Abstract
Breast cancer is the most commonly diagnosed cancer in females; thus, there is an urgent requirement to identify precise biomarkers for the diagnosis and treatment of the disease. Mucin 1 (MUC1) is a glycoprotein that has been demonstrated to be involved in the metastasis and invasion of multiple tumor types. Bioinformatics analyses were conducted to indicate the prognostic value of MUC1 in breast cancer. Additionally, the expression level of MUC1 was assessed using Oncomine analysis. Furthermore, PrognoScan was used to analyze the prognostic value of MUC1 in breast cancer. Mutations of MUC1 were analyzed by the Catalogue of Somatic Mutations in Cancer and cBioPortal databases. In addition, University of California, Santa Cruz (UCSC) was used to examine the methylation status of MUC1. Co-expression of MUC1 mRNA was detected with the cBioPortal, UCSC and Breast Cancer Gene-Expression Miner v4.0 datasets. The results demonstrated that MCU1 is frequently overexpressed in breast cancer and is negatively associated with CpG sites. Furthermore, pooled data indicated that abnormally high expression of MUC1 indicates poor prognosis. Additionally, upregulation of MUC1 expression is associated with estrogen receptor- and progesterone receptor-positive disease, aging and increased Scarff, Bloom and Richardson grade, but is not associated with triple-negative and basal-like status. Subsequent data mining across multiple large databases demonstrated a positive association between MUC1 mRNA expression and cyclic AMP-responsive element-binding protein 3-like 4 (CREB3L4) in breast cancer tissues. The present data indicated that the overexpression of MUC1 indicates a poor prognosis in patients with breast cancer and is associated with MUC1 promoter methylation status. Additionally, MUC1 positively correlated with CREB3L4 and may serve as a potential prognostic factor and therapy target for breast cancer.
Keywords: breast cancer, prognosis, mucin 1, cyclic AMP-responsive element-binding protein 3-like 4, biomarker
Introduction
Breast cancer is the most common malignant tumor type among females globally, with >1.3 million cases diagnosed and ~0.5 million associated mortalities annually globally, according to the data of World Health Organization in 2011 (1,2). Although progress in the early detection, diagnosis and treatment of breast cancer has been achieved in recent decades, the disease remains a significant global health burden (3,4). Accurate biomarkers for early diagnosis and more accurate prognosis could improve the efficiency of current treatments for breast cancer and represent molecular markers for targeted therapy (5). Therefore, the identification of specific and sensitive molecular biomarkers involved in breast cancer has a crucial clinical significance.
Mucin 1 (MUC1) is a transmembrane glycoprotein and is the most thoroughly researched tumor-associated antigen (6–8). As a cell membrane glycoprotein, MUC1 is normally expressed at low levels on the apical surfaces of epithelial cells, including in the pancreas, breast, lung and gastrointestinal tract (9). Additionally, MUC1 has become a topic of concern in the treatment of cancer due its upregulation possibly affecting the invasion, proliferation and survival of tumor cells by reducing cell-cell adhesion and cell-extracellular matrix adhesion (10–12). MUC1 has also been demonstrated to be associated with epidermal growth factor receptors (EGFRs), β-catenin and nuclear factor (NF)-κb signaling in the regulation of the progression and invasiveness of cancer (13,14). In addition, aberrant overexpression of MUC1 is associated with angiogenesis and chemoresistance in cancer (15,16). Thus, MUC1 may have roles in tumorigenesis, progression and metastasis, and may serve as an underlying prognostic factor for tumors. Nevertheless, the role of MUC1 in breast cancer and the potential molecular mechanisms have not yet been elucidated and warrant further investigation.
In the present study, Oncomine microarray datasets were mined to evaluate the expression profile of MUC1 in human breast cancer. Subsequently, the association between MUC1 expression and clinical pathological parameters, including prognostic value, was investigated, and the biological function and mechanism of action of MUC1 in patients with breast cancer was examined by mining publicly accessible databases.
Materials and methods
Oncomine database analysis
The Oncomine database (https://www.oncomine.org/resource/login.html), a publicly accessible online cancer microarray database containing 715 datasets and 86,733 samples, was searched to determine the transcription level of MUC1 gene in breast cancer (17,18). The expression levels of MUC1 mRNA (log2-transformed) in breast cancer and normal tissues were retrieved and statistically compared. To obtain the most significant MUC1 probes, the following parameters were used: P<1×10−4, fold-change >2 and gene ranking in the top 10%.
University of California, Santa Cruz (UCSC) Cancer Genomics Browser analysis
The UCSC Cancer Genomics Browser (http://xena.ucsc.edu/) (19–22) was searched to verify and analyze the heatmap of MUC1 expression and the correlation between MUC1 and cyclic AMP-responsive element-binding protein 3-like 4 (CREB3L4) expression. To investigate the mechanism of the dysregulation of MUC1, the methylation status of MUC1 was analyzed with TCGA Breast Cancer (dataset ID: TCGA.BRCA.sampleMap/HumanMethylation450) (23).
Catalogue of Somatic Mutations in Cancer (COSMIC) analysis for MUC1 mutations
The COSMIC database (http://cancer.sanger.ac.uk), a high-resolution resource for investigating the influence of somatic mutations in all forms of human tumors, was used to analyze the mutations of MUC1 (24,25). An overview of the distribution and substitutions on the coding strand in breast cancer were depicted in a pie chart.
Breast Cancer Gene-Expression Miner v4.0 (bc-GenExMiner v4.0)
The expression of MUC1 and its prognostic value in breast cancer were evaluated using Breast Cancer Gene-Expression Miner v4.0 online data set (http://bcgenex.centregauducheau.fr), which is a statistical mining tool that contains published annotated genomic data, including 36 annotated genomic datasets and 5,861 patients with breast cancer (26,27). Subsequently, the correlation between MUC1 and CREB3L4 genes was estimated by a Pearson's correlation module of bc-GenExMiner v4.0.
cBioPortal database analysis
Cancer genomics analysis was performed by querying the online cBioPortal for Cancer Genomics (http://www.cbioportal.org/; date last accessed, December 4, 2017) (28–37). The cBioPortal for Cancer genomics is attached to the Memorial Sloan Kettering Cancer Center and provides comprehensive analyses of complex tumor genomics and clinical profiles from research into 105 cancer types in The Cancer Genome Atlas (TCGA) (28,29) (study ID, brca_tcga_pub2015) (32). cBioPortal was used to investigate the genes positively associated with MUC1 expression in breast cancer and the RNA sequencing data with the default setting and copy-number variance from Genomic Identification of Significant Targets in Cancer 2.0 supplied by The Cancer Genome Analysis group (https://cancergenome.nih.gov/). cBioPortal was also used to analyze the alteration frequency of MUC1 mutations in breast cancer. The data used included: The Metastatic Breast Cancer Project (38); breast (Molecular Taxonomy of Breast Cancer International Consortium) (30); breast (BC Cancer Research Centre Xenograft; British Columbia) (31); breast cancer (TCGA; provisional); breast invasive carcinoma (TCGA) (32); MBL (33); breast invasive carcinoma (TCGA; provisional); Memorial Sloan Kettering Cancer Center (MSKCC)/Breast 2015: Adenoid cystic carcinoma of the breast (MSKCC) (34); BCCRC 2012: Breast invasive carcinoma (British Columbia) (35); Broad 2012: Breast invasive carcinoma (Broad) (36); and Sanger: Breast invasive carcinoma (Sanger) (37).
PrognoScan database analysis
PrognoScan (http://www.prognoscan.org/) is an integrative online database for investigating underlying tumor indicators and therapeutic targets (39). The PrognoScan database was searched to confirm the prognostic significance of MUC1 and CREB3L4 mRNA expression in patients with breast cancer, and the threshold was adjusted to a corrected P-value. This tool allowed the expression of MUC1 and CREB3L4 to be divided into ‘high’ or ‘low’, according to the median expression of the genes, which was included in the low group. Blue curve corresponds to low MUC1 or CREB3L4 expression, and red curves to high MUC1 or CREB3L4 expression.
Statistical analysis
mRNA expression differences between breast cancer and normal group were assessed using unpaired Welch's t-test and an one-way analysis of variance with Bonferroni's post hoc analysis for comparison between multiple groups. Data are depicted as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference. The association between relative gene expression values was performed by Pearson's correlation and Spearman's correction analysis. Survival curves were plotted according to the Kaplan-Meier method and the log-rank test was used to compare groups. Statistical analyses were performed using SPSS 19.0 software (IBM Corp., Armonk, NY, USA).
Results
MUC1 mRNA expression and methylation status in human breast cancer
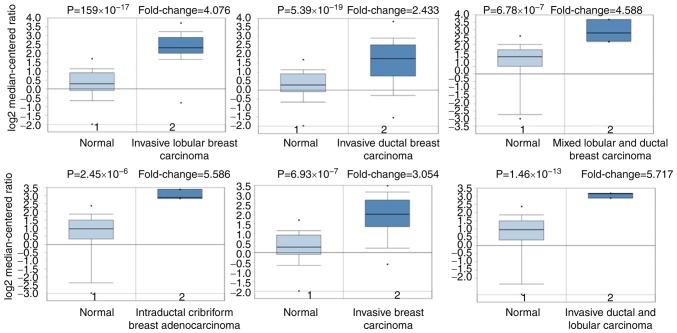
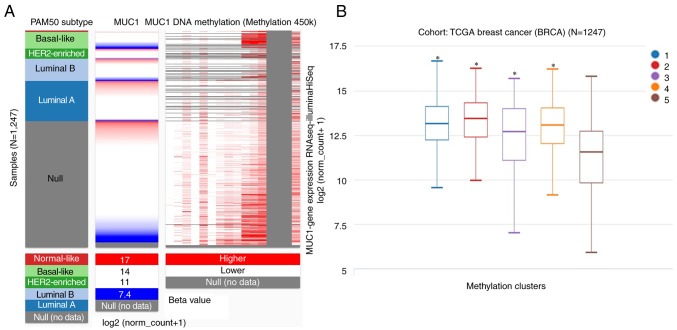
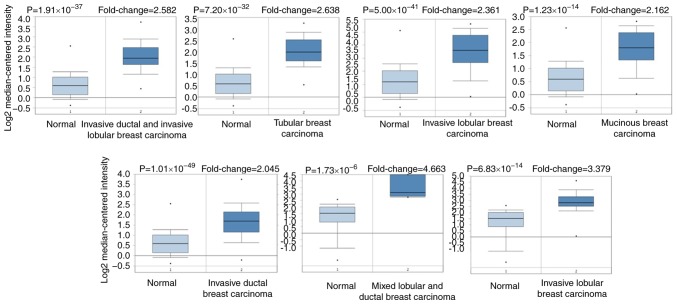
As a membrane glycoprotein, the expression profile of MUC1 was investigated by searching the Oncomine database. MUC1 has been determined in various human cancer types, including hematological malignancies and solid tumors (Fig. 1). MUC1 was determined to be elevated in multiple breast cancer types, compared with the normal tissue (P<1×10−4), including tubular breast carcinoma (P=1.59×10−17), invasive lobular breast carcinoma (P=5.39×10−19), invasive ductal and lobular breast carcinoma (P=1.46×10−13), mucinous breast carcinoma (1.11×10−12), invasive breast carcinoma (6.39×10−19), mixed ductal and lobular carcinoma (6.78×10−7), intraductal cribriform breast adenocarcinoma (2.45×10−6) and invasive ductal breast cancer (5.39×10−19) (Table I and Fig. 2). By comparing the MUC1 mRNA expression heatmap and the DNA methylation status, it was confirmed that MUC1 expression gradually decreased with increasing DNA methylation (P<0.01), which was determined that the transcript expression of MUC1 may be negatively associated with a number of CpG sites (blank frame) (Fig. 3).
Figure 1.
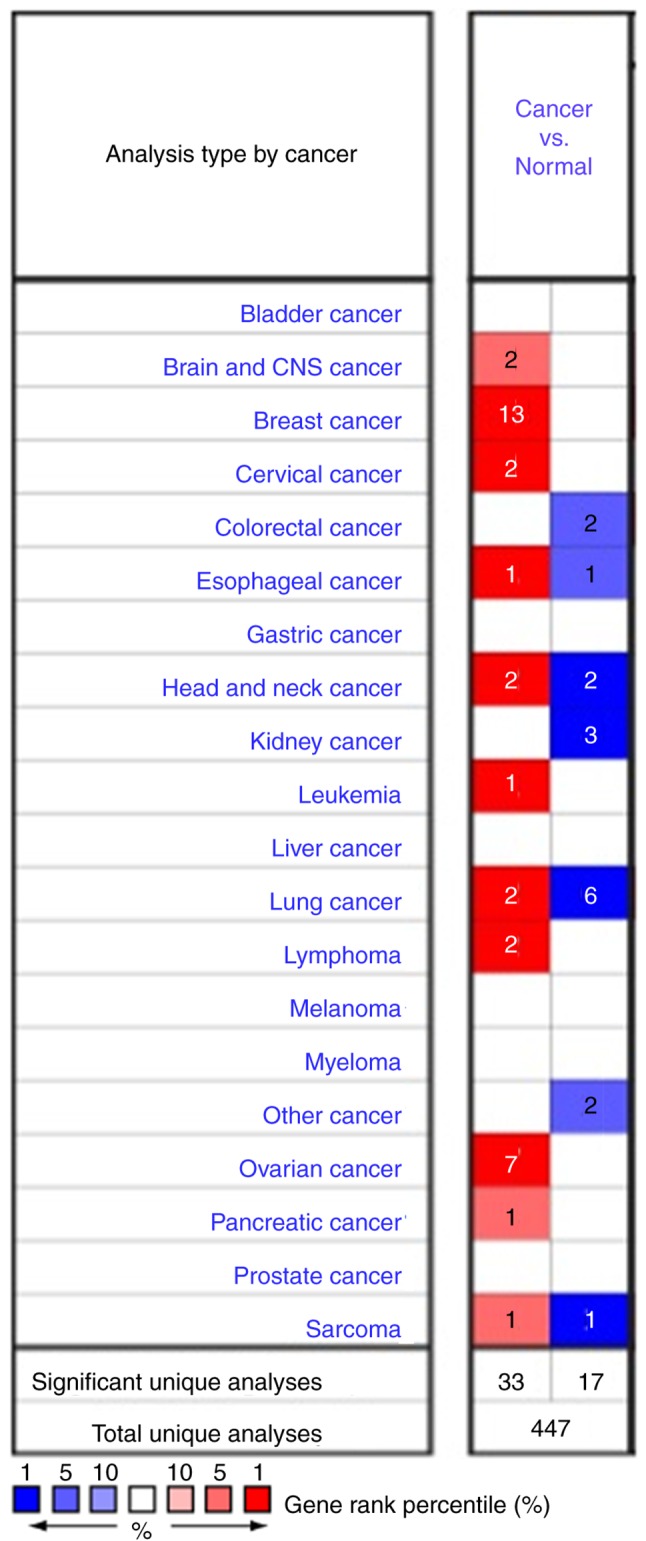
Pooled analyses on the mRNA expression of MUC1 in various carcinoma types. The mRNA expression of MUC1 (cancer vs. corresponding normal tissue) was evaluated using the Oncomine database (red represents significant overexpression and blue represents reduced expression). The following parameters were used as thresholds: P<1×10−4, fold-change >2 and gene ranking in the top 10%. CNS, central nervous system; MUC1, mucin 1.
Table I.
Significant changes in mucin 1 expression at the transcription level between different types of breast cancer and normal tissues (Oncomine database).
| Subtype of breast cancer | P-value | Fold-change | Rank (%) | Sample | (Refs.) |
|---|---|---|---|---|---|
| Tubular breast carcinoma | 1.50×10−35 | 5.767 | 1 | 211 | (28) |
| Invasive lobular breast carcinoma | 3.18×10−39 | 5.581 | 2 | 292 | (28) |
| Invasive ductal and invasive lobular breast carcinoma | 2.12×10−24 | 2.866 | 4 | 234 | (28) |
| Mucinous breast carcinoma | 1.11×10−12 | 3.570 | 6 | 190 | (28) |
| Invasive breast carcinoma | 3.41×10−5 | 3.021 | 7 | 165 | (28) |
| Invasive lobular breast carcinoma | 1.59×10−17 | 4.076 | 1 | 97 | (63) |
| Invasive ductal and lobular carcinoma | 1.46×10−13 | 5.717 | 1 | 64 | (63) |
| Mixed lobular and ductal breast carcinoma | 6.78×10−7 | 4.588 | 1 | 68 | (63) |
| Intraductal cribriform breast adenocarcinoma | 2.45×10−6 | 5.586 | 2 | 64 | (63) |
| Invasive breast carcinoma | 6.93×10−19 | 3.054 | 4 | 137 | (63) |
| Invasive ductal breast carcinoma | 5.39×10−19 | 2.433 | 9 | 450 | (63) |
| Invasive ductal breast carcinoma | 1.77×10−7 | 3.776 | 4 | 41 | (29) |
| Lobular breast carcinoma | 1.97×10−5 | 3.929 | 4 | 24 | (29) |
Figure 2.
Analysis of MUC1 gene expression of the patients with breast cancer using the Oncomine database. Box plot derived from gene expression data in the Oncomine database comparing the expression of specific MCU1 in normal tissue and cancer tissue. Invasive breast carcinoma, invasive ductal breast carcinoma, mixed lobular and ductal breast carcinoma, invasive lobular breast carcinoma, intraductal cribriform breast adenocarcinoma, and invasive ductal and lobular carcinoma were included in the box plots. MUC1, mucin 1.
Figure 3.
(A) MCU1 gene expression heatmap and its DNA methylation status. (B) MUC1 expression in different breast cancer DNA methylation clusters (1–5 represents different methylation clusters). Results were generated using the UCSC Xena browser based on data in TCGA. MUC1, mucin 1; HER2, human epidermal growth factor receptor 2. *P<0.01 vs. cluster 5 group.
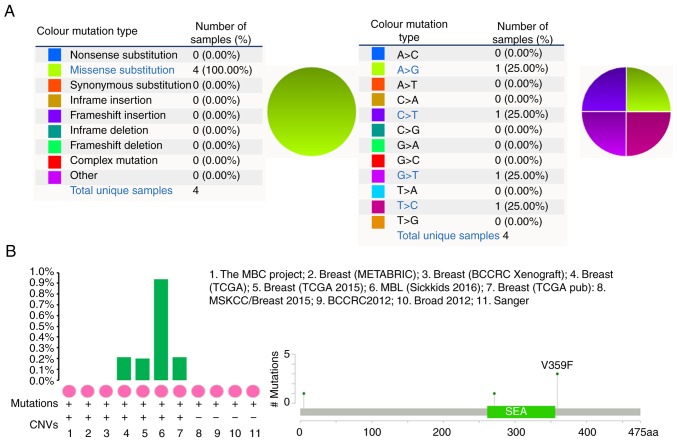
MUC1 mutation in human breast cancer
The pie chart depicts that the mutant types of breast cancer were all missense substitutions (Fig. 4A). The breast cancer data contained A>G, C>T, G>T and T>C mutations, each accounting for 25% of the MUC1 coding strand (Fig. 4A). cBioPortal was used to evaluate the alteration frequency of MUC1 mutations in breast cancer, and <1.0% of the mutations were identified in patients with breast cancer (Fig. 4B).
Figure 4.
MCU1 mutation in human breast cancer. (A) The percentages of mutation types of MUC1 in breast cancer were revealed in a pie chart generated from the Catalogue of Somatic Mutations in Cancer database. (B) cBioPortal was used to analyze the alteration frequency of MUC1 mutations in breast cancer. The data used included: The MBC Project (38); Breast (METABRIC) (30); breast (BCCRC Xenograft; British Columbia) (31); breast cancer (TCGA; provisional); breast invasive carcinoma (TCGA) (32); MBL (33); breast invasive carcinoma (TCGA; provisional); MSKCC/Breast 2015: Adenoid cystic carcinoma of the breast (MSKCC) (34); BCCRC 2012: Breast invasive carcinoma (British Columbia) (35); Broad 2012: Breast invasive carcinoma (Broad) (36); Sanger: Breast invasive carcinoma (Sanger) (37). MUC1, mucin 1; CNVs, copy number variations; MBC, metastatic breast cancer; METBRIC, Molecular Taxonomy of Breast Cancer International Consortium; TCGA, The Cancer Genome Atlas; BCCRC, BC Cancer Research Centre; MSKCC, Memorial Sloan Kettering Cancer Center.
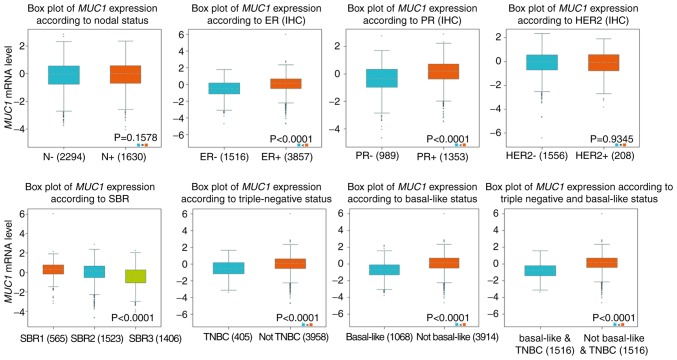
Genetic alterations of MUC1 and clinicopathological parameters in patients with breast cancer
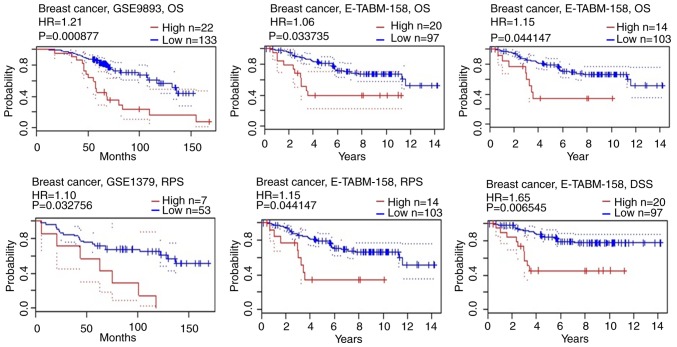
The expression profiles of MUC1 were examined across PAM50 breast cancer subtypes using 5,861 patients with breast cancer cohorts in bc-GenExMiner 4.0, based on different clinical pathological indicators. The age criterion demonstrated significantly increased expression of MUC1 mRNA levels in tumors of patients aged >51 years, compared with those aged ≤51 years (P=0.0037) (Table II). Additionally, the MUC1 mRNA expression was significantly increased in estrogen receptor (ER) (+) group (P<0.0001) and progesterone receptor (PR) (+) group (P<0.0001), compared with the corresponding negative group (Table II and Fig. 5). However, there was no significant differences between the positive and negative statuses of human EGFR 2 (HER2) and nodal status (Table II and Fig. 5). Triple-negative breast cancer (TNBC) is a breast cancer type that is negative for ER (−), PR (−) and HER2 (−) (40). It was determined that MUC1 mRNA expression was significantly downregulated in patients with TNBC (P<0.0001), compared with the not TNBC group (Table II and Fig. 5). Furthermore, patients with negative basal-like characteristics also exhibited significantly increased MUC1 expression, compared with patients with basal-like characteristics (P<0.0001) (Table II, Fig. 5). In the Scarff, Bloom and Richardson (SBR) grade (41) status criterion, an increased SBR grade was significantly associated with a decreased MCU1 transcript level (P<0.0001), compared with the SBR1 group (Fig. 5). The prognostic value of MUC1 expression has been reported by the PrognoScan database (42). In the present study, it was determined that increased expression of MUC1 mRNA is significantly associated with decreased overall survival (OS), disease specific survival (DSS) and relapse-free survival (RFS) time in breast cancer (Fig. 6).
Table II.
Association between mucin 1 mRNA expression and the clinicopathological parameters of breast carcinoma.
| SDC1 | |||
|---|---|---|---|
| Variables | Number | mRNA | P-value |
| Age, years | |||
| ≤51 | 1,436 | − | 0.0037 |
| >51 | 1,988 | ↑ | |
| Nodal status | |||
| − | 2,294 | − | 0.1578 |
| + | 1,630 | − | |
| ER | |||
| − | 1,516 | − | <0.0001 |
| + | 3,857 | ↑ | |
| PR | |||
| − | 989 | − | <0.0001 |
| + | 1,353 | ↑ | |
| HER2 | |||
| − | 1,556 | − | 0.9345 |
| + | 208 | − | |
| Basal-like status | |||
| None | 3,914 | ↑ | <0.0001 |
| Basal-like | 1,068 | − | |
| Triple-negative status | |||
| None | 3,322 | ↑ | <0.0001 |
| TNBC | 242 | − | |
↑, upregulated; TNBC, triple-negative breast cancer; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Figure 5.
Association between MUC1 gene expression and clinical pathological parameters in patients with breast cancer. Notable global differences between the groups were evaluated by Welch's t-test to generate the P-value. MUC1, mucin 1; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; SBR, Scarff, Bloom and Richardson; TNBC, triple-negative breast cancer; IHC, immunohistochemistry.
Figure 6.
Prognostic significance of mucin 1 gene expression in patients with breast cancer (OS, RFS and DSS time in the PrognoScan database). OS, overall survival; RFS, relapse-free survival; DSS, disease-specific survival; HR, hazard ratio.
Co-expression of MUC1 mRNA
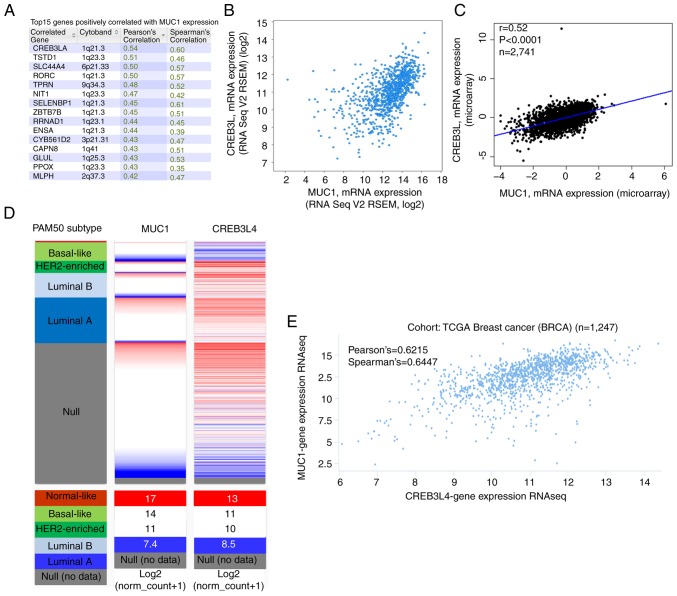
To further investigate the underlying regulation of MUC1 in breast cancer, data mining was conducted in a breast cancer cohort (34) using cBioPortal. CREB3L4 is a principal correlated gene (Fig. 7A) that is abundantly expressed in prostate and breast cancer cell lines (43–45). Regression analysis revealed that MUC1 and CREB3L4 had high relevant coefficients (Spearman's correlation=0.60; Pearson's correlation=0.54) (Fig. 7B). The positive correlation between MUC1 and CREB3L4 transcript expression was verified by the bc-GenExMiner 4.0 database (Fig. 7C). By investigating patient with breast cancer data in TCGA database using UCSC Xena, the positive correlation was confirmed (Fig. 7D and E). These data indicated that MUC1 could be associated with the CREB3L4 signaling pathways in breast cancer.
Figure 7.
MCU1 gene expression is associated with CREB3L4 gene expression in breast cancer. (A) The top 20 genes positively associated with MCU1 transcript level based on TCGA among ~482 patients with breast cancer. (B) Through regression analysis, it was determined that MUC1 and CREB3L4 were highly correlated. (C) Data mining in Breast Cancer Gene-Expression Miner v4.0 further confirmed the positive correlation of MCU1 and CREB3L4 mRNA expression. (D) A heatmap derived from University of California, Santa Cruz Xena revealed the MUC1 and CREB3L4 mRNA expression levels among PAM50 breast cancer subtypes in TCGA database. (E) Association between MUC1 and CREB3L4 gene expression in TCGA database. TCGA, The Cancer Genome Atlas; MUC1, mucin 1; HER2, human epidermal growth factor receptor 2; CREB3L4, cyclic AMP-responsive element-binding protein 3-like 4.
CREB3L4 mRNA expression and prognosis in patients with breast cancer
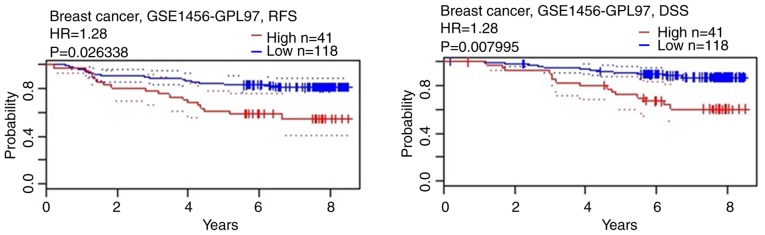
To confirm the genetic alterations of CREB3L4, the Oncomine database was searched to investigate the expression profiles of CREB3L4. The results of Oncomine analysis of tissues (tumor vs. normal) demonstrated that CREB3L4 expression was expressed at a significantly increased level in invasive ductal and invasive lobular breast carcinoma, tubular breast carcinoma, invasive lobular breast carcinoma, mucinous breast carcinoma, invasive ductal breast carcinoma, and mixed lobular and ductal breast carcinoma (Fig. 8). Subsequently, the prognostic value of CREB3L4 in breast cancer was investigated via the PrognoScan database. It was determined that a high expression of CREB3L4 mRNA was significantly associated with reduced RFS and disease-specific survival (DSS) time in breast cancer (Fig. 9).
Figure 8.
CREB3L4 analysis in breast cancer (Oncomine database). Box plot derived from gene expression data in Oncomine comparing the specific CREB3L4 expression levels in normal and cancer tissues. There are two invasive lobular breast carcinoma datasets from different databases included. CREB3L4, cyclic AMP-responsive element-binding protein 3-like 4.
Figure 9.
Prognostic value of mRNA level of cyclic AMP-responsive element-binding protein 3-like 4 in patients with breast cancer (RFS and DSS time in the PrognoScan database). RFS, relapse-free survival; DSS, disease-specific survival; HR, hazard ratio.
Discussion
MUC1, a transmembrane glycoprotein that is altered in various cancer types, can increase the invasive and metastatic properties of adenocarcinomas by reducing cell-cell adhesion and cell-extracellular matrix adhesion (11,46,47). Previous research demonstrated that MCU1 has prognostic value in multiple tumor types, including lung (47,48), colorectal (49,50), gastric (51) and prostate cancer (52). In particular, overexpression of MUC1 is associated with a reduced prognosis, and more malignant and increased grade cancerous tissues in patients with breast cancer (53,54). Furthermore, MUC1 can promote tumor progression and invasiveness through the activation of β-catenin, NF-κB, pyruvate kinase muscle isozyme M2, EGFR and other pathways (13,14,55). However, the distinct roles of MUC1 as a diagnostic marker of poor prognosis and a target for therapeutic intervention in breast cancer remain unknown.
The present analyses were conducted based on the expression of numerous genes with well-defined parameters in breast cancer and normal tissues. Following analyzing the Oncomine database, it was determined that the MUC1 transcriptional level was significantly upregulated in tubular breast carcinoma, invasive lobular breast carcinoma, invasive ductal and invasive lobular breast carcinoma, mucinous breast carcinoma, invasive breast carcinoma, mixed ductal and lobular carcinoma, intraductal cribriform breast adenocarcinoma and invasive ductal breast carcinoma. Furthermore, bc-GenExMiner 4.0 was used to investigate the expression profile of MUC1 across PAM50 breast cancer subtypes based on different clinicopathological parameters. It was determined that the overexpression of MUC1 associated with increased age and risk of ER (+), PR (+), negative basal-like characteristics and SBR grade status. However, the MUC1 mRNA expression was significantly downregulated in patients with TNBC. Subsequently, the frequencies of alterations and mutations of MUC1 were analyzed through the COSMIC and cBioPortal databases. The present results revealed that the only type of mutation of MUC1 in the breast cancer data were missense mutations. Furthermore, the alteration frequency of MUC1 in breast cancer is notably low. For the sake of investigating the mechanism of the dysregulation of MUC1, the methylation status of MUC1 was analyzed with TCGA Breast Cancer (dataset ID: TCGA.BRCA.sampleMap/HumanMethylation450) (23). The results indicated a negative association between MUC1 expression and methylation status. Survival analysis revealed that overexpressed MUC1 associated significantly with reduced OS, RFS, and DSS time, which indicated that the mRNA level of MUC1 may be a valuable biomarker for the prognosis of patients with breast carcinoma.
By mining co-expression and correlation data, it was determined that CREB3L4 was co-upregulated with MUC1 in breast cancer. CREB3L4 is a member of the CREB/ATF transcription factor family, which regulates various processes, including cell proliferation, differentiation and apoptosis, by regulating gene expression through the cAMP-responsive element (56–59). Furthermore, as an ER membrane-bound bZIP domain-containing transcription factor, CREB3L4 has been reported to be associated with a variety of cancer types, including prostate and hepatocellular carcinomas (43–45,60–62). Thus, the mRNA expression of CREB3L4 was analyzed in breast cancer. It was determined that MUC1 mRNA was significantly increased in invasive ductal and invasive lobular breast carcinoma, tubular breast carcinoma, invasive lobular breast carcinoma, mucinous breast carcinoma, invasive ductal breast carcinoma, and mixed lobular and ductal breast carcinoma, compared with normal samples. The survival results revealed that high expression of CREB3L4 mRNA was associated with reduced RFS and DSS time in breast cancer. These results indicated that MUC1 transcript expression may regulate tumor invasion and metastasis associated with CREB3L4 transcription.
The present study was hypothetically driven and performed using experimental generated data available in public databases. Therefore, this emphasizes the requirement for future experimental verification of the MUC1 prognostic value and downstream targets identified in the present study.
Acknowledgements
Not applicable.
Funding
This study was supported by the Scientific Research Project of Shanxi Provincial Department of Health (grant no. 201601070) and Initial Scientific Research Fund of PhD in Shanxi Provincial People's Hospital (grant no. b201635).
Availability of data and materials
All data generated or analyzed during this study are included in the published article.
Author's contributions
XJ and XC conducted the data acquisition, data analyses and figure/table preparations. XJ, HL, CH and XY provided material input, data analysis, and assisted with revising the manuscript. XC supervised the experimental design and manuscript writing. All authors read and approved the final manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Li XJ, Ren ZJ, Tang JH, Yu Q. Exosomal microRNA miR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem. 2017;44:1741–1748. doi: 10.1159/000485780. [DOI] [PubMed] [Google Scholar]
- 2.Benson JR, Jatoi I. The global breast cancer burden. Future Oncol. 2012;8:697–702. doi: 10.2217/fon.12.61. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 4.Qiu N, He Y, Zhang S, Hu X, Chen M, Li H. Cullin 7 is a predictor of poor prognosis in breast cancer patients and is involved in the proliferation and invasion of breast cancer cells by regulating the cell cycle and microtubule stability. Oncol Rep. 2018;39:603–610. doi: 10.3892/or.2017.6106. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Rice M, Tworoger SS, Rosner BA, Eliassen AH, Tamimi RM, Joshi AD, Lindstrom S, Qian J, Colditz GA, et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: A nested case-control study. PLoS Med. 2018;15:e1002644. doi: 10.1371/journal.pmed.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanafi-Bojd MY, Moosavian Kalat SA, Taghdisi SM, Ansari L, Abnous K, Malaekeh-Nikouei B. MUC1 aptamer-conjugated mesoporous silica nanoparticles effectively target breast cancer cells. Drug Dev Ind Pharm. 2018;44:13–18. doi: 10.1080/03639045.2017.1371734. [DOI] [PubMed] [Google Scholar]
- 7.Merikhian P, Ghadirian R, Farahmand L, Mansouri S, Majidzadeh-AK MUC1 induces tamoxifen resistance in estrogen receptor-positive breast cancer. Expert Rev Anticancer Ther. 2017;17:607–613. doi: 10.1080/14737140.2017.1340837. [DOI] [PubMed] [Google Scholar]
- 8.Goode G, Gunda V, Chaika NV, Purohit V, Yu F, Singh PK. MUC1 facilitates metabolomic reprogramming in triple-negative breast cancer. PLoS One. 2017;12:e0176820. doi: 10.1371/journal.pone.0179098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou D, Xu L, Huang W, Tonn T. Epitopes of MUC1 tandem repeats in cancer as revealed by antibody crystallography: Toward glycopeptide signature-guided therapy. Molecules. 2018;23(pii):E1326. doi: 10.3390/molecules23061326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patriarca C, Colombo P, Pio Taronna A, Wesseling J, Franchi G, Guddo F, Naspro R, Macchi RM, Giunta P, Di Pasquale M, et al. Cell discohesion and multifocality of carcinoma in situ of the bladder: New insight from the adhesion molecule profile (e-cadherin, Ep-CAM, and MUC1) Int J Surg Pathol. 2009;17:99–106. doi: 10.1177/1066896908326918. [DOI] [PubMed] [Google Scholar]
- 11.Wesseling J, van der Valk SW, Vos HL, Sonnenberg A, Hilkens J. Episialin (MUC1) overexpression inhibits integrin-mediated cell adhesion to extracellular matrix components. J Cell Biol. 1995;129:255–265. doi: 10.1083/jcb.129.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suwa T, Hinoda Y, Makiguchi Y, Takahashi T, Itoh F, Adachi M, Hareyama M, Imai K. Increased invasiveness of MUC1 and cDNA-transfected human gastric cancer MKN74 cells. Int J Cancer. 1998;76:377–382. doi: 10.1002/(SICI)1097-0215(19980504)76:3<377::AID-IJC15>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Bernier AJ, Zhang J, Lillehoj E, Shaw AR, Gunasekara N, Hugh JC. Non-cysteine linked MUC1 cytoplasmic dimers are required for Src recruitment and ICAM-1 binding induced cell invasion. Mol Cancer. 2011;10:93. doi: 10.1186/1476-4598-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansouri N, Movafagh A, Soleimani S, Taheri M, Hashemi M, Heidary Pour A, Alizadeh Shargh S, Mosavi-Jarahi A, Sasaninejad Z, Zham H, et al. Overexpression of the MUC1 gene in iranian women with breast cancer micrometastasis. Asian Pac J Cancer Prev. 2016;17:275–278. doi: 10.7314/APJCP.2016.17.S3.275. [DOI] [PubMed] [Google Scholar]
- 15.Pillai K, Pourgholami MH, Chua TC, Morris DL. MUC1 as a potential target in anticancer therapies. Am J Clin Oncol. 2015;38:108–118. doi: 10.1097/COC.0b013e31828f5a07. [DOI] [PubMed] [Google Scholar]
- 16.Apostolopoulos V, Stojanovska L, Gargosky SE. MUC1 (CD227): A multi-tasked molecule. Cell Mol Life Sci. 2015;72:4475–4500. doi: 10.1007/s00018-015-2014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 18.Viala M, Alexandre M, Thezenas S, Lamy PJ, Maran-Gonzalez A, Gutowski M, Colombo PE, Romieu G, Jacot W, Guiu S. Prognostic impact of the inclusion of uPA/PAI-1 for adjuvant treatment decision-making in ER+/Her2-pN0 early breast cancers. Breast Cancer Res Treat. 2017;165:611–621. doi: 10.1007/s10549-017-4373-7. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Lv QL, Huang YT, Zhang LH, Zhou HH. Akt/FoxM1 signaling pathway-mediated upregulation of MYBL2 promotes progression of human glioma. J Exp Clin Cancer Res. 2017;36:105. doi: 10.1186/s13046-017-0573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X, Shang F, Zhu W, Lin Q. CXCR4 expression varies significantly among different subtypes of glioblastoma multiforme (GBM) and its low expression or hypermethylation might predict favorable overall survival. Expert Rev Neurother. 2017;17:941–946. doi: 10.1080/14737175.2017.1351299. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Li T, Liu XL. DDA1 is induced by NR2F6 in ovarian cancer and predicts poor survival outcome. Eur Rev Med Pharmacol Sci. 2017;21:1206–1213. [PubMed] [Google Scholar]
- 22.Qu LP, Zhong YM, Zheng Z, Zhao RX. CDH17 is a downstream effector of HOXA13 in modulating the Wnt/beta-catenin signaling pathway in gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:1234–1241. [PubMed] [Google Scholar]
- 23.Daniel JW, David VDB, Fei P, Berman BP, Laird PW. Comprehensive DNA methylation analysis on the Illumina® infinium® assay platform. Illumina. 2008 [Google Scholar]
- 24.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes SA, Beare D, Bindal N, Bamford S, Ward S, Cole CG, Jia M, Kok C, Boutselakis H, De T, et al. COSMIC: High-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr Protoc Hum Genet. 2016;91:10.11.1–10.11.37. doi: 10.1002/cphg.21. [DOI] [PubMed] [Google Scholar]
- 26.Jezequel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 27.Jezequel P, Frenel JS, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database. 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang C, Wang T, Deng K, Li B, Hu S, Qin Q, Xiao T, Zhang S, Meyer CA, He HH, et al. High-dimensional genomic data bias correction and data integration using MANCIE. Nat Commun. 2016;7:11305. doi: 10.1038/ncomms11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shlien A, Raine K, Fuligni F, Arnold R, Nik-Zainal S, Dronov S, Mamanova L, Rosic A, Ju YS, Cooke SL, et al. Direct transcriptional consequences of somatic mutation in breast cancer. Cell Rep. 2016;16:2032–2046. doi: 10.1016/j.celrep.2016.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martelotto LG, De Filippo MR, Ng CK, Natrajan R, Fuhrmann L, Cyrta J, Piscuoglio S, Wen HC, Lim RS, Shen R, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–189. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Metastatic Breast Cancer Project. https://www.mbcproject.org/ [Jun 10;2018 ]; Provisional April, 2018.
- 39.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karaayvaz M, Cristea S, Gillespie SM, Patel AP, Mylvaganam R, Luo CC, Specht MC, Bernstein BE, Michor F, Ellisen LW. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun. 2018;9:3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dabakuyo TS, Bonnetain F, Roignot P, Poillot ML, Chaplain G, Altwegg T, Hedelin G, Arveux P. Population-based study of breast cancer survival in Cote d'Or (France): Prognostic factors and relative survival. Ann Oncol. 2008;19:276–283. doi: 10.1093/annonc/mdm491. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell P, Thatcher N, Socinski MA, Wasilewska-Tesluk E, Horwood K, Szczesna A, Martín C, Ragulin Y, Zukin M, Helwig C, et al. Tecemotide in unresectable stage III non-small-cell lung cancer in the phase III START study: Updated overall survival and biomarker analyses. Ann Oncol. 2015;26:1134–1142. doi: 10.1093/annonc/mdv104. [DOI] [PubMed] [Google Scholar]
- 43.Kim TH, Park JM, Kim MY, Ahn YH. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci Rep. 2017;7:45300. doi: 10.1038/srep45300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inagaki Y, Yasui K, Endo M, Nakajima T, Zen K, Tsuji K, Minami M, Tanaka S, Taniwaki M, Itoh Y, et al. CREB3L4INTS3, and SNAPAP are targets for the 1q21 amplicon frequently detected in hepatocellular carcinoma. Cancer Genet Cytogenet. 2008;180:30–36. doi: 10.1016/j.cancergencyto.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, El-Alfy M, Labrie C. AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res. 2002;62:721–733. [PubMed] [Google Scholar]
- 46.Wesseling J, van der Valk SW, Hilkens J. A mechanism for inhibition of E-cadherin-mediated cell-cell adhesion by the membrane-associated mucin episialin/MUC1. Mol Biol Cell. 1996;7:565–577. doi: 10.1091/mbc.7.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang X, Sun Q, Chen C, Zhang Y, Kang X, Zhang JY, Ma DW, Xia L, Xu L, Xu XY, Ren BH. MUC1 overexpression predicts worse survival in patients with non-small cell lung cancer: Evidence from an updated meta-analysis. Oncotarget. 2017;8:90315–90326. doi: 10.18632/oncotarget.19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue M, Tao W. Upregulation of MUC1 by its novel activator 14-3-3ζ promotes tumor invasion and indicates poor prognosis in lung adenocarcinoma. Oncol Rep. 2017;38:2637–2646. doi: 10.3892/or.2017.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Betge J, Schneider NI, Harbaum L, Pollheimer MJ, Lindtner RA, Kornprat P, Ebert MP, Langner C. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: Expression profiles and clinical significance. Virchows Arch. 2016;469:255–265. doi: 10.1007/s00428-016-1970-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Khayal K, Abdulla M, Al-Obaid O, Zubaidi A, Vaali-Mohammed MA, Alsheikh A, Ahmad R. Differential expression of mucins in Middle Eastern patients with colorectal cancer. Oncol Lett. 2016;12:393–400. doi: 10.3892/ol.2016.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang XT, Kong FB, Mai W, Li L, Pang LM. MUC1 Immunohistochemical expression as a prognostic factor in gastric cancer: Meta-analysis. Dis Markers. 2016;2016:9421571. doi: 10.1155/2016/9421571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Genitsch V, Zlobec I, Thalmann GN, Fleischmann A. MUC1 is upregulated in advanced prostate cancer and is an independent prognostic factor. Prostate Cancer Prostatic Dis. 2016;19:242–247. doi: 10.1038/pcan.2016.11. [DOI] [PubMed] [Google Scholar]
- 53.Dai D, Chen B, Tang H, Wang B, Zhao Z, Xie X, Wei W. Nomograms for predicting the prognostic value of pre-therapeutic CA15-3 and CEA serum levels in TNBC patients. PLoS One. 2016;11:e0161902. doi: 10.1371/journal.pone.0161902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darlix A, Lamy PJ, Lopez-Crapez E, Braccini AL, Firmin N, Romieu G, Thezenas S, Jacot W. Serum HER2 extra-cellular domain, S100β and CA 15-3 levels are independent prognostic factors in metastatic breast cancer patients. BMC Cancer. 2016;16:428. doi: 10.1186/s12885-016-2448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, Bajaj E, Shanmugam K, Lee YY, Hwang SI, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Velpula KK, Rehman AA, Chigurupati S, Sanam R, Inampudi KK, Akila CS. Computational analysis of human and mouse CREB3L4 protein. Bioinformation. 2012;8:574–577. doi: 10.6026/97320630008574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iguchi H, Mitsui T, Ishida M, Kanba S, Arita J. cAMP response element-binding protein (CREB) is required for epidermal growth factor (EGF)-induced cell proliferation and serum response element activation in neural stem cells isolated from the forebrain subventricular zone of adult mice. Endocr J. 2011;58:747–759. doi: 10.1507/endocrj.K11E-104. [DOI] [PubMed] [Google Scholar]
- 58.Lee YW, Park HJ, Son KW, Hennig B, Robertson LW, Toborek M. 2,2′,4,6,6′-pentachlorobiphenyl (PCB 104) induces apoptosis of human microvascular endothelial cells through the caspase-dependent activation of CREB. Toxicol Appl Pharmacol. 2003;189:1–10. doi: 10.1016/S0041-008X(03)00084-X. [DOI] [PubMed] [Google Scholar]
- 59.Velmurugan K, Balamurugan AN, Loganathan G, Ahmad A, Hering BJ, Pugazhenthi S. Antiapoptotic actions of exendin-4 against hypoxia and cytokines are augmented by CREB. Endocrinology. 2012;153:1116–1128. doi: 10.1210/en.2011-1895. [DOI] [PubMed] [Google Scholar]
- 60.Kim TH, Park JM, Jo SH, Kim MY, Nojima H, Ahn YH. Effects of low-fat diet and aging on metabolic profiles of Creb3l4 knockout mice. Nutr Diabetes. 2015;5:e179. doi: 10.1038/nutd.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi J, Djebbar S, Fournier A, Labrie C. The co-chaperone DNAJC12 binds to Hsc70 and is upregulated by endoplasmic reticulum stress. Cell Stress Chaperones. 2014;19:439–446. doi: 10.1007/s12192-013-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Labrie C, Lessard J, Ben Aicha S, Savard MP, Pelletier M, Fournier A, Lavergne E, Calvo E. Androgen-regulated transcription factor AIbZIP in prostate cancer. J Steroid Biochem Mol Biol. 2008;108:237–244. doi: 10.1016/j.jsbmb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 63.The Cancer Genome Atlas. https://tcga-data.nci.nih.gov/tcga/ [Jun 10;2018 ];
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the published article.