Abstract
Glioma originates from the glial cells of the spine or brain, and promoter methylation of O6-methylguanine-DNA methyltransferase (MGMT) can promote the chemosensitivity of glioma. The present study aimed to reveal the key genes implicated in MGMT promoter methylation in patients with glioma. RNA-sequencing data and methylation data for glioma were extracted from The Cancer Genome Atlas database. Following expression characteristic analysis and differential expression analysis using unsupervised hierarchical clustering and a rank sum test, the feature genes were identified between high and low methylation groups. Furthermore, multivariate survival analysis for the feature genes was performed using the survival package in R. Additionally, the independent glioma RNA expression datasets GSE7696 and GSE42669 were used to validate the prognostic efficiency of the gene combination. The results indicated that the prognosis of the low methylation group was significantly worse than that of the high methylation group. The ten genes corresponding to the cut-off value of 0.56 (Rho GTPase-activating protein 21, CECR2, histone acetyl-lysine reader, endosulfine α, G-patch domain-containing 8, KIAA1109, MGMT, protocadherin β 13, selenoprotein M, sperm-associated antigen 9 and WD repeat domain 6) were able to significantly predict prognosis and were differentially expressed between the two groups. Multivariate survival analysis suggested that the ten genes were effective for sample classification and prognostic prediction. Furthermore, the validation datasets confirmed the correlation of the ten genes with prognosis. In conclusion, these 10 genes may be mediated by MGMT promoter methylation in glioma. In addition, the ten-gene combination may be associated with the prognosis of patients with glioma.
Keywords: glioma, promoter methylation, feature genes, differential expression analysis, multivariate survival analysis
Introduction
Glioma occurs in the glial cells of the spine or brain, and is classified according to cell type, location and grade (1). The incidence rate of glioblastoma multiforme is the highest (3.20 per 100,000 population) of all malignant central nervous system tumors in the United States (2). Gliomas in different locations exhibit distinct symptoms, and the tumor metastasizes via the cerebrospinal fluid, rather than the bloodstream (3). The majority of gliomas are incurable; in particular, the prognosis for older patients with advanced glioma is poorer (4). The O6-methylguanine-DNA methyltransferase (MGMT) gene, which is located on chromosome 10q26 and codes for a DNA repair enzyme, can eliminate the efficacy of alkylating chemotherapy (5). Promoter methylation of MGMT has been detected in 51–66% of cases of glioblastoma; this methylation inactivates the MGMT gene and promotes the chemosensitivity and survival of patients with glioblastoma (6,7). Therefore, exploring the key genes associated with MGMT promoter methylation in glioma is important.
In recent years, the mechanisms underlying glioma prognosis have been investigated. For example, enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) overexpression is associated with tumor grade and predicts a short survival; therefore, EZH2 is a promising prognostic factor and therapeutic target in patients with glioblastoma (8). Chitinase 3-like 1 (CHI3L1) functions in mediating local invasiveness and malignant transformation; therefore, CHI3L1 may be considered a potential target for the treatment of glioma (9,10). Furthermore, large tumor suppressor kinase 1 is a critical tumor suppressor, the inhibition of which may promote the progression of glioma (11). Conversely, the 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4) gene serves an important role in maintaining cancer stem-like cells in the brain, and high PFKFB4 expression is correlated with unfavorable survival of patients with glioblastoma (12). Nevertheless, the mechanisms underlying the effects of MGMT promoter methylation on the prognosis of patients with glioma have not been thoroughly studied.
Expression profile analysis is widely used for selecting meaningful and important genes from large amounts of data via the evaluation of gene expression levels (13,14). In the present study, RNA-sequencing (RNA-seq) data and methylation data of glioma were downloaded, and were then analyzed through a series of bioinformatics analyses. The present study aimed to reveal the key genes involved in MGMT promoter methylation in patients with glioma, in order to provide potential targets for promoting MGMT promoter methylation and improving the chemosensitivity of patients with glioma.
Materials and methods
Data source and preprocessing
For glioma, RNA-seq data (platform: IlluminaHiSeq), methylation data (platform: HumanMethylation27) and clinical follow-up data were obtained from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) database (updated March 1, 2017). From a total of 295 glioma samples, 77 samples (61 samples from patients that had succumbed, 15 samples from patients that had survived, and 1 sample from a patient for whom there was no survival information) with RNA-seq data and methylation data were selected, and their clinical follow-up data were downloaded. RNA-seq data and methylation data were normalized using the quantile normalization method (15).
Identification of feature genes between high and low methylation samples
The methylation levels of CpG islands in the promoter region of MGMT in the 77 samples were analyzed and ranked. The median methylation level of the CpG sites in CpG islands was determined, and samples with methylation levels >0.35 were defined as high methylation samples, whereas those with methylation levels <0.35 were considered low methylation samples. Methylation levels in the high and low methylation groups were compared using independent samples t-test; P<0.05 was considered to indicate a statistically significant difference. High and low methylation levels indicated patients with high and low MGMT promoter methylation; these patients have differences in prognosis (16). To identify the feature genes that could differentiate high and low methylation samples, the expression levels of each gene (i) in high (H) and low (L) methylation samples were calculated to obtain the difference index (Fi) using R software (https://www.r-project.org/). The formula used was as follows:
To identify the optimal gene combination that could distinguish between high and low methylation samples, the permTS function in the R package perm (https://cran.r-project.org/web/packages/perm/) (17) was used to calculate the significant P-value of each gene in the two groups of samples. The step size for setting the threshold k for Fi was 0.01. The gene combination with |Fi| > k and P<0.01 was selected, and its corresponding expression profile was obtained. Afterwards, the following classifier models were used for performing sample classification:
Bi refers to the mean value between high and low methylation samples; ei represents the expression level of gene i; Vi is the product of Fi and the difference between ei and Bi; N indicates the number of genes under the present threshold; PSj represents the classification score of sample j. Finally, the samples were divided into two groups according to their positive or negative scores. samples with positive scores were divided into a new high methylation group, whereas samples with negative scores were divided into a new low methylation group. Meanwhile, the consistency ratio of the new groups and the previous groups (high and low methylation groups) was calculated. The gene combination that corresponded to the highest consistency ratio was selected as the feature gene set.
Expression characteristic analysis and differential expression analysis of the feature genes
To determine the stability of the feature gene set for classifying samples, five-fold cross validation (18) was performed 10 times. Under each cross validation, the consistency ratio of the new high and low methylation groups, and the previous high and low methylation groups was calculated. Based on the expression levels of the feature genes in each sample, unsupervised hierarchical clustering was performed to observe the expression characteristics of the feature genes using heatmap function (https://stat.ethz.ch/R-manual/R-devel/library/stats/html/heatmap.html) in R package. In addition, the prognostic differences of the samples in different clusters were analyzed using the R package Kaplan Meier (19). Furthermore, Wilcoxon rank sum test (20) was performed in R software to observe the differential expression of the feature genes in high and low methylation samples.
Multivariate survival analysis
Multivariate survival analysis was performed for the feature genes to examine the effects of the feature genes on prognosis. Using the R package survivalROC (https://cran.r-project.org/web/packages/survivalroc/index.html) (21), a receiver operating characteristic (ROC) curve was generated.
Validation of the feature genes using independent datasets
To confirm that the feature genes were repeatable, the validation datasets GSE7696 (22) (platform: Affymetrix Human Genome U133 Plus 2.0 Array; comprised 84 samples, including 78 glioma samples with clinical follow-up data) and GSE42669 (23) (platform: Affymetrix Human Gene 1.0 ST Array; comprised 58 samples, including 55 glioma samples with clinical follow-up data) were downloaded from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) database. Using the R package survival (http://cran.r-project.org/package=survival) (24), analysis of the two datasets was conducted with Cox regression analysis.
Results
Identification of feature genes between high and low methylation samples
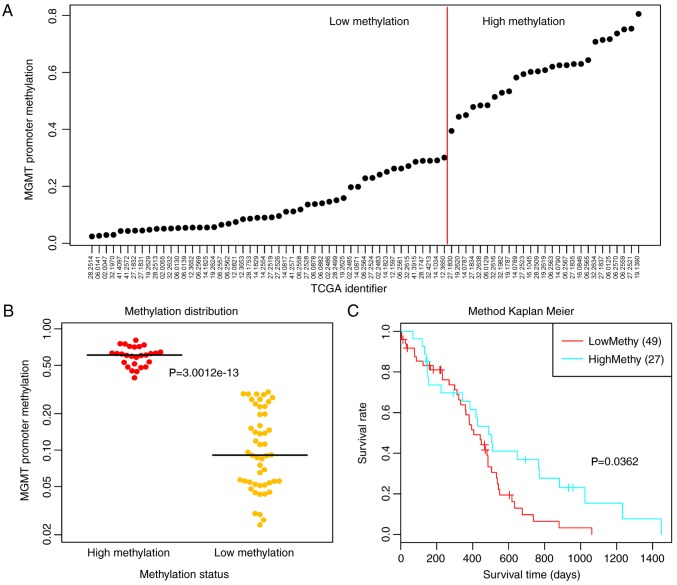
The methylation levels of CpG islands in the promoter region of MGMT were markedly varied in the different samples, and the 77 samples were ranked according to methylation levels (Fig. 1A). Subsequently, the samples were divided into high and low methylation groups using 0.35 as the cut-off point for methylation levels. The results demonstrated that the two groups of samples had significant MGMT methylation differences (P<0.0001; Fig. 1B). Meanwhile, univariate survival analysis revealed that the low methylation group had a significantly poorer prognosis (P=0.0362; Fig. 1C).
Figure 1.
Classification and differential analysis of the 77 samples. (A) High and low methylation groups were divided according to the methylation levels of CpG islands in the promoter region of MGMT (red line indicates the cut-off point of methylation levels, 0.35); (B) Methylation differences of the high and low methylation groups. (C) Prognostic differences of high and low methylation groups. MGMT, O6-methylguanine-DNA methyltransferase; TCGA, The Cancer Genome Atlas.
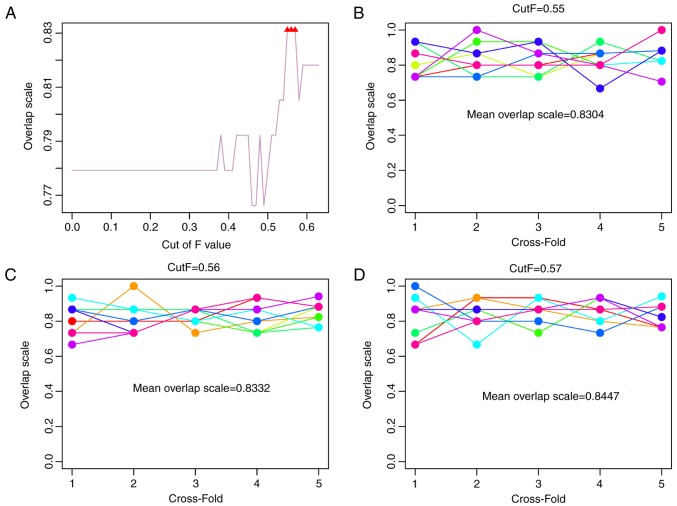
There were 24,991 genes expressed in the 77 samples. A total of 19,659 genes were expressed in >50% of samples and were selected for the following analysis. Briefly, the difference index and P-value of the permutation test were calculated for each gene. The fold change between high and low methylation groups and P-value for MGMT were 1.23 and 9.26×10−11, respectively. Combined with different classifier models, sample classification was performed using gene combinations with different cut-off values. Subsequently, the consistency ratios of the new groups (divided according to the positive and negative PSj classification scores) and the previous groups (divided according to high and low methylation) were calculated. The results revealed that the consistency ratios under different cut-off values were different. In particular, the consistency ratios were higher when the cut-off values were 0.55, 0.56 and 0.57 (Fig. 2A).
Figure 2.
(A) Consistency ratio of the new groups, and the previous high and low methylation groups. The results of five-fold cross validation under the cut-off values (B) 0.55, (C) 0.56 and (D) 0.57.
Furthermore, five-fold cross validation was performed 10 times to detect the consistency ratios under the three cut-off values. The results indicated that the consistency ratios under the cut-off values of 0.55 (Fig. 2B), 0.56 (Fig. 2C) and 0.57 (Fig. 2D) were high and were all >0.83.
Expression characteristic analysis and differential expression analysis of the feature genes
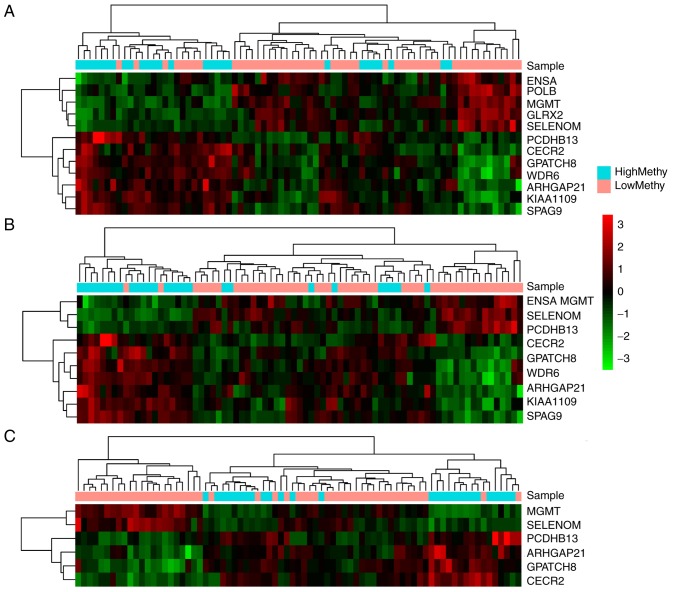
Based on the three cut-off values, three feature gene sets were selected. Subsequently, unsupervised hierarchical clustering analysis was performed for the feature gene sets, which corresponded to the cut-off values of 0.55 (Fig. 3A), 0.56 (Fig. 3B) and 0.57 (Fig. 3C). As shown in Fig. 3A, 11 genes corresponding to cut-off values of 0.55 had high distinction degrees for the high and low methylation groups. Meanwhile, there were 10 genes corresponding to cut-off values of 0.56 (Fig. 3B), which were able to clearly distinguish between the high and low methylation groups. However, the six genes corresponding to cut-off values of 0.57 (Fig. 3C) could not efficiently differentiate between the two groups.
Figure 3.
Clustering heatmaps for the feature gene sets corresponding to the cut-off values of (A) 0.55, (B) 0.56 and (C) 0.57. The horizontal and vertical axes refer to Euclidean distance and Pearson correlation coefficients, respectively.
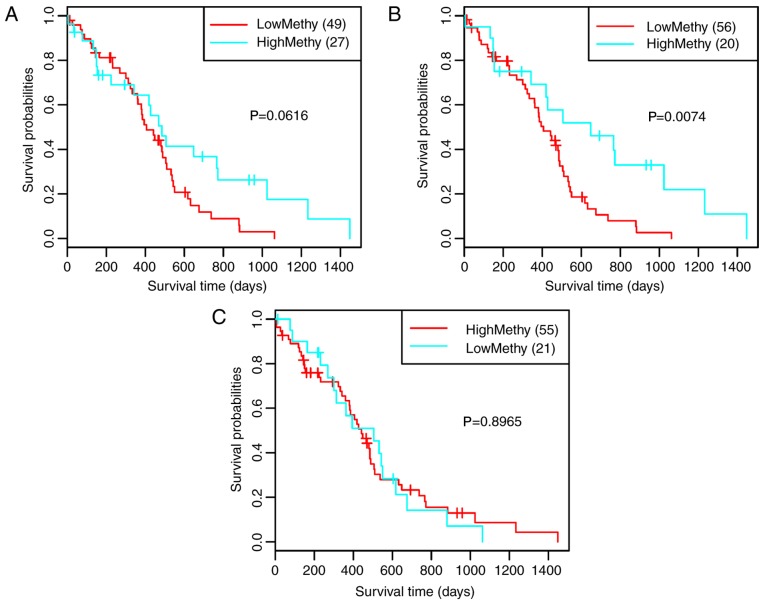
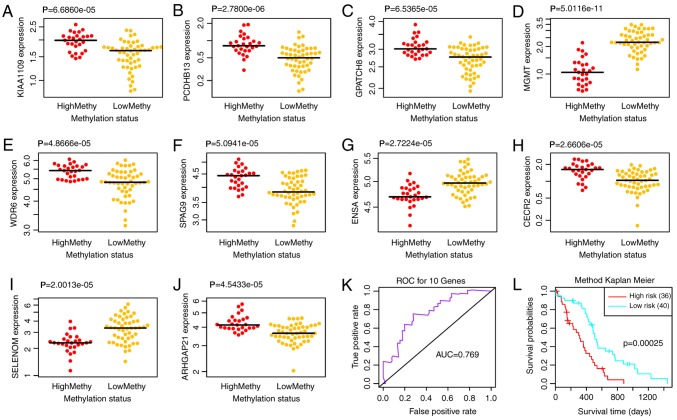
According to the clustering results, the samples were divided into two clusters: High methylation and low methylation. Survival analysis for the clustering results revealed that only the 10 genes corresponding to cut-off values of 0.56 exhibited significant correlation with prognosis (Fig. 4B, P=0.0074). Therefore, the 10 genes were selected as feature genes [Rho GTPase-activating protein 21 (ARHGAP21), CECR2, histone acetyl-lysine reader (CECR2), endosulfine α (ENSA), G patch domain-containing 8 (GPATCH8), KIAA1109, MGMT, protocadherin β 13 (PCDHB13), selenoprotein M (SELM) sperm-associated antigen 9 (SPAG9) and WD repeat domain 6 (WDR6)]. Subsequently, the differential expression of the 10 feature genes in high and low methylation samples was analyzed. As a result, all of the 10 feature genes exhibited significant differential expression between the high and low methylation samples (Fig. 5A-J).
Figure 4.
Survival analysis of the clustering results corresponding to the cut-off values of (A) 0.55, (B) 0.56 and (C) 0.57.
Figure 5.
Differential expression of (A) KIAA1109, (B) PCDHB13, (C) GPATCH8, (D) MGMT, (E) WDR6, (F) SPAG9, (G) ENSA, (H) CECR2, (I) SELM and (J) ARHGAP21 in high and low methylation samples. (K) ROC curve and (L) prognostic differences of the two risk groups. AUC, area under the curve; ROC, receiver operating characteristic.
Multivariate survival analysis
To examine the effects of the 10 feature genes on prognosis, a multivariate survival analysis was conducted. Based on the feature genes, the samples were classified into high and low risk groups. The area under the ROC curve (AUC) was 0.769 (Fig. 5K) and the two risk groups had a significant difference in survival probability (Fig. 5L; P=0.0003), thus indicating that the 10 feature genes could effectively perform classification and prognostic prediction for the samples.
Validation of the feature genes using independent datasets
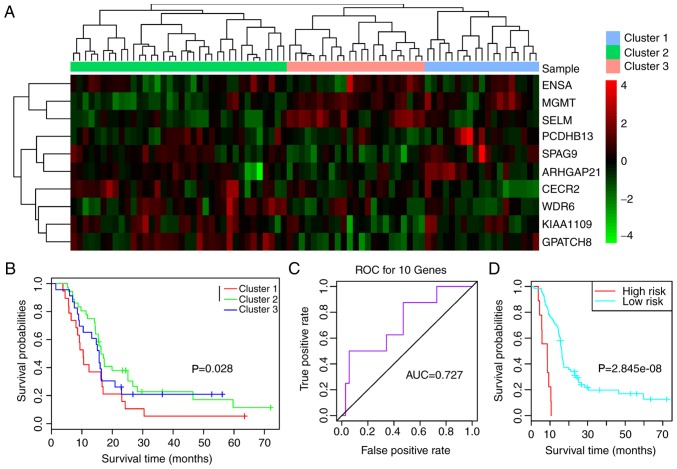
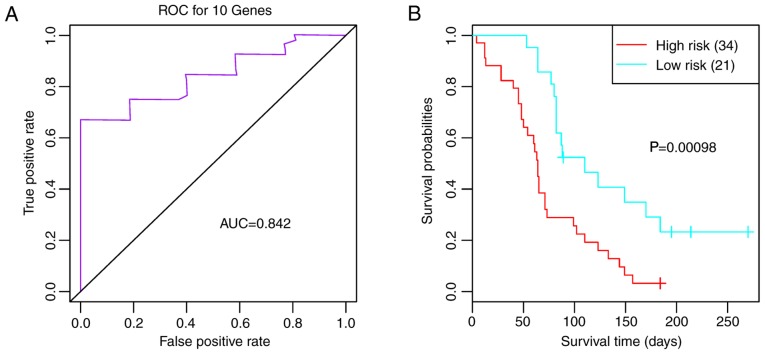
The validation datasets GSE7696 and GSE42669 were downloaded to confirm that the feature genes were repeatable. Combined with the 10 feature genes, survival analysis was performed for GSE7696. As shown in Fig. 6A, the 10 feature genes divided the samples into three clusters (clusters 1, 2 and 3) (Fig. 6A). Survival analysis for the three clusters suggested that prognosis was significantly different between clusters 1 and 2 (P=0.0280; Fig. 6B). The AUC was 0.727 (Fig. 6C) and the two risk groups had a significant difference (Fig. 6D; P<0.0001), suggesting that the 10 feature genes had good classification and prognostic effects for the samples. For the validation dataset GSE42669, the AUC was 0.842 (Fig. 7A), and the high and low risk groups had a significant difference (Fig. 7B; P=0.0010). These findings indicated that the 10 feature genes were key genes that may affect the prognosis of glioma.
Figure 6.
(A) Clustering heatmap of the samples in the GSE7696 dataset based on the 10 feature genes. (B) Survival curves for the three clusters. (C) ROC curve and (D) survival curves for the two risk groups. AUC, area under the curve; ROC, receiver operating characteristic.
Figure 7.
(A) ROC curve and (B) survival curves for the two risk groups in the GSE42669 validation dataset. AUC, area under the curve; ROC, receiver operating characteristic.
Discussion
In the present study, 77 samples were divided into high and low methylation groups. Subsequently, 11, 10 and six genes were revealed to have high distinction degrees for the high and low methylation groups. However, the results of a survival analysis revealed that only the gene set containing 10 genes had a significant result; therefore, the 10 genes were selected as feature genes (including ARHGAP21, CECR2, PCDHB13, MGMT, SELM, SPAG9 and WDR6). Multivariate survival analysis indicated that the 10 feature genes were effective in performing sample classification and prognostic prediction. Furthermore, the 10 feature genes were confirmed by the validation datasets GSE7696 and GSE42669.
ARHGAP21 is considered a potential tumor suppressor gene that may act in mediating the migration of various glial tumor types (25). Chromatin remodeling complexes are important for development, and the transcription factor CECR2 is associated with neurulation and is a composition of the chromatin remodeling complex CECR2-containing remodeling factor (26). Protocadherins (PCDHs) belong to the cadherin superfamily, and are transmembrane proteins that affect brain development and are associated with some neuronal diseases (27). PCDH dysregulation has been detected in numerous malignant tumors, and their downregulation or absence is correlated with tumor progression (28,29). These results suggested that ARHGAP21, CECR2 and PCDHB13 may be implicated in the development and progression of glioma.
Glioma stem-like cells (GSCs) are associated with the recurrence and chemoresistance of glioma, and MGMT overexpression can promote the resistance of GSCs to temozolomide (30,31). Selenoproteins (SELs) are highly expressed in astrocytes prior to brain injury, and their impaired biosynthesis can lead to neurological dysfunction (32). SELs can resist oxidative stress through neutralizing reactive oxygen species (ROS), and some SELs are highly expressed in the brain and are necessary for brain development (33). As a selenium-rich plasma protein, selenoprotein P serves an antioxidative role in selenium-deficient astrocytes (34,35). The role of SELM in the pathogenesis of glioma has not been reported; however, the Sec-to-Cys mutant SELM is capable of binding transition metal ions and modulating Zn2+-mediated Aβ aggregation, ROS production and neurotoxicity (36). Therefore, MGMT and SELM may serve roles in the pathogenesis of glioma.
SPAG9 is overexpressed in astrocytomas and can act as a critical oncoprotein by mediating cell proliferation and invasion (37). SPAG9 contributes to the invasion of astrocytoma cells through improving the expression of podocalyxin-like via a c-Jun N-terminal kinase-dependent mechanism (38,39). WDR1 overexpression is an independent predictor of unfavorable prognosis for the patients with primary glioblastoma, indicating that WDR1 is a promising prognostic marker and a candidate therapeutic target for the disease (40). WDR6 is implicated in the cell growth inhibitory pathway of threonine kinase 11 via regulation of p27 (Kip1) (41). These findings indicated that SPAG9 and WDR6 may be associated with the prognosis of patients with glioma. In the present study, the expression levels of SPAG9 and WDR6 were reduced in the low methylation group, thus indicating that the mechanism regulating the expression of these genes is not limited to MGMT promoter methylation; therefore, the complexity of these gene regulatory mechanisms should be considered.
At present, the role of epigenetic mechanisms in carcinogenesis is well documented; CpG island hypomethylation promotes the transcriptional activation of oncogenes and induces chromosomal instability, whereas hypermethylation silences tumor suppressor genes (42,43). Etcheverry et al (44) used array technology for quantitative expression and methylation profiling in a well-characterized cohort of patients with glioblastoma. This previous study identified frequent tumor-specific methylation alterations in glioblastoma, some of which directly affected gene expression. In the present study, the 10 feature genes were selected based on MGMT promoter methylation, thus suggesting that associations may exist between MGMT promoter methylation and these genes; however, to the best of our knowledge, there is currently no report regarding these relationships. Given the role of DNA methylation in gene expression, it was hypothesized that MGMT promoter methylation may influence the survival of patients with glioma by regulating the expression levels of these genes; however, this requires further investigation.
There are numerous limitations to the present study. The present results were acquired using bioinformatics analyses, not experimental research. In addition, data heterogeneities and platform differences among the datasets may affect the accuracy of the results. In the future, comprehensive experiments should be designed and performed to validate these findings.
In conclusion, 10 feature genes (ARHGAP21, CECR2, ENSA, GPATCH8, KIAA1109, MGMT, PCDHB13, SELM, SPAG9 and WDR6) that corresponded to a cut-off value of 0.56 were selected as the optimal gene combination in the present study. The 10-gene combination may be mediated by MGMT promoter methylation and could be associated with the prognosis of patients with glioma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YZ performed data analyses and wrote the manuscript. JZ conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mamelak AN, Jacoby DB. Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601) Expert Opin Drug Deliv. 2007;4:175–186. doi: 10.1517/17425247.4.2.175. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen PY, Reardon DA. Neuro-oncology in 2015: Progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12:69–70. doi: 10.1038/nrneurol.2015.242. [DOI] [PubMed] [Google Scholar]
- 4.McGuire S. World cancer report 2014. Geneva, Switzerland: World health organization, international agency for research on cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, Hegi ME. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 6.Wick W, Weller M, van den Bent M, Sanson M, Weiler M, von Deimling A, Plass C, Hegi M, Platten M, Reifenberger G. MGMT testing - the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10:372–385. doi: 10.1038/nrneurol.2014.100. [DOI] [PubMed] [Google Scholar]
- 7.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Chen L, Han L, Shi Z, Zhang J, Pu P, Kang C. EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in glioblastoma. Cancer Lett. 2015;356:929–936. doi: 10.1016/j.canlet.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Ku BM, Lee YK, Ryu J, Jeong JY, Choi J, Eun KM, Shin HY, Kim DG, Hwang EM, Yoo JC, et al. CHI3L1 (YKL-40) is expressed in human gliomas and regulates the invasion, growth and survival of glioma cells. Int J Cancer. 2011;128:1316–1326. doi: 10.1002/ijc.25466. [DOI] [PubMed] [Google Scholar]
- 10.Steponaitis G, Skiriutė D, Kazlauskas A, Golubickaitė I, Stakaitis R, Tamašauskas A, Vaitkienė P. High CHI3L1 expression is associated with glioma patient survival. Diagn Pathol. 2016;11:42. doi: 10.1186/s13000-016-0492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji T, Liu D, Shao W, Yang W, Wu H, Bian X. Decreased expression of LATS1 is correlated with the progression and prognosis of glioma. J Exp Clin Cancer Res. 2012;31:67. doi: 10.1186/1756-9966-31-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goidts V, Bageritz J, Puccio L, Nakata S, Zapatka M, Barbus S, Toedt G, Campos B, Korshunov A, Momma S, et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene. 2012;31:3235–3243. doi: 10.1038/onc.2011.490. [DOI] [PubMed] [Google Scholar]
- 13.Li PCH. Overview of microarray technology. In: Li PCH, Sedighi A, Wang L, editors. Microarray Technology: Methods and Applications. 1st. Vol. 1368. Humana Press, Inc.; Totowa, NJ: 2016. pp. 3–4. [DOI] [Google Scholar]
- 14.Naidu CN, Suneetha Y. Review article: Current knowledge on microarray technology-an overview. Trop J Pharm Res. 2012;11:153. doi: 10.4314/tjpr.v11i1.20. [DOI] [Google Scholar]
- 15.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandes AA, Franceschi E, Paccapelo A, Tallini G, De Biase D, Ghimenton C, Danieli D, Zunarelli E, Lanza G, Silini EM, et al. Role of MGMT methylation status at time of diagnosis and recurrence for patients with glioblastoma: Clinical implications. Oncologist. 2017;22:432–437. doi: 10.1634/theoncologist.2016-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pesarin F, Salmaso L. The permutation testing approach: A review. Statistica. 2013;70:481–509. [Google Scholar]
- 18.Wiens TS, Dale BC, Boyce MS, Kershaw GP. Three way k-fold cross-validation of resource selection functions. Ecological Modelling. 2008;212:244–255. doi: 10.1016/j.ecolmodel.2007.10.005. [DOI] [Google Scholar]
- 19.Lacny S, Wilson T, Clement F, Roberts DJ, Faris PD, Ghali WA, Marshall DA. Kaplan-meier survival analysis overestimates the risk of revision arthroplasty: A meta-analysis. Clin Orthop Relat Res. 2015;473:3443–3445. doi: 10.1007/s11999-015-4235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perolat J, Couso I, Loquin K, Strauss O. Generalizing the Wilcoxon rank-sum test for interval data. Int J Approximate Reasoning. 2015;56:108–121. doi: 10.1016/j.ijar.2014.08.001. [DOI] [Google Scholar]
- 21.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 22.Lambiv WL, Vassallo I, Delorenzi M, Shay T, Diserens AC, Misra A, Feuerstein B, Murat A, Migliavacca E, Hamou MF, et al. The Wnt inhibitory factor 1 (WIF1) is targeted in glioblastoma and has a tumor suppressing function potentially by induction of senescence. Neuro Oncol. 2011;13:736–747. doi: 10.1093/neuonc/nor036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, et al. Stem cell-related ‘self-renewal’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 24.Mizukami A, Matsue Y, Naruse Y, Kowase S, Kurosaki K, Suzuki M, Matsumura A, Nogami A, Aonuma K, Hashimoto Y. Kaplan-Meier survival analysis and Cox regression analyses regarding right ventricular septal pacing: Data from Japanese pacemaker cohort. Data Brief. 2016;8:1303–1307. doi: 10.1016/j.dib.2016.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bigarella CL, Borges L, Costa FF, Saad ST. ARHGAP21 modulates FAK activity and impairs glioblastoma cell migration. Biochim Biophys Acta. 2009;1793:806–816. doi: 10.1016/j.bbamcr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Banting GS, Barak O, Ames TM, Burnham AC, Kardel MD, Cooch NS, Davidson CE, Godbout R, McDermid HE, Shiekhattar R. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum Mol Genet. 2005;14:513–524. doi: 10.1093/hmg/ddi048. [DOI] [PubMed] [Google Scholar]
- 27.Hirabayashi T, Yagi T. Protocadherins in neurological diseases. Adv Neurobiol. 2014;8:293–314. doi: 10.1007/978-1-4614-8090-7_13. [DOI] [PubMed] [Google Scholar]
- 28.Shan M, Su Y, Kang W, Gao R, Li X, Zhang G. Aberrant expression and functions of protocadherins in human malignant tumors. Tumor Biol. 2016;37:12969–12981. doi: 10.1007/s13277-016-5169-9. [DOI] [PubMed] [Google Scholar]
- 29.Sui X, Wang D, Geng S, Zhou G, He C, Hu X. Methylated promoters of genes encoding protocadherins as a new cancer biomarker family. Mol Biol Rep. 2012;39:1105–1111. doi: 10.1007/s11033-011-0837-8. [DOI] [PubMed] [Google Scholar]
- 30.Villalva C, Cortes U, Wager M, Tourani JM, Rivet P, Marquant C, Martin S, Turhan AG, Karayan-Tapon L. O6-Methylguanine-methyltransferase (MGMT) promoter methylation status in glioma stem-like cells is correlated to temozolomide sensitivity under differentiation-promoting conditions. Int J Mol Sci. 2012;13:6983–6994. doi: 10.3390/ijms13066983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiano V, Trevisan M, Trevisan E, Senetta R, Castiglione A, Sacerdote C, Gillio-Tos A, De Marco L, Grasso C, Magistrello M, et al. MGMT promoter methylation in plasma of glioma patients receiving temozolomide. J Neurooncol. 2014;117:347–357. doi: 10.1007/s11060-014-1395-4. [DOI] [PubMed] [Google Scholar]
- 32.Steinbrenner H, Sies H. Selenium homeostasis and antioxidant selenoproteins in brain: Implications for disorders in the central nervous system. Arch Biochem Biophys. 2013;536:152–157. doi: 10.1016/j.abb.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 33.Pitts MW, Byrns CN, Ogawa-Wong AN, Kremer P, Berry MJ. Selenoproteins in nervous system development and function. Biol Trace Elem Res. 2014;161:231–245. doi: 10.1007/s12011-014-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinbrenner H, Alili L, Bilgic E, Sies H, Brenneisen P. Involvement of selenoprotein P in protection of human astrocytes from oxidative damage. Free Radic Biol Med. 2006;40:1513–1523. doi: 10.1016/j.freeradbiomed.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Hill KE, Maguire MJ, Burk RF. Synthesis and secretion of selenoprotein P by cultured rat astrocytes. Biochim Biophys Acta. 2000;1474:390–396. doi: 10.1016/S0304-4165(00)00035-0. [DOI] [PubMed] [Google Scholar]
- 36.Du X, Li H, Wang Z, Qiu S, Liu Q, Ni J. Selenoprotein P and selenoprotein M block Zn2+ -mediated Aβ42 aggregation and toxicity. Metallomics. 2013;5:861–870. doi: 10.1039/c3mt20282h. [DOI] [PubMed] [Google Scholar]
- 37.Yi F, Ni W, Liu W, Pan X, Han X, Yang L, Kong X, Ma R, Chang R. SPAG9 is overexpressed in human astrocytoma and promotes cell proliferation and invasion. Tumour Biol. 2013;34:2849–2855. doi: 10.1007/s13277-013-0845-5. [DOI] [PubMed] [Google Scholar]
- 38.Jiang J, Liu Y, Fang W, Liu F. Sperm-associated antigen 9 promotes astrocytoma cell invasion through the upregulation of podocalyxin. Mol Med Rep. 2014;10:417–422. doi: 10.3892/mmr.2014.2168. [DOI] [PubMed] [Google Scholar]
- 39.Ando K, Uemura K, Kuzuya A, Maesako M, Asada-Utsugi M, Kubota M, Aoyagi N, Yoshioka K, Okawa K, Inoue H, et al. N-cadherin regulates p38 MAPK signaling via association with JNK-associated leucine zipper protein: Implications for neurodegeneration in Alzheimer disease. J Biol Chem. 2011;286:7619–7628. doi: 10.1074/jbc.M110.158477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, Chen Y, Tan C, Xu T, Yan Y, Qin R, Huang Q, Lu C, Liang C, Lu Y, et al. High expression of WDR1 in primary glioblastoma is associated with poor prognosis. Am J Transl Res. 2016;8:1253–1264. [PMC free article] [PubMed] [Google Scholar]
- 41.Xie X, Wang Z, Chen Y. Association of LKB1 with a WD-repeat protein WDR6 is implicated in cell growth arrest and p27Kip1 induction. Mol Cell Biochem. 2007;301:115–122. doi: 10.1007/s11010-006-9402-5. [DOI] [PubMed] [Google Scholar]
- 42.Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karpf AR, Matsui S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005;65:8635–8639. doi: 10.1158/0008-5472.CAN-05-1961. [DOI] [PubMed] [Google Scholar]
- 44.Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L, Menei P, et al. DNA methylation in glioblastoma: Impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.