Abstract
Long non-coding RNAs (lncRNAs) have been generally considered to serve important roles in various types of cancer, including gastric cancer. However, a comprehensive understanding of lncRNAs in gastric cancer requires further study. The present study performed an in-depth study revealed 50 differently expressed lncRNAs. The changed cellular pathways and biological process in gastric cancer were determined. To further confirm the functions of the differently expressed lncRNAs, co-expression networks were constructed between the lncRNAs and mRNA; this lead to the identification of 6 modules, which participated in various cellular pathways. In addition, 2 lncRNAs were identified which were associated with clinical outcome. The biological analysis and experimental evidence suggested that LINC00982 inhibited, while LL22NC03-N14H11.1 promoted the proliferation of gastric cancer cells. These lncRNAs may be considered as potential prognostic factor in gastric cancer.
Keywords: gastric cancer, lncRNA, Gene Set Enrichment Analysis, weighted gene co-expression network analysis, LL22NC03-N14H11.1, LINC00982, survival analysis, proliferation
Introduction
Gastric cancer is one of the malignant tumors with highest mortality all over the world, especially in East Asia (1). Many patients have missed the best time for diagnosis and treatment when they are certainly diagnosed, resulting in the metastasis of tumor cells and developing into the terminal stage of cancer. Nowadays, lack of efficient biomarkers for early diagnosis, comprehensive treatment and cancer monitoring has been considered as one of the main obstacles for better prognosis of gastric cancer (2). As a result, it is of great importance to further explore the molecular mechanism during the occurrence and development of gastric cancer, hoping to provide new strategy for diagnosis, prognosis and treatment (3).
During the recent years, non-coding RNAs have been generally concerned because of their diverse roles in the post-transcriptional regulation and they are considered to have great influence on human diseases (4). Long non-coding RNAs (lncRNA) are one of the functional non-coding RNAs, which have more than 200 nucleotides, and they are usually lack of open reading frames and the ability of coding proteins (5). LncRNAs are located in both cell nucleus and cytoplasm, and involved in the regulation of several cellular events, such as cell development, proliferation, apoptosis and so on (6).
More and more lncRNAs have been reported to be tightly connected with the occurrence and development of malignant tumors (7). These lncRNAs are proved to be engaged in the imbalanced gene regulation and aberrant biological processes (BPs) that contribute to malignant transformation. Especially, the functions and therapeutic potential of cancer-related lncRNAs have been greatly focused in the past few years. To this end, we consider that the overall knowledge about lncRNAs in gastric cancer needs to be fully elucidated.
In our study, we analyzed the qualified public data from He et al study (8) and identified 50 differentially expressed lncRNAs in gastric cancer. We adopted the systems biology-based approach of weighted gene co-expression network analysis (WGCNA) to construct a co-expression network about these lncRNAs and mRNAs, and identified 6 significant co-expression modules. Next, we carried out Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to predict the possible functions of the lncRNAs in modules. Besides, to further confirm the roles of these lncRNAs in gastric cancer, we performed survival analysis and found two of the lncRNAs could be considered as prognostic factors, one of which was novel. In all, our study identified several gastric cancer-related lncRNAs, confirmed their biological functions, and most importantly, we found LINC00982 and LL22NC03-N14H11.1 were associated with patients' survival time and could be considered as potential prognosis for gastric cancer. Besides, we performed cis, trans or ceRNA regulation analysis and found the potential target genes of these two lncRNAs were mainly involved in the proliferation of gastric cancer cells. For further validation, we overexpressed or downregulated the lncRNAs in gastric cancer cell lines and found that LINC00982 inhibited while LL22NC03-N14H11.1 promoted the proliferation of gastric cancer cells. As a result they can be considered as hopeful prognostic factors in gastric cancer.
Materials and methods
Collection of gene expression datasets
Data preprocessing and microarray data were downloaded from Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) under the accession number GSE79973 (8). This dataset was acquired from the study by He et al (8). Totally, 20 samples were included in this dataset, which consisted of 10 gastric cancer samples and 10 normal samples. We used array Quality package to quality control and limma package to apply raw data in R software. The normalization criteria were quantile normalization. Genes having fold-changes ≥2 and FDR <0.05 were selected as of significantly differential expression (9).
Enrichment analysis
To conduct enrichment analyses, the package clusterProfiler (version 3.5.5) of R (version 3.4.0) was used for analyzing the KEGG pathways and GO processes, and DOSE (version 3.3.2) of R (version 3.4.0) as was used for Gene Set Enrichment Analysis (GSEA) (10,11).
WGCNA
WGCNA is a typical systemic biological method for describing the correlation patterns among genes and identifying modules of highly correlated genes by using average linkage hierarchical clustering coupled with the topological overlap dissimilarity measure based on high-throughput chip data or RNA-Seq data (12). In current study, WGCNA package (version 1.60) in R was used to construct lncRNA-mRNA co-expression network and identify modules based on the expression levels of the differentially expressed mRNAs and lncRNAs. The gene modules were signified by different colors and the grey module showed the genes that cannot be merged (13).
LncRNAs function prediction
Cis-regulation target genes were predicted based on the nearby genes of lncRNAs and we chose the promoters that located in the 1M bp regions around the lncRNAs as potential target genes by Bedtools and hg19 as reference genome (14). Trans-regulation target genes were predicted by the sequence of lncRNAs based on RBPDP, and relative score >80% was used to select potential target genes (15). Co-expression target genes were predicted by the psych package in R (version 3.4.0) to calculate Pearson correlation coefficient between genes and two lncRNAs. The absolute value of co-expression coefficient >0.7 and P-value <0.05 were used to select co-expression genes. The construction of ceRNA network was as followings: i) We predicted the potential target microRNAs of the lncRNAs through RegRNA 2.0 (16); ii) Then, we analyzed the differential expression of the microRNAs that got from 1 in gastric cancers through The Cancer Genome Atlas (TCGA); iii) Next, we found the potential target genes of the microRNAs based on miRTarBase (17); and iv) Finally, the target genes and lncRNAs were used to construct ceRNA network when the co-expression coefficient was >0.7 (9).
Cell culture
The SGC-7901 human gastric carcinoma cell line were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and incubated in an atmosphere of 5% CO2 at 37°C. SGC-7901 cell line was obtained from the cell bank of the Committee on Type Culture Collection of the Chinese Academy of Sciences.
Cell transfection
For overexpression of LINC00982, the coding region was PCR-amplified from cDNA generated from the SGC-7901 cell line and was subcloned into a PINCO retroviral vector. For knocking down LL22NC03-N14H11.1, we firstly designed the lncRNA specific small interfering (si)RNAs (the sequence is 5′-GCACUCACCUACACGUUUAGG-3′), and cloned it into plko.1 vector. SGC-7901 cells (1×105 per well) were inoculated in six-well plates and cultured for 24 h. The cells were transfected with LINC00982-overexpression and the corresponding negative control, or the siRNAs targeting LL22NC03-N14H11.1 and the corresponding negative control retroviral vectors using Lipofectamine 3000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer's instructions.
Cell proliferation assay
An MTT assay was used to determine the cell proliferative capacity after overexpression of LINC00982 or down-regulation of LL22NC03-N14H11.1. In brief, transfected cells (2×104 cells/well) were seeded into 96-well culture plates and cultured in RPMI-1640 medium containing 10% FBS. After culturing the seeding cells for 12, 24, 48 or 72 h, MTT reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well, followed by incubation at 37°C for an additional 4 h. Subsequently, 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to dissolve the crystals for 10 min at 37°C. The spectrometric absorbance at 490 nm was measured by an EnSpire Multimode Plate Reader (PerkinElmer, Inc., Waltham, MA, USA).
Flow cytometry
Anti-mouse-ki-67-PE flow cytometric antibody (eBioscience; Thermo Fisher Scientific, Inc.) was purchased from eBioscience. And the intracellular staining of ki-67 was performed by the eBioscience™ Foxp3/Transcription Factor Staining Buffer Set (eBioscience; Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. Briefly, cells were incubated with the mixture of fixation/permeabilization concentrate and diluent (at the ratio of 1:3) at 4°C for 2 h. Then after washing with fixation/permeabilization buffer, cells were incubated with anti-mouse-ki-67-PE flow cytometric antibody for 0.5 h in 4°C. Finally, the cells were washed and analyzed by BD FACSCanto™.
Reverse transcription-quantified polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from cells with GenElute™ Total RNA Purification kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's instructions. Then the RNA was reversely transcribed into cDNA with PrimeScript™ RT reagent kit (Takara Bio, Inc., Otsu, Japan). RT-PCR reactions were carried out with SYBR® Premix Ex Taq™ II (Takara Bio, Inc.) using an ABI Prism 7700 Sequence Detector. Relative mRNA expression levels were calculated by normalizing the relative cycle threshold value to the control group after normalization to the internal control, β-actin and then the results of the semi-quantitative RT-PCR were quantified (18). The primer pairs used are as follows: Human-LL22NC03-N14H11.1-Foward: 5′-GAGTCTGGGGATCAGCATCG-3′, Human-LL22NC03-N14H11.1-Reverse: 5′-TCCAGGGGGCTGGATAATGA-3′; Human-LINC00982-Forward: 5′-AAGTCGTGCTGAGTGTCTGG-3′, Human-LINC00982-Reverse: 5′-CACAACGTGCCACGAACAAT-3′; Human-β-actin-Forward: 5′-CAGGGCGTGATGGTGGGCA-3′, Human-β-actin- Reverse: 5′-CAAACATCATCTGGGTCATCTTCTC-3′.
Statistical analysis
Statistical analyses were performed using R (version 3.4.0; http://www.r-project.org/). The GSE15459 dataset, which includes 200 gastric cancer samples, together with lncRNA and mRNA expression and clinical information, were used to analyze the associations between lncRNA expression signatures and the corresponding overall survival in patients with gastric cancer (19). Survival curves were constructed with the Kaplan-Meier method and the log-rank test was used to determine survival differences between groups. The survival data were evaluated by univariate and multivariate Cox regression analyses to search for independent prognostic factors. P<0.05 was considered to indicate a statistically significant difference (20). When two independent groups were compared, if the data was normally distributed, an unpaired Student's t-test was used.
Results
Differentially expressed mRNAs and lncRNAs in gastric cancer
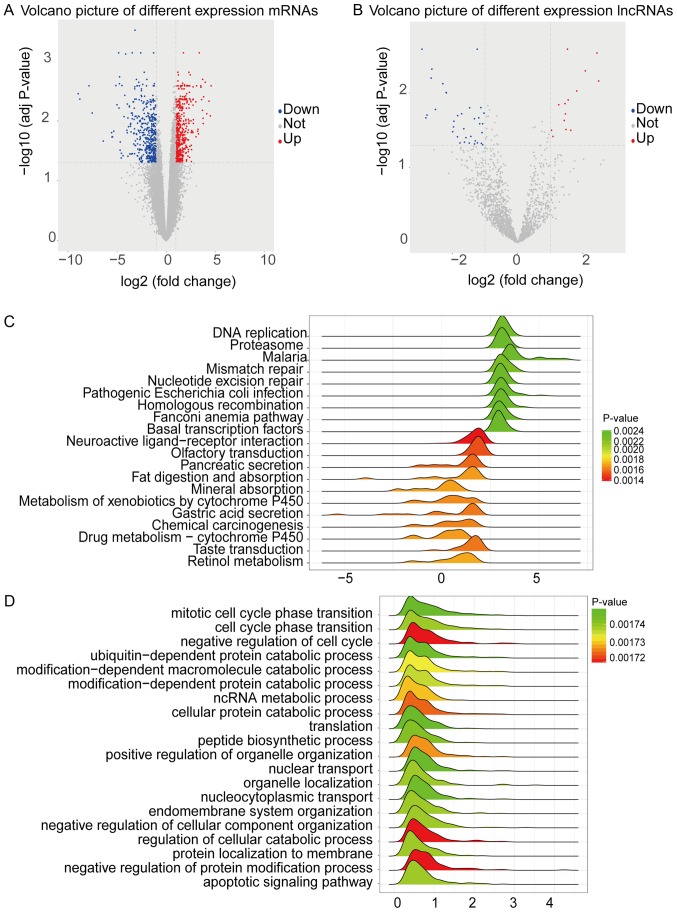
Based on the qualified data from He et al study (8), we identified the lncRNAs related to gastric cancer through GenCode V24 (21). By the criteria of FDR <0.05 and fold-change >2, we identified 953 mRNAs and 50 lncRNAs differentially expressed in gastric cancer compared with para-carcinoma tissues. As a result, 499 mRNAs and 14 lncRNAs were upregulated, and 454 mRNAs and 36 lncRNAs were downregulated (Fig. 1A and B).
Figure 1.
Volcano plot of differentially expressed mRNAs and lncRNAs in gastric cancer and GSEA analysis of all genes. (A) Volcano plot of differentially expressed mRNAs. (B) Volcano plot of differentially expressed lncRNAs. mRNAs or lncRNAs with log2FC >2 and FDR <0.05 were shown in red; mRNAs or lncRNAs with log2FC <-2 and FDR <0.05 were in blue. (C) Joyplot of GSEA KEGG pathway analysis. (D) Joyplot of GSEA GO biological process analysis. The x-axis was ranked by the fold-change of different expression gene. The peak represents the genes which enrich in the pathway or GO term, and the area of peak showed the gene number which enrich in the pathway or GO term. The color represents the significance and the 20 most significant GO and Kyoto Encyclopedia of Genes and Genomes pathways are shown. FC, fold-change; GSEA, Gene Set Enrichment Analysis; GO, Gene Ontology; lncRNA, long non-coding RNA; FDR, false discovery rate.
GSEA identified important pathway and GO in gastric cancer
We assumed that the 50 differently expressed lncRNAs we identified might be involved in various cellular pathways in gastric cancer. To figure out the issue, we firstly run GSEA to identify the dysregulated pathways and BP in gastric adenocarcinoma. Through KEGG pathways, we found 9 upregulated enrichment pathways, including DNA replication, Proteasome, Nucleotide excision repair and 11 downregulated enrichment pathways, including Neuroactive ligand-receptor interaction, Pancreatic secretion, Gastric acid secretion and so on (Fig. 1C). Further, we also conducted GO BP analysis to assign pathways and functionally classified the dysregulated genes. As shown in Fig. 1D, 20 GO BP term, such as mitotic cell cycle phase transition, and ncRNA metabolic process, were significantly enriched.
Construct lncRNA-mRNA co-expression network and predict the function of key lncRNAs
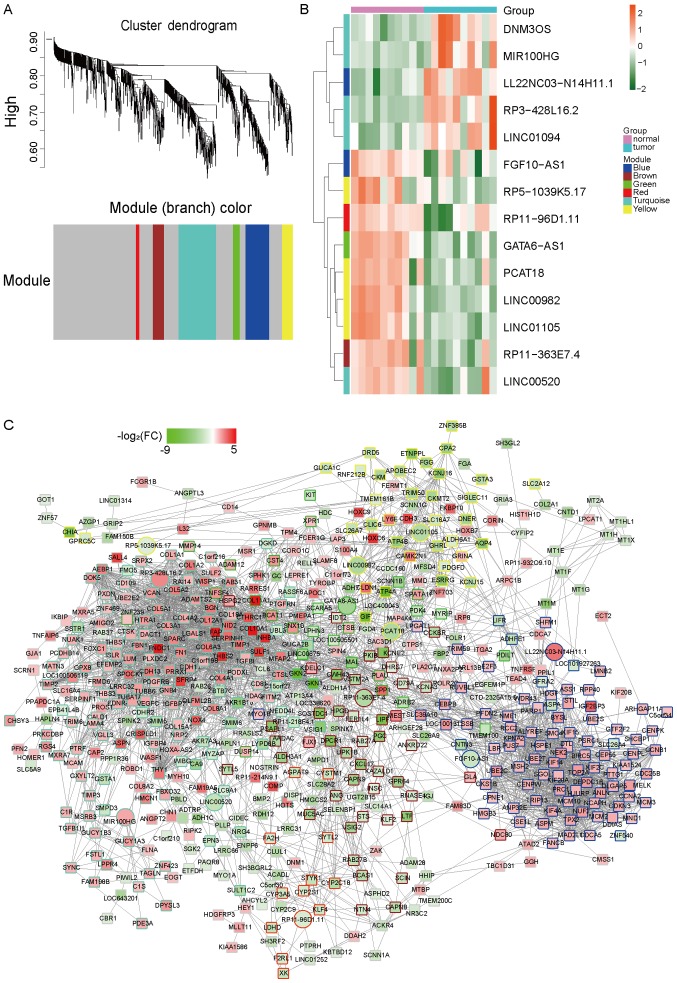
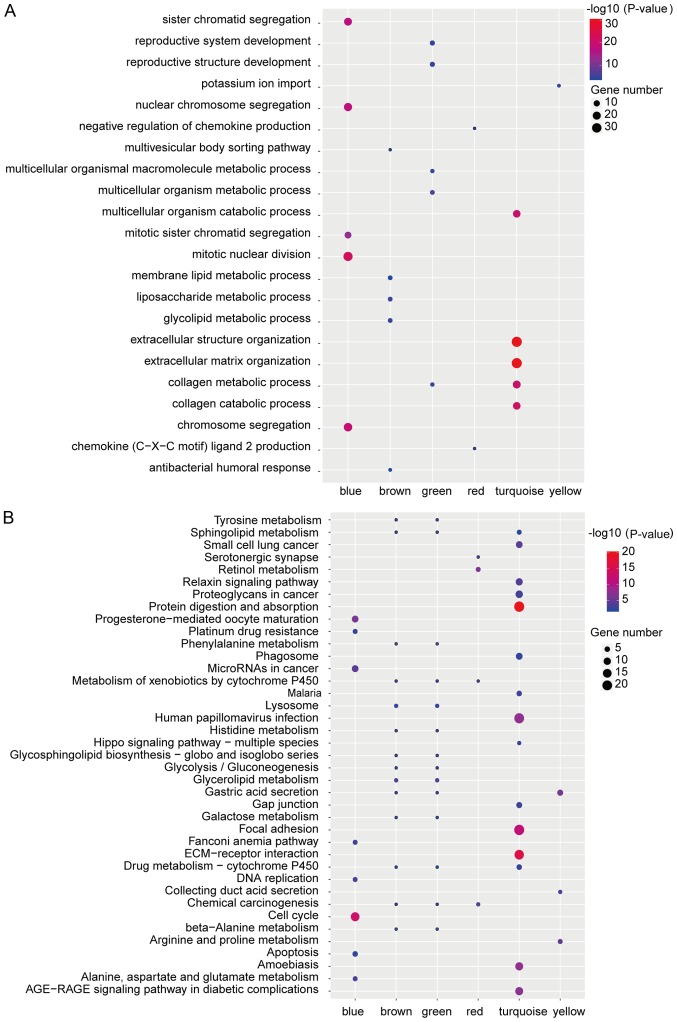
Co-expression analyses of protein-coding RNAs and lncRNAs reflect the potential function of lncRNAs (22). To further confirm the functions of lncRNAs in gastric cancer, we used WGCNA to construct a co-expression network for both mRNAs and lncRNAs that were identified as differentially expressed. We identified 6 co-expression modules in which the highly co-expressed mRNAs and lncRNAs were clustered in the same module (Fig. 2A) and we specifically identified 14 differently expressed lncRNAs in all the modules (Fig. 2B and Table I). The co-expression network from WGCNA as showed in Fig. 2C and hub-genes in different modules were listed in Table II. In order to further explore the functions of the module, we performed GO enrichment and KEGG pathway analysis and found several dysregulated pathways, such as mitotic nuclear division, extracellular structure organization, extracellular matrix organization, ECM-receptor interaction or cell cycle, which were key signal pathways in cancer generation or development (Fig. 3A and B).
Figure 2.
LncRNA-mRNA co-expression network identified by WGCNA. (A) Hierarchical clustering of differentially expressed mRNAs and lncRNAs based on gene co-expression pattern across GSE79973. The different colors represent different co-expression network modules for the significant genes, except the grey color, which was assigned to genes that were not part of any module. (B) Heat map of the lncRNAs which were involved in the modules expressed between cancer and normal tissues, with red indicating higher expression and green indicating lower expression. The color of the vertical axis stands for which module the lncRNAs were from. (C) Co-expression network in the different modules. The fill color showed the log2FC of each gene; the border color showed the module the gene was from; the shape showed the type of gene: The ellipse indicates the mRNA and the triangle indicates the lncRNA. FC, fold-change; lncRNA, long non-coding RNA; WGNCA, weighted gene co-expression network analysis.
Table I.
lncRNAs in different modules.
| Gene symbol | Chromosome | Ensembl ID | FC | FDR | Module |
|---|---|---|---|---|---|
| LL22NC03-N14H11.1 | chr22:15,823,197–15,823,890 | ENST00000608286.1 | 5.37 | 0.0029 | Blue |
| FGF10-AS1 | chr5:44,388,732–44,413,989 | ENST00000502457.1 | 0.29 | 0.0466 | Blue |
| RP11-363E7.4 | chr9:19,453,209–19,455,173 | ENST00000563205.1 | 0.15 | 0.0197 | Brown |
| GATA6-AS1 | chr18:22,166,898–22,168,968 | ENST00000583490.1 | 0.10 | 0.0015 | Green |
| RP11-96D1.11 | chr16:68,225,969–68,229,145 | ENST00000571197.1 | 0.42 | 0.0464 | Red |
| RP3-428L16.2 | chr6:160,990,318–160,992,342 | ENST00000608721.1 | 4.20 | 0.0051 | Turquoise |
| DNM3OS | chr1:172,136,531–172,144,794 | ENST00000417354.2 | 2.39 | 0.0143 | Turquoise |
| LINC01094 | chr4:78,646,186–78,682,392 | ENST00000504675.5 | 2.68 | 0.0233 | Turquoise |
| LINC00520 | chr14:55,782,067–55,796,688 | ENST00000560336.6 | 0.26 | 0.0239 | Turquoise |
| MIR100HG | chr11:122,180,338–122,367,973 | ENST00000527474.5 | 2.79 | 0.0308 | Turquoise |
| LINC00982 | chr1:3059615-3062531 | ENST00000606861.1 | 0.13 | 0.0026 | Yellow |
| LINC01105 | chr2:5,982,571–6,001,275 | ENST00000450794.1 | 0.16 | 0.0063 | Yellow |
| PCAT18 | chr18:26,687,621–26,703,638 | ENST00000579458.1 | 0.14 | 0.0215 | Yellow |
| RP5-1039K5.17 | chr22:37,950,965–37,951,778 | ENST00000609976.1 | 0.46 | 0.0472 | Yellow |
lncRNA, long non-coding RNA; FC, fold-change; FDR, false discovery rate.
Table II.
Hub-genes in different modules.
| Node name | FC | Module | Type | Node number |
|---|---|---|---|---|
| THY1 | 2.410 | Turquoise | mRNA | 74.000 |
| KPNA2 | 1.313 | Blue | mRNA | 59.000 |
| PSAPL1 | −4.757 | Brown | mRNA | 40.000 |
| ATP4A | −7.860 | Yellow | mRNA | 38.000 |
| BCAT1 | 1.863 | Green | mRNA | 36.000 |
FC, fold-change.
Figure 3.
Results of functional enrichment analysis of protein-coding genes in the different modules. (A) Gene Ontology biological process analysis results of the modules. (B) Kyoto Encyclopedia of Genes and Genomes pathway analysis results of the modules. The colors indicate the significance (-log10 transferred P-value) and the circle size represents the gene number of genes enriching the corresponding annotation.
Identify lncRNAs that is associated with apparent clinical outcome
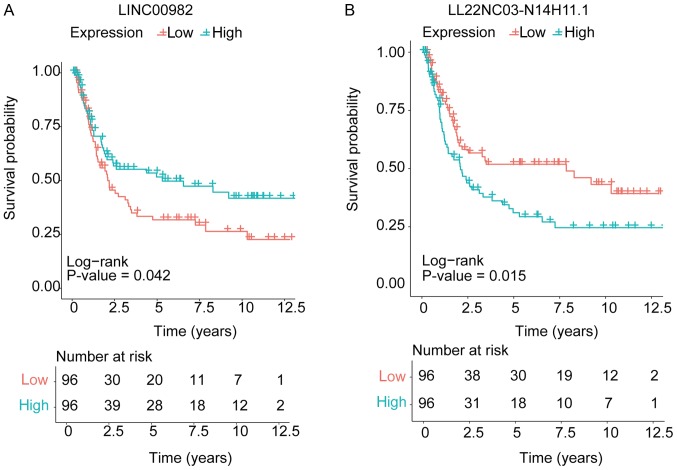
To further investigate the role of these lncRNAs in gastric cancer, we compared the expression of the lncRNAs from different modules, and chose the lncRNAs which changed similarly in different datasets for further analysis (Table III). Then we used GSE15459 to identify lncRNAs associated with clinical outcome (19). Univariate cox survival analysis was performed to analyze the association of clinico-pathological variables, including sex, age, clinical stage, and the lncRNAs' expression with clinical prognosis. Moreover, further multivariate cox analysis showed that LINC00982 expression (P=0.026) and LL22NC03-N14H11.1 expression (P=0.015) were independent prognostic indicators for gastric cancer patients' overall survival (Table IV). Kaplan-Meier survival analysis was used to show the relationship of the two lncRNAs expression level and pathology grade on patients' survival time (Fig. 4A and B).
Table III.
The expression of lncRNA in different datastes.
| GSE19826 | GSE13911 | GSE79973 | ||||
|---|---|---|---|---|---|---|
| Name | FC | P-value | FC | P-value | FC | P-value |
| DNM3OS | 1.828 | 0.009 | 1.108 | 0.461 | 2.389 | <0.001 |
| FGF10-AS1 | 0.722 | 0.453 | 0.710 | 0.149 | 0.293 | 0.004 |
| GATA6-AS1 | 0.249 | 0.001 | 0.166 | <0.001 | 0.100 | <0.001 |
| LINC00520 | 0.524 | 0.001 | 0.700 | <0.001 | 0.258 | 0.001 |
| LINC00982 | 0.187 | <0.001 | 0.171 | <0.001 | 0.131 | <0.001 |
| LINC01094 | 1.822 | 0.139 | 1.608 | 0.005 | 2.684 | 0.001 |
| LINC01105 | 0.262 | 0.007 | 0.270 | <0.001 | 0.159 | <0.001 |
| LL22NC03-N14H11.1 | 2.642 | 0.027 | 3.064 | <0.001 | 5.375 | <0.001 |
| MIR100HG | 2.597 | 0.002 | 1.388 | 0.202 | 2.786 | 0.002 |
| PCAT18 | 0.263 | 0.001 | 0.410 | <0.001 | 0.142 | 0.001 |
| RP11-363E7.4 | 0.234 | 0.003 | 0.146 | <0.001 | 0.146 | 0.001 |
| RP11-96D1.11 | 0.489 | <0.001 | 0.662 | <0.001 | 0.425 | 0.004 |
| RP3-428L16.2 | 4.998 | <0.001 | 3.849 | <0.001 | 4.198 | <0.001 |
| RP5-1039K5.17 | 0.323 | 0.002 | 0.433 | <0.001 | 0.463 | 0.004 |
lncRNA, long non-coding RNA; FC, fold-change.
Table IV.
Univariate and multivariate analyses of the association between overall survival of 193 patients with gastric cancer and prognostic factors by Cox proportional hazard models.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age | 1 | 0.98–1.02 | 0.97 | 1 | 0.99–1.02 | 0.706 |
| Gender (male vs. female) | 0.713 | 0.46–1.10 | 0.127 | 1.13 | 0.71–1.81 | 0.606 |
| Clinical stage (III+IV vs. I+II) | 6.521 | 3.60–11.82 | <0.001c | 7.69 | 4.07–14.54 | <0.001c |
| LL22NC03-N14H11.1 | 1.29 | 1.11–1.50 | <0.001c | 1.23 | 1.04–1.46 | 0.015a |
| RP3-428L16.2 | 1.2 | 1.06–1.35 | 0.004b | 0.95 | 0.79–1.14 | 0.594 |
| LINC01105 | 0.7 | 0.54–0.92 | 0.010a | 0.97 | 0.71–1.34 | 0.859 |
| LINC00982 | 0.74 | 0.59–0.93 | 0.011a | 0.7 | 0.51–0.96 | 0.026a |
| LINC01094 | 1.32 | 1.06–1.64 | 0.013a | 0.98 | 0.76–1.27 | 0.889 |
| MIR100HG | 1.15 | 1.03–1.28 | 0.014a | 1.05 | 0.84–1.3 | 0.672 |
| DNM3OS | 1.2 | 1.03–1.41 | 0.020a | 1.2 | 0.88–1.63 | 0.258 |
| GATA6-AS1 | 0.85 | 0.73–0.98 | 0.028a | 1 | 0.81–1.24 | 0.980 |
| RP11-363E7.4 | 0.86 | 0.75–0.99 | 0.032a | 1.06 | 0.87–1.3 | 0.560 |
HR, hazard ratio; CI, confidence interval.
P<0.05.
P<0.01
P<0.001.
Figure 4.
Kaplan-Meier analysis for the influences of the two differently expressed lncRNAs on the survival of gastric cancer patienta. Kaplan-Meier survival analysis for (A) LINC00982 and (B) LL22NC03-N14H11.1. The log-rank test was performed to evaluate the survival differences between the two curves. lncRNA, long non-coding RNA.
Potential mechanisms of LINC00982 and LL22NC03-N14H11.1 in the regulation of gastric cancer cells
Next, we explored the potential mechanisms of LINC00982 and LL22NC03-N14H11.1 in the regulation of gastric cancer cells. Firstly, we forecasted the two lncRNAs potential target genes by cis or trans regulation analysis (Tables V and VI). We found LINC00982 could regulate target genes through potential ceRNA network (Fig. 5), but not the same with LL22NC03-N14H11.1. The microRNAs that could bind to LL22NC03-N14H11.1 were not differently expressed in gastric cancers, and these microRNAs were not co-expressed with the target genes of LL22NC03-N14H11.1. This was not accord with the principles of ceRNA network and suggested that LL22NC03-N14H11.1 could not regulate gastric cancer through ceRNA regulation. Then we used enrichment analysis based on the potential target genes to forecast potential pathways that the two lncRNAs may be involved in. The results showed that both LINC00982 and LL22NC03-N14H11.1 may be involved in drug metabolism, ECM-receptor interaction, cell cycle, focal adhesion and other pathway in cancer (Table VII). According to previous studies, cell cycle, ECM-receptor interaction and focal adhesion are important pathways to regulate cell proliferation which play an important role in different cancers, so we further inquiried the ability of the two lncRNAs in regulating tumor cell proliferation (23,24).
Table V.
Potential cis-regulation target genes.
| lncRNA | Target genes |
|---|---|
| LL22NC03-N14H11.1 | TPTEP1, CECR7, OR11H1, POTEH, POTEH-AS1, LINC01297, DUXAP8, BMS1P18, BMS1P17, BMS1P22, CCT8L2, ANKRD62P1-PARP4P3, LOC101929350, XKR3, HSFY1P1, GAB4, IL17RA, CECR6, LOC100996342, CECR5 |
| LINC00982 | PRKCZ, PRDM16, CFAP74, MORN1, PLCH2, MMEL1, TTC34, MEGF6, TP73, GABRD, LOC105378591, FAAP20, SKI, LOC100129534, RER1, PEX10, PANK4, HES5, LOC115110, TNFRSF14, LOC100996583, FAM213B, ACTRT2, LINC00982, MIR4251, ARHGEF16, MIR551A, TPRG1L, WRAP73, TP73-AS1, CCDC27, SMIM1, LRRC47, CEP104, DFFB, C1orf174, LINC01134, LINC01346 |
Table VI.
Potential trans-regulation target genes.
| lncRNA | Target genes |
|---|---|
| LINC00982 | NONO, PABPC1, EIF4B, FUS, SNRPA, Pum2, ACO1, SFRS9, MBNL1, KHSRP, YTHDC1, Vts1, QKI, RBMX, SFRS13A, SFRS1, ELAVL1, KHDRBS3 |
| LL22NC03-N14H11.1 | HNRNPA1, NONO, sap-49, PABPC1, a2bp1, EIF4B, FUS, SFRS9, MBNL1, Vts1, RBMX, SFRS1, KHDRBS3, ELAVL1, SFRS13A |
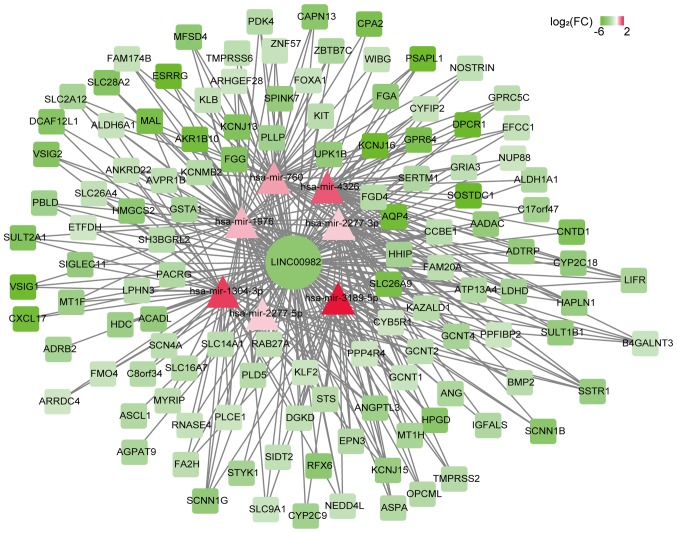
Figure 5.
CeRNA-network of LINC00982. The color revealed the log2FC of each gene. The shape revealed the type of gene; rectangle indicates mRNAs, ellipse indicates lncRNAs and triangle indicates microRNAs. FC, fold-change; lncRNA, long non-coding RNA.
Table VII.
Correlation pathway of LINC00982 and LL22NC03-N14H11.1.
| A, Positive correlation KEGG pathway of LINC00982 | |||
|---|---|---|---|
| Term | Count | P-value | Genes |
| hsa00982: Drug metabolism | 10 | 1.35E-07 | FMO4, GSTA1, FMO5, GSTA3, CYP2C18, CYP2C9, ADH1C, ADH7, ADH1A, UGT2B15, ALDH3A1 |
| hsa00980: Metabolism of xenobiotics by cytochrome P450 | 9 | 1.39E-06 | GSTA1, GSTA3, CYP2C18, CYP2C9, ADH1C, ADH7, ADH1A, UGT2B15, AKR1C1, ALDH3A1 |
| hsa00830: Retinol metabolism | 7 | 9.11E-05 | ALDH1A1, RDH12, CYP2C18, CYP2C9, ADH1C, ADH7, ADH1A, UGT2B15 |
| hsa00591: Linoleic acid metabolism | 4 | 0.006654721 | CYP2C18, CYP2C9, AKR1B10, PLA2G1B |
| hsa00350: Tyrosine metabolism | 4 | 0.022960524 | GOT1, ADH1C, ADH7, ADH1A, ALDH3A1 |
| B, Negative correlation KEGG pathway of LINC00982 | |||
| Term | Count | P-value | Genes |
| hsa04512: ECM-receptor interaction | 11 | 3.99E-08 | COL4A2, COL4A1, COMP, COL6A3, COL3A1, COL1A2, HSPG2, ITGA2, COL5A2, THBS2, SPP1 |
| hsa04510: Focal adhesion | 10 | 5.72E-04 | COL4A2, COL4A1, COMP, COL6A3, COL3A1, COL1A2, ITGA2, COL5A2, THBS2, SPP1 |
| hsa04110: Cell cycle | 6 | 0.016438397 | CCNB1, MAD2L1, PTTG1, CCNA2, CDC25A, CDC25B |
| hsa04914: Progesterone-mediated oocyte maturation | 5 | 0.019154758 | CCNB1, MAD2L1, CCNA2, CDC25A, CDC25B |
| hsa05130: Pathogenic escherichia coli infection | 4 | 0.030393085 | ARPC1B, CLDN1, TUBB6, CD14 |
| C, Positive correlation KEGG pathway of LL22NC03-N14H11.1 | |||
| Term | Count | P-value | Genes |
| hsa04512: ECM-receptor interaction | 9 | 6.49E-06 | COL4A2, COL4A1, COL6A3, COL3A1, COL1A2, ITGA2, COL5A2, THBS2, SPP1 |
| hsa04110: Cell cycle | 9 | 1.18E-04 | CCNB1, E2F3, MAD2L1, MCM2, PTTG1, MCM3, CCNA2, CDC25A, CDC25B |
| hsa04510: Focal adhesion | 9 | 0.002793081 | COL4A2, COL4A1, COL6A3, COL3A1, COL1A2, ITGA2, COL5A2, THBS2, SPP1 |
| hsa05222: Small cell lung cancer | 5 | 0.018703322 | CKS1B, E2F3, COL4A2, COL4A1, ITGA2 |
| hsa04914: Progesterone-mediated oocyte maturation | 5 | 0.020218016 | CCNB1, MAD2L1, CCNA2, CDC25A, CDC25B |
| D, Negative correlation KEGG pathway of LL22NC03-N14H11.1 | |||
| Term | Count | P-value | Genes |
| hsa00982: Drug metabolism | 7 | 4.21E-05 | CYP3A43, FMO4, FMO5, CYP2C18, CYP2C9, ADH7, ADH1A |
| hsa00830: Retinol metabolism | 6 | 2.38E-04 | CYP3A43, ALDH1A1, CYP2C18, CYP2C9, ADH7, ADH1A |
| hsa04960: Aldosterone-regulated sodium reabsorption | 5 | 8.60E-04 | NR3C2, NEDD4L, SCNN1G, SCNN1B, SCNN1A |
| hsa00591: Linoleic acid metabolism | 4 | 0.003074157 | CYP3A43, CYP2C18, CYP2C9, PLA2G1B |
| hsa00980: Metabolism of xenobiotics by cytochrome P450 | 5 | 0.00356804 | CYP3A43, CYP2C18, CYP2C9, ADH7, ADH1A |
Negative and positive correlation pathway revealed the enrichment analysis result of the lncRNA's negative and positive correlation target genes, respectively. KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extra cellular matrix.
LINC00982 and LL22NC03-N14H11.1 have opposite roles in proliferation of gastric cancer cells
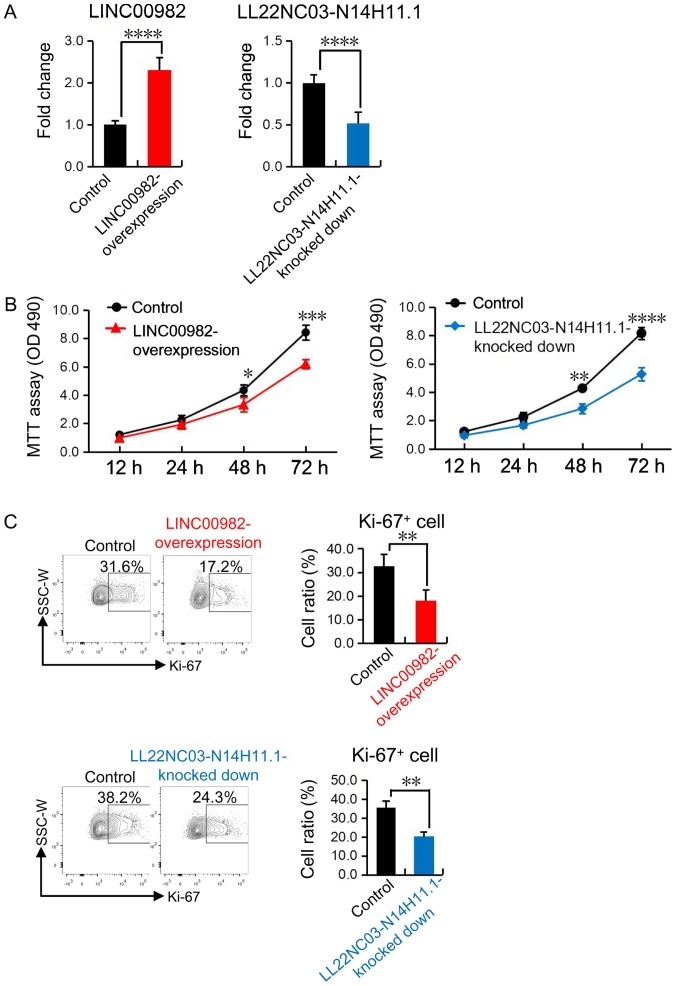
To further confirm the functions of LINC00982 and LL22NC03-N14H11.1 in the regulation of gastric cancer cells' proliferation, we decided to construct overexpressed or down-regualted cell lines for the two lncRNAs. As our previous bioinformatics analysis revealed that LINC00982 was reduced while LL22NC03-N14H11.1 was increased in gastric cancer cells (Table III), we overexpressed LINC00982 and knocked down LL22NC03-N14H11.1 in gastric cell lines SGC-7901 respectively (Fig. 6A). Then we performed MTT assay and flow cytometric analysis and found that the proliferation as well as the expression of ki-67, which acts as a key marker for cell mitosis, were impaired in LINC00982 overexpressed and LL22NC03-N14H11.1 down-regulated SGC-7901 cell lines respectively (Fig. 6B and C). These experimental results provided solid evidence to support our conclusion that LINC00982 inhibited while LL22NC03-N14H11.1 promoted the proliferation of gastric cancer cells.
Figure 6.
LINC00982 and LL22NC03-N14H11.1 have opposite roles in the proliferation of gastric cancer cells. (A) Reverse transcription-quantitative polymerase chain reaction revealed that the expression of LINC00982 in the LINC00982-overexpressed SGC-7901 cell line (left) and the expression of LL22NC03-N14H11.1 in the LL22NC03-N14H11.1-down-regulated SGC-7901 cell line (right). (B) The MTT assay revealed the proliferative status of the SGC-7901 cell line after LINC00982 being overexpressed or LL22NC03-N14H11.1 being downregulated. (C) Flow cytometric analysis of the expression of Ki-67 in the SGC-7901 cell line after LINC00982 was overexpressed or LL22NC03-N14H11.1 was downregulated. All the experiments were replicated a minimum of 3. The error bars represent the standard deviation. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Discussion
LncRNAs are endogenous, lncRNAs that participate in the regulation of diverse cellular process and a great many evidence demonstrates that the abnormal expression of lncRNAs might serve as potential diagnostic and prognostic factors for cancers (25,26).
In this study, we made a deep analysis of the data from He et al study (8), and identified 50 differently expressed lncRNAs in gastric cancer cells. Then we carried out the GSEA and found that several cellular pathways were changed in gastric cancer, which suggested that the differently expressed lncRNAs might be involved in these pathways.
In order to further verify the functions of these differently expressed lncRNAs, we used WGCNA to construct a co-expression network including these lncRNAs and the associated mRNA, and figured out 6 co-expression modules, which involved 14 lncRNAs. The co-expression network revealed that these 14 lncRNAs were functional, for they were related with various protein-encoding mRNAs. To further elucidate the functions of these lncRNAs, we performed KEGG pathway analyses and GO enrichment analyses, and found the modules were tightly associated with mitotic nuclear division, extracellular structure organization, extracellular matrix organization, ECM-receptor interaction or cell cycle, which were the key pathways involved in cancer generation and development.
LncRNAs can be considered as independent indicators for prognosis and we also identified two lncRNAs, LINC00982 and LL22NC03-N14H11.1, associated with apparent clinical outcome in gastric cancer. LINC00982 is located on chromosome 1p36.32 and has two transcript variants. In previous study, Fei et al found that LINC00982 functioned as an inhibitor of cancer cell's proliferation and arrested cell cycle partly via regulating P15 and P16 protein expressions (27). As a result, they identified LINC00982 as a prognostic biomarker in gastric cancer, which was consistent with our current results. This also proved that our bioinformatics analysis process was highly reliable.
On the other hand, we also identified LL22NC03-N14H11.1 as a potential prognostic marker for gastric cancer. LL22NC03-N14H11.1 is located on chromosome 22 and encodes a transcript of 694 bp, however it has never been reported in any study. Here in our study, we found it was highly expressed in gastric cancer and formed the blue module in the co-expression network we constructed, which was involved in the cell cycle related cellular process, including sister chromatid segregation, nuclear chromosome segregation, mitotic nuclear division, chromosome segregation and so on. As a result, we assumed that LL22NC03-N14H11.1 might promote the proliferation of cancer cells, leading to the generation or development of gastric cancer.
To further explore the mechanisms of LINC00982 and LL22NC03-N14H11.1 in the regulation of gastric cancer cells, we performed in-depth analysis on the cis, trans or ceRNA regulation of the lncRNAs in gastric cancer cells. It turned out that the potential target genes of the two lncRNAs were mainly involved in the proliferation of gastric cancer cells, which was consistent with our previous analysis. Besides, in order to provide solid evidence for their roles in the proliferation, we further reversed the expression of LINC00982 and LL22NC03-N14H11.1 in gastric cell linces SGC-7901 with genetic interference. The results completely supported our conclusions that LINC00982 inhibited while LL22NC03-N14H11.1 promoted the proliferation of gastric cancer cells. We believe our findings possess important value, because we may control the proliferation of gastric cancer cells artificially by interfering the expression of LINC00982 and LL22NC03-N14H11.1.
In all, our study contributed a comprehensive knowledge about lncRNAs in gastric cancer and hopefully, LL22NC03-N14H11.1 and LINC00982 might be potential prognostic indicators and clinical targets for gastric cancer.
Acknowledgements
The authors would like to thank Dr Yao Yang (Army Medical University, Chongqing, China) for technology consulting.
Glossary
Abbreviations
- lncRNA
Long non-coding RNAs
- WGNCA
weighted gene co-expression network analysis
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GEO
Gene Expression Omnibus
- BP
biological process
- FC
fold-change
- FDR
false discovery rate
- TCGA
The Cancer Genome Atlas
Funding
No funding was received.
Authors' contributions
DQ and XZ conceived and designed the experiments. DQ, QW, MW and XZ performed the experiments. DQ, QW, MW and XZ analyzed the data. DQ and XZ supervised the study and wrote the paper.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shin HR, Carlos MC, Varghese C. Cancer control in the Asia Pacific region: Current status and concerns. Jpn J Clin Oncol. 2012;42:867–881. doi: 10.1093/jjco/hys077. [DOI] [PubMed] [Google Scholar]
- 2.Ilson DH. Advances in the treatment of gastric cancer. Curr Opin Gastroenterol. 2017;33:473–476. doi: 10.1097/MOG.0000000000000395. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki K, Onodera S, Otsuka K, Satomura H, Kurayama E, Kubo T, Takahashi M, Ito J, Nakajima M, Yamaguchi S, et al. Validity of neoadjuvant chemotherapy with docetaxel, cisplatin, and S-1 for resectable locally advanced gastric cancer. Med Oncol. 2017;34:139. doi: 10.1007/s12032-017-0997-z. [DOI] [PubMed] [Google Scholar]
- 4.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 5.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Fu H, Wu Y, Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci. 2013;56:876–885. doi: 10.1007/s11427-013-4553-6. [DOI] [PubMed] [Google Scholar]
- 7.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He J, Jin Y, Chen Y, Yao HB, Xia YJ, Ma YY, Wang W, Shao QS. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther. 2016;9:6099–6109. doi: 10.2147/OTT.S110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen Q, Yang Y, Chen XH, Pan XD, Han Q, Wang D, Deng Y, Li X, Yan J, Zhou J. Competing endogenous RNA screening based on long noncoding RNA-messenger RNA co-expression profile in Hepatitis B virus-associated hepatocarcinogenesis. J Tradit Chin Med. 2017;37:510–521. doi: 10.1016/S0254-6272(17)30158-9. [DOI] [PubMed] [Google Scholar]
- 10.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu G, Wang LG, Yan GR, He QY. DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2015;31:608–609. doi: 10.1093/bioinformatics/btu684. [DOI] [PubMed] [Google Scholar]
- 12.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Peng L, Zhang Y, Liu Z, Li W, Chen S, Li G. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med Oncol. 2017;34:101. doi: 10.1007/s12032-017-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell RD, III, Wallace AD, Hodgson E, Roe RM. Differential expression profile of lncRNAs from primary human hepatocytes following DEET and fipronil exposure. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18102104. pii: E2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: A database of RNA-binding specificities. Nucleic Acids Res. 2011;39:D301–D308. doi: 10.1093/nar/gkq1069. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Deng Y, Lai W, Guan X, Sun X, Han Q, Wang F, Pan X, Ji Y, Luo H, et al. Maternal inflammation activated ROS-p38 MAPK predisposes offspring to heart damages caused by isoproterenol via augmenting ROS generation. Sci Rep. 2016;6:30146. doi: 10.1038/srep30146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5:e1000676. doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YS, Ma LN, Sun JX, Liu N, Wang H. Long non-coding RNA CPS1-IT1 is a positive prognostic factor and inhibits epithelial ovarian cancer tumorigenesis. Eur Rev Med Pharmacol Sci. 2017;21:3169–3175. [PubMed] [Google Scholar]
- 21.Jalali S, Gandhi S, Scaria V. Navigating the dynamic landscape of long noncoding RNA and protein-coding gene annotations in GENCODE. Hum Genomics. 2016;10:35. doi: 10.1186/s40246-016-0090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz MA, Assoian RK. Integrins and cell proliferation: Regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J Cell Sci. 2001;114:2553–2560. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- 24.Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, Tian X, Yu C, Shen C, Yan T, Hong J, Wang Z, Fang JY, Chen H. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15:60. doi: 10.1186/s12943-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fei ZH, Yu XJ, Zhou M, Su HF, Zheng Z, Xie CY. Upregulated expression of long non-coding RNA LINC00982 regulates cell proliferation and its clinical relevance in patients with gastric cancer. Tumour Biol. 2016;37:1983–1993. doi: 10.1007/s13277-015-3979-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.