2 (6) | 8 (23) | 1 (3) | 8 (23) | 1 (3) | 6 (17) | 3 (9) | 5 (14) | 4 (11) | ||||||
| IVB | 4 (11) | 4 (11) | 0 | 3 (9) | 1 (3) | 4 (11) | 0 | 4 (11) | 0 | 3 (9) | 1 (3) | 3 (9) | 1 (3) | ||||||
| Liver metastatic | NS | NS | NS | NS | NS | NS | |||||||||||||
| Simultaneous | 11 (31) | 11 (31) | 0 | 8 (23) | 3 (9) | 10 (29) | 1 (3) | 10 (29) | 1 (3) | 8 (23) | 3 (9) | 7 (20) | 4 (11) | ||||||
| Metachronous | 24 (69) | 20 (57) | 4 (11) | 13 (37) | 11 (31) | 21 (88) | 3 (9) | 21 (88) | 7 (20) | 14 (40) | 10 (29) | 15 (43) | 9 (26) | ||||||
NS, not significant; CRC, colorectal cancer; PRL, phosphatase of regenerating liver; EGFR, epidermal growth factor receptor.
Based on the clinical data for the liver metastases, the genomic gains in PRL-3 and EGFR were relatively independent, while that of c-myc was redundant with that of PRL-3 in the liver metastases. The former observation reflects the central role of the PRL-3/EGFR pathway in tumour progression through PRL-3-induced EGFR oncogene addiction (16), while the latter may reflect the proximity of c-myc to the PRL-3 genomic locus (21).
Immunohistochemical experiments for liver metastasis
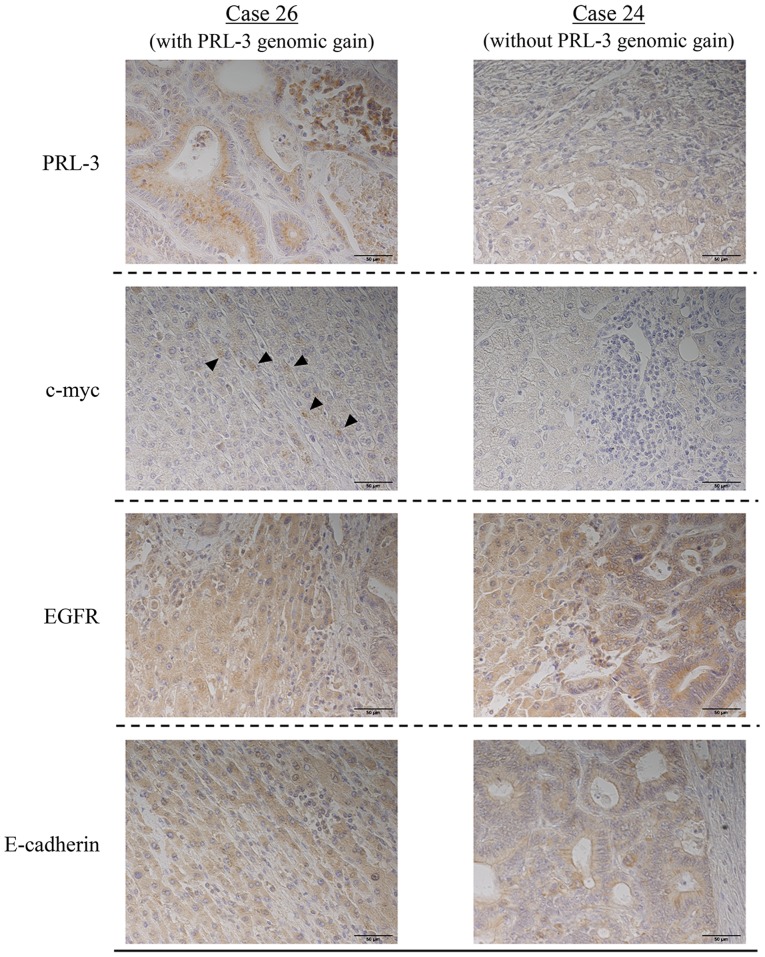
Cases 26 and 24, which were representative of the presence and absence of a PRL-3 genomic gain, respectively, were used to investigate changes in the protein expression of c-myc or EGFR according to PRL-3 genomic gain (Fig. 3). Neither of these cases exhibited a c-myc or an EGFR genomic gain. PRL-3 protein was observed in case 26 but not in case 24, suggesting a correlation with its gene amplification. The expression of EGFR protein was observed in both case 26 and 24, while c-myc protein was only expressed in case 26.
Figure 3.
Immunostaining of liver metastases from CRC. Staining for PRL-3 protein was positive in case 26 and negative in case 24. Staining for c-myc protein was positive in case 26 (arrowhead) and negative in case 24. Staining for EGFR and for E-cadherin protein were both positive in cases 26 and 24. (magnification, ×400). CRC, colorectal cancer; EGFR, epidermal growth factor receptor; PRL-3, phosphatase of regenerating liver-3.
Moreover, the expression of E-cadherin protein was examined in liver metastases to clarify the relationship between EMT markers and amplification of the PRL-3 gene. E-cadherin protein was observed in both cases 26 and 24, suggesting that the expressions of PRL-3 protein and E-cadherin protein were not correlated.
Discussion
Gene copy number changes are major aberrations that occur mostly upstream of cancer cells, and their potential role as molecular targets in cancer therapy has been reported (27). Nakayama et al (26) showed, for the first time, that a genomic gain in PRL-3 is associated with lymph node metastasis and liver metastasis in CRC. In the current study, Q-PCR analyses for the c-myc gene and the EGFR gene were additionally performed using both liver metastases and the corresponding primary tumours, and the results were compared with the genomic gain status of PRL-3 to investigate the relationship among the 3 genes during liver metastasis from CRC.
In our previous experiment, the genomic gain in PRL-3 in a primary tumour was associated with the liver metastasis of CRC, progression, a poor tumour grade, and a poor prognosis (26); in the present study, however, no significant correlations between a genomic gain in PRL-3 and clinicopathological factors were observed except for liver metastasis because of the relatively small number of samples that were tested. Nevertheless, the genomic copy numbers of PRL-3 in the liver metastases were significantly higher than those in the corresponding primary tumours (P=0.004) (Fig. 2C). Because of this result, we additionally examined the genomic gains in the c-myc and EGFR genes in the same patients.
Both PRL-3 and c-myc are located adjacently on chromosome 8q24. Buffart et al (21) reported that the gene amplification of PRL-3 and c-myc in advanced CRC was observed more frequently than that in early-stage CRC, but in their heat map, the copy number ratio of these two genes did not necessarily coincide in the same patient. Moreover, Saha et al (28) showed that gene amplification of c-myc was absent in cases with PRL-3 amplified on chromosome 8q24.3, and Zimmerman et al (29) confirmed an increase in c-myc protein in a PRL-3-knockout mouse model. Hence, these two genes are not necessarily synchronised for genomic amplification even though they are located on the same chromosome, indicating a complementary role in liver metastasis. Our results supported this hypothesis, since PRL-3 and c-myc genomic gene gains were both present in the primary tumour tissue in only once case (Fig. 1C). On the other hand, the gains in these two genes were often redundant in liver metastases. In terms of gene amplification, the relationship between the PRL-3 and c-myc genes was considered to be complementary prior to liver metastasis and synergistic thereafter. Gains in both the PRL-3 and the c-myc genes might occur together at the time of liver metastasis, while genomic gains in these two genes might occur separately in primary CRC. The PRL-3 genomic gain is likely to contribute more to the liver metastatic capacity than a gain in c-myc because 1 case with liver metastasis from a stage I primary cancer exhibited the highest genomic gain in PRL-3, but the genomic gain in c-myc was relatively low in the corresponding liver metastasis (Fig. 1C).
In this study, the genomic gain in EGFR was correlated with the tumour size (P=0.04) in both the liver metastases and the corresponding primary tumours (Table I). EGFR gene activation is known to be important for tumour progression, and EGFR-targeting therapies have been rigorously established for CRC and non-small cell lung cancer in clinical practice. Overexpression of the EGFR gene is frequently recognised in CRC (25–80%) and is associated with aggressive disease and a poor prognosis (30,31). In our study, a genomic gain in EGFR was not associated with the acquisition of liver metastatic potential but was useful as a predictor of liver metastasis, since it was consistent between the liver metastases and the corresponding primary tumours (P=0.0000008).
In a combination analysis of the 3 genes, their positive gains were exclusive of each other in primary CRC. The PRL-3 gene exerts various functions, such as proliferation and the growth and inhibition of apoptosis signals, through the activation of EGFR and c-myc. In the present study, 23 cases (66%) exhibited gains in any of the 3 genes; moreover, the genomic gains in c-myc and EGFR in liver metastases were significantly associated with vascular invasion factor and tumour growth, respectively. The PRL-3, c-myc, and EGFR genes are likely to have complementary roles in the proliferation of liver metastases in CRC. In liver metastasis, a PRL-3 genomic gain is correlated with its own protein expression, and c-myc protein was induced by the PRL-3 protein or a genomic gain (Fig. 3). PRL-3 protein might increase the expression of c-myc protein in liver metastasis independently of the EGFR protein. PRL-3-targeted therapy, which targets a point furthest upstream in the signalling cascade, may be effective against CRC with liver metastasis as a new treatment to improve prognosis in patients with a PRL-3 genomic gain.
Anti-EGFR monoclonal antibodies have been used in patients with unresectable advanced or recurrent CRC who are negative for RAS mutations. The K-ras gene wild type is seen in 60–70% of CRC with metastasis; however, the actual response rate to anti-EGFR monoclonal antibody was reported to be 10–40% (32). Moreover, some reports have indicated that only about 10% of patients with CRC and metastasis who are resistant to chemotherapy responded to treatment with P-mab or cetuximab (33,34). Therefore, it is necessary to predict responders to anti-EGFR monoclonal antibody therapy. Balin-Gauthier et al (35) reported that a response to cetuximab was correlated with the level of phosphorylated EGFR but not with the level of EGFR expression. On the other hand, another report indicated that the response to anti-EGFR treatment is influenced by the EGFR amplification status (15). Al-Aidaroos et al (16) showed that in mice, PRL-3 activates EGFR leading to EGFR oncogene addiction and is a molecular marker that is indicative of a response to anti-EGFR monoclonal drugs. Intriguingly, there have been reports describing the effects of monoclonal antibodies against EGFR, if the EGFR gene is amplified, even in the presence of a K-ras exon 2 mutation (16). Whether the expression of PRL-3 or EGFR has a greater effect on anti-EGFR treatments in CRC cell lines remains unclear. However, considering that PRL-3 is located upstream of EGFR, the PRL-3 gene could be a superior therapeutic target than EGFR in anti-EGFR monoclonal antibody therapy. In a preliminary investigation performed at our hospital, anti-EGFR antibody treatment tended to have a good therapeutic effect (partial response) in cases with a high expression of PRL-3 (data not shown), similar to a recent report that actually demonstrated this effect in a prospective cohort study (16).
Limitations
First, the number of target cancer tissues was limited, and the tissues had been archived. Several effects arising from the long-term preservation of tissues used for DNA extraction and tissue immunostaining are conceivable. In the future, we would like to perform additional studies using a larger number of freshly obtained CRC tissues and corresponding liver metastases. Second, all the experiments were performed in vitro. Since in vivo experiments are necessary for the development of clinical applications of PRL-3 as a biomarker, we plan to perform prospective research involving such investigations in the near future.
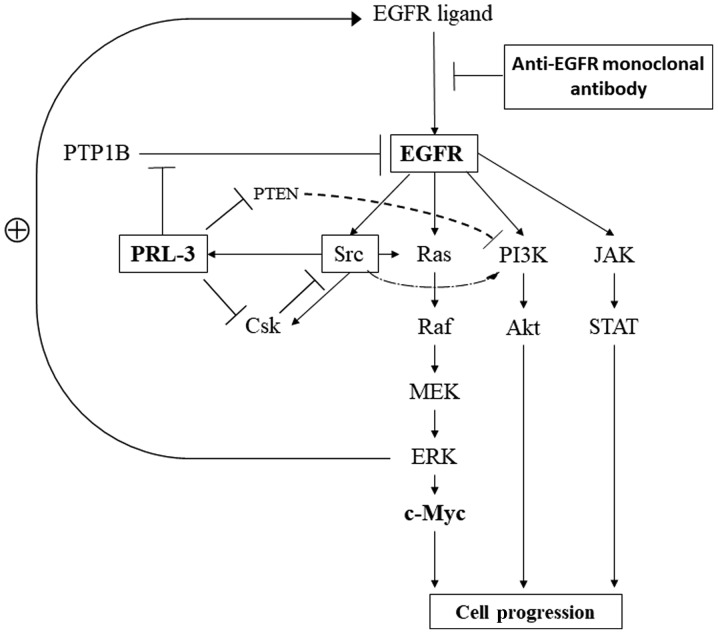
Finally, we summarised the PRL-3/EGFR/K-ras pathway based on previous reports (Fig. 4). Numerous reports have indicated that EGFR is involved in cell proliferation and survival through individual critical pathways, such as MAPK, PI3K/Akt, and JAK/STAT, as well as those of feedback mechanisms (36,37). PRL-3 has been reported to be involved in the activation of EGFR through the inhibition of PTP1B (12), the promotion of cell motility and invasion related to Src and Csk (38,39), and the up-regulation of the PI3K/Akt pathway through the down-regulation of phosphatase and tensin homologue deleted chromosome 10 (PTEN), which are inhibitors of the PI3K pathway for the dephosphorylation of PI3K products (40).
Figure 4.
Signalling pathways related to PRL-3/EGFR/c-myc genes in cancer cells. EGFR, epidermal growth factor receptor; PTP1B, protein tyrosine phosphatase 1B; PTEN, phosphatase and tensin homolog; PRL-3, phosphatase of regenerating liver-3; PI3K, phospho-inositide 3 kinase; JAK, Janus kinase; Csk, C-terminal Src kinase; STAT, signal transducers and activators of transcription; MEK, mitogen-activated protein kinase kinase; ERK, extracellular signal-regulated kinase; EGFR, epidermal growth factor receptor.
Genomic gain aberrations in the copy numbers of PRL-3, c-myc and EGFR are frequently observed in liver metastases from CRC. These results may be beneficial for the treatment of CRC patients with liver metastasis and may be useful for the identification of patients who are likely to respond well to anticancer therapies in the near future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during this study are included in the published article.
Authors' contributions
Conception and design: TT, KYa and MW undertook study conception and design and TT, TK, KYo, SI, NN, HKaw, HKat, TS and TN were responsible for the acquisition and analysis and interpretation of data. TT and KYa drafted the article and revised it critically for important intellectual content. TT, KYa, TK, KYo, SI, NN, HKaw, HKat, TS, TN and MW gave final approval for this version to be published.
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Research Ethics Committee of Kitasato University School of Medicine. All patients agreed to the use of their samples in scientific research. And, written informed consents for pathological investigation were obtained from all patients.
Patient consent for publication
Informed consent for publication was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Watanabe T, Itabashi M, Shimada Y, Tanaka S, Ito Y, Ajioka Y, Hamaguchi T, Hyodo I, Igarashi M, Ishida H, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2014 for treatment of colorectal cancer. Int J Clin Oncol. 2015;20:207–39. doi: 10.1007/s10147-015-0801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katoh H, Yamashita K, Wang G, Sato T, Nakamura T, Watanabe M. Anastomotic leakage contributes to the risk for systemic recurrence in stage II colorectal cancer. J Gastrointest Surg. 2011;15:120–129. doi: 10.1007/s11605-010-1379-4. [DOI] [PubMed] [Google Scholar]
- 3.Katoh H, Yamashita K, Wang G, Sato T, Nakamura T, Watanabe M. Prognostic significance of preoperative bowel obstruction in stage III colorectal cancer. Ann Surg Oncol. 2011;18:2432–2441. doi: 10.1245/s10434-011-1625-3. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita K, Watanabe M. Clinical significance of tumor markers and an emerging perspective on colorectal cancer. Cancer Sci. 2009;100:195–199. doi: 10.1111/j.1349-7006.2008.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katoh H, Yamashita K, Sato T, Ozawa H, Nakamura T, Watanabe M. Prognostic significance of peritoneal tumour cells identified at surgery for colorectal cancer. Br J Surg. 2009;96:769–777. doi: 10.1002/bjs.6622. [DOI] [PubMed] [Google Scholar]
- 6.Kato T, Yasui K, Hirai T, Kanemitsu Y, Mori T, Sugihara K, Mochizuki H, Yamamoto J. Therapeutic results for hepatic metastasis of colorectal cancer with special reference to effectiveness of hepatectomy: Analysis of prognostic factors for 763 cases recorded at 18 institutions. Dis Colon Rectum. 2003;46(10 Suppl):S22–S31. doi: 10.1097/01.DCR.0000089106.71914.00. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura T, Yamashita K, Sato T, Ema A, Naito M, Watanabe M. Neoadjuvant chemoradiation therapy using concurrent S-1 and irinotecan in rectal cancer: Impact on long-term clinical outcomes and prognostic factors. Int J Radiat Oncol Biol Phys. 2014;89:547–555. doi: 10.1016/j.ijrobp.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Wong SL, Mangu PB, Choti MA, Crocenzi TS, Dodd GD, III, Dorfman GS, Eng C, Fong Y, Giusti AF, Lu D, et al. American Society of Clinical Oncology 2009 clinical evidence review on radiofrequency ablation of hepatic metastases from colorectal cancer. J Clin Oncol. 2010;28:493–508. doi: 10.1200/JCO.2009.23.4450. [DOI] [PubMed] [Google Scholar]
- 9.Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, Bechstein WO, Primrose JN, Walpole ET, Finch-Jones M, et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): A randomised controlled trial. Lancet. 2008;371:1007–1016. doi: 10.1016/S0140-6736(08)60455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Final results from PRIME: Randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann Oncol. 2014;25:1346–1355. doi: 10.1093/annonc/mdu141. [DOI] [PubMed] [Google Scholar]
- 11.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmuller C, Kahl C, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 12.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 15.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M, Siena S, Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: A cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 16.Al-Aidaroos AQ, Yuen HF, Guo K, Zhang SD, Chung TH, Chng WJ, Zeng Q. Metastasis-associated PRL-3 induces EGFR activation and addiction in cancer cells. J Clin Invest. 2013;123:3459–3471. doi: 10.1172/JCI72968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogelstein B, Kinzler KW. The path to cancer-three strikes and you're out. N Engl J Med. 2015;373:1895–1898. doi: 10.1056/NEJMp1508811. [DOI] [PubMed] [Google Scholar]
- 18.Ashton-Rickardt PG, Dunlop MG, Nakamura Y, Morris RG, Purdie CA, Steel CM, Evans HJ, Bird CC, Wyllie AH. High frequency of APC loss in sporadic colorectal carcinoma due to breaks clustered in 5q21-22. Oncogene. 1989;4:1169–1174. [PubMed] [Google Scholar]
- 19.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 20.Linsalata M, Notarnicola M, Caruso MG, Di Leo A, Guerra V, Russo F. Polyamine biosynthesis in relation to K-ras and P-53 mutations in colorectal carcinoma. Scand J Gastroenterol. 2004;39:470–477. doi: 10.1080/0036552031008755. [DOI] [PubMed] [Google Scholar]
- 21.Buffart TE, Coffa J, Hermsen MA, Carvalho B, van der Sijp JR, Ylstra B, Pals G, Schouten JP, Meijer GA. DNA copy number changes at 8q11-24 in metastasized colorectal cancer. Cell Oncol. 2005;27:57–65. doi: 10.1155/2005/401607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugimachi K, Niida A, Yamamoto K, Shimamura T, Imoto S, Iinuma H, Shinden Y, Eguchi H, Sudo T, Watanabe M, et al. Allelic imbalance at an 8q24 oncogenic SNP is involved in activating MYC in human colorectal cancer. Ann Surg Oncol. 2014;21(Suppl 4):S515–S521. doi: 10.1245/s10434-013-3468-6. [DOI] [PubMed] [Google Scholar]
- 23.Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Watanabe M. Phosphatase of regenerating liver-3 as a convergent therapeutic target for lymph node metastasis in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:543–554. doi: 10.1002/ijc.25082. [DOI] [PubMed] [Google Scholar]
- 24.Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Waraya M, Kawamata H, Nishimiya H, Nakamura K, Watanabe M. Therapeutic potential of PRL-3 targeting and clinical significance of PRL-3 genomic amplification in gastric cancer. BMC Cancer. 2011;11:122. doi: 10.1186/1471-2407-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatate K, Yamashita K, Hirai K, Kumamoto H, Sato T, Ozawa H, Nakamura T, Onozato W, Kokuba Y, Ihara A, Watanabe M. Liver metastasis of colorectal cancer by protein-tyrosine phosphatase type 4A, 3 (PRL-3) is mediated through lymph node metastasis and elevated serum tumor markers such as CEA and CA19-9. Oncol Rep. 2008;20:737–743. [PubMed] [Google Scholar]
- 26.Nakayama N, Yamashita K, Tanaka T, Kawamata H, Ooki A, Sato T, Nakamura T, Watanabe M. Genomic gain of the PRL-3 gene may represent poor prognosis of primary colorectal cancer, and associate with liver metastasis. Clin Exp Metastasis. 2016;33:3–13. doi: 10.1007/s10585-015-9749-7. [DOI] [PubMed] [Google Scholar]
- 27.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St Croix B, Romans KE, Choti MA, Lengauer C, Kinzler KW, Vogelstein B. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman MW, Homanics GE, Lazo JS. Targeted deletion of the metastasis-associated phosphatase Ptp4a3 (PRL-3) suppresses murine colon cancer. PLoS One. 2013;8:e58300. doi: 10.1371/journal.pone.0058300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JC, Wang ST, Chow NH, Yang HB. Investigation of the prognostic value of coexpressed erbB family members for the survival of colorectal cancer patients after curative surgery. Eur J Cancer. 2002;38:1065–1071. doi: 10.1016/S0959-8049(02)00004-7. [DOI] [PubMed] [Google Scholar]
- 31.Porebska I, Harlozińska A, Bojarowski T. Expression of the tyrosine kinase activity growth factor receptors (EGFR, ERB B2, ERB B3) in colorectal adenocarcinomas and adenomas. Tumour Biol. 2000;21:105–115. doi: 10.1159/000030116. [DOI] [PubMed] [Google Scholar]
- 32.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 33.Saltz LB, Meropol NJ, Loehrer PJ, Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 35.Balin-Gauthier D, Delord JP, Rochaix P, Mallard V, Thomas F, Hennebelle I, Bugat R, Canal P, Allal C. In vivo and in vitro antitumor activity of oxaliplatin in combination with cetuximab in human colorectal tumor cell lines expressing different level of EGFR. Cancer Chemother Pharmacol. 2006;57:709–718. doi: 10.1007/s00280-005-0123-3. [DOI] [PubMed] [Google Scholar]
- 36.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 38.Fiordalisi JJ, Dewar BJ, Graves LM, Madigan JP, Cox AD. Src-mediated phosphorylation of the tyrosine phosphatase PRL-3 is required for PRL-3 promotion of Rho activation, motility and invasion. PLoS One. 2013;8:e64309. doi: 10.1371/journal.pone.0064309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang F, Liang J, Wang WQ, Sun JP, Udho E, Zhang ZY. PRL3 promotes cell invasion and proliferation by down-regulation of Csk leading to Src activation. J Biol Chem. 2007;282:5413–5419. doi: 10.1074/jbc.M608940200. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 2007;67:2922–2926. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in the published article.