Abstract
Intestinal bacteria are symbiotic microbiota within the human gut and are implicated in the occurrence and development of colorectal cancer (CRC). The current study investigated the changes in bacterial composition prior to and following surgery, as well as the differences in the bacterial community structure between cancer tissue and adjacent normal tissue. The diversity of the bacterial community and the composition of the bacteria were assessed. In addition, phylogenetic analysis and principle component analysis (PCA) were performed. The results revealed that cancer tissue and adjacent normal tissue exhibited similar bacterial compositions. However, a significant difference was identified in the composition of intestinal bacteria in stool samples collected from patients following surgery compared with stool samples collected prior to surgery. Each patient had their own unique intestinal bacterial community, likely due to a number of factors, including diet, genetic factors and health status. In addition, phylogenetic trees revealed that the most abundant operational taxonomic unit, 0001, was associated with Escherichia coli in all samples. Finally, PCA suggested that the bacterial community structure in all patient stools was similar following surgery. The current study provides information regarding the diversity of the intestinal bacterial community of patients with CRC and provides a basis for postoperative intestinal assessments.
Keywords: colorectal cancer, surgery, bacterial composition, stool, tissue
Introduction
Approximately 1014 colony-forming units of bacteria colonize the human intestinal tract, particularly within the distal intestine, which contains 1,000–1,150 prevalent bacterial species (1,2). These bacteria degrade various indigestible polysaccharides, including plant-derived pectin, cellulose, hemicellulose and resistant starches, which is mutualistic with host health (1). Intestinal bacteria serve an important role in human health and dysbiosis of these bacteria is associated with obesity, type II diabetes, inflammatory bowel disease and colorectal cancer (CRC) (3–7).
CRC is one of the most common cancer types, with the third highest incidence rate and the fourth highest mortality rate of all cancer types worldwide (8). CRC initiation and development are associated with diet, environment and inheritance; however, only 20–25% of cases are associated with genetic factors (9). Previous studies have suggested that the western lifestyle is the most important risk factor for CRC, particularly improper diet, including the consumption of fat, alcohol and red meat (10,11). Improper diet leads to changes in intestinal microbiota, which is associated with CRC. CRC then causes further changes in intestinal microbiota (12). It has been reported that specific bacterial species serve important roles in CRC initiation and development. Enterotoxigenic Bacteroides fragilis can secrete B. fragilis toxin that induces colonic signal transducer and activator of transcription 3 activation, which activates the T helper 17 cell mucosal immune response and enhances colonic tumor formation (13). Fusobacterium nucleatum and Escherichia coli are implicated in CRC development and metastasis, respectively (14,15). Certain studies have confirmed that the occurrence of CRC affects the gut microbiota community structure. The diversity of microbiota is lower in CRC tissues compared with corresponding colorectal mucosal tissues from healthy individuals (3). Furthermore, Firmicutes and Fusobacteria are more highly abundant in patients with CRC compared with healthy individuals, whereas Proteobacteria are less abundant in patients with CRC (3). Lower levels of core species of bacteria have been identified in stool samples from patients with CRC compared with healthy individuals, including Bifidobacterium longum, Clostridium clostridioforme and Ruminococcus species (16). However, to the best of our knowledge, the similarity of bacterial composition between cancer tissue and adjacent normal tissue remains unknown.
Colorectal tumors are typically surgically excised (17), however, it is not understood whether resection may induce changes in intestinal bacteria. In addition, the use of antibiotics following surgery may lead to changes in the intestinal bacteria (18), however, the bacterial community structure following surgery is unknown.
The aim of the current study was to determine the differences in the bacterial community structure between CRC tissue and adjacent normal tissue, as well as changes in intestinal bacteria in patients with CRC following surgery. Stool samples were collected prior to and following surgery, cancer tissue and adjacent normal tissue were obtained from patients with CRC, and the bacterial community structures were analyzed.
Materials and methods
Sample collection
The current study was reviewed and approved by the Ethics Committee of The First People's Hospital of Yunnan Province (Yunnan, China) and informed written consent was obtained from all participants. A total of 16 stool samples and 16 tissue samples were obtained from 8 patients with CRC at The First People's Hospital of Yunnan Province between January 2015 and December 2016. The patients' information is presented in Table I. Stool samples were collected ~7 days prior to surgery when a normal diet was received and ~10 days following surgery when a normal postoperative diet was received. All patients received cephamycin treatment for 3 days following surgery, consisting of 2 g twice a day. Tissue samples were collected during surgery, including cancerous tissue and adjacent normal tissue. The cancerous tissue consisted of the tumor and the corresponding intestinal wall, including the mucosal layer, the submucosal layer, the muscular layer and the serous layer. The adjacent normal tissue consisted of the normal intestinal wall, including the mucosal layer, the submucosal layer, the muscular layer and the serous layer, with a distance from the tumor of 2 cm. All samples were collected in a sterile sample box and stored at −80°C until further experiments were performed.
Table I.
Information regarding the 8 patients with colorectal cancer.
| No. | Age, years | Sex | BMI | Location of tumor | Type of antibiotics following surgery | Duration of antibiotics following surgery | History of diabetes, hypertension or angiocardiopathy |
|---|---|---|---|---|---|---|---|
| Patient 1 | 74 | F | 17.78 | Right colon | Cephamycin | 3 days, 2 g two times a day | No |
| Patient 2 | 78 | M | 18.03 | Sigmoid colon | Cephamycin | 3 days, 2 g two times a day | No |
| Patient 3 | 51 | F | 22.31 | Right colon | Cephamycin | 3 days, 2 g two times a day | No |
| Patient 4 | 63 | M | 22.31 | Rectum | Cephamycin | 3 days, 2 g two times a day | Hypertension |
| Patient 5 | 59 | M | 21.95 | Right colon | Cephamycin | 3 days, 2 g two times a day | No |
| Patient 6 | 56 | M | 21.95 | Rectum | Cephamycin | 3 days, 2 g two times a day | No |
| Patient 7 | 50 | M | 22.71 | Transverse colon | Cephamycin | 3 days, 2 g two times a day | No |
| Patient 8 | 59 | F | 30.84 | Right colon | Cephamycin | 3 days, 2 g two times a day | Hypertension |
F, female; M, male; BMI, body mass index.
Total DNA extraction
The layer of mucous membranes that surrounds tissues may affect the efficiency of DNA extraction. To minimize this, tissue samples were ground in liquid nitrogen. Then, DNA was extracted using a cetyltrimethylammonium bromide-based method (19), with slight modifications. Briefly, 0.2 g of sample was placed in a 2 ml sterile centrifugal tube and resuspended with 10 µl (50 mg/ml) lysozyme. Subsequently, 25 µl 20% SDS and 5 µl (20 mg/ml) protease K were added to the solution, which was then incubated in a water bath at 55°C for 2 h. Finally, DNA was extracted and purified, as previously described (19). DNA samples were stored at −20°C until subsequent experiments.
The genomic DNA of the stool samples was extracted using the QIAamp Fast DNA Stool Mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. DNA samples were stored at −20°C until subsequent experiments.
Bacterial 16s ribosomal RNA (rRNA) gene amplification and analysis
The bacterial 16s rRNA gene was amplified using polymerase (Tiangen Biotech, Co., Ltd., Beijing, China) and the following barcoded primers: 343F, 5′-TACGGRAGGCAGCAGTCG-3′ and 798R 5′-AGGGTATCTAATCCTGTCT-3′ (20). The thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min followed by 30 cycles of 94°C for 30 sec, 52°C for 1 min and 72°C for 1 min, and a final elongation step of 10 min at 72°C. To eliminate heteroduplexes, the amplified reaction was diluted 10-fold into a fresh reaction mixture of the same composition, which was cycled five times using the aforementioned conditions (21). Polymerase chain reaction (PCR) products were purified using a Gel Extraction kit (BioTeke Corporation, Beijing, China) according to the manufacturer's protocol. Each purified PCR product was sequenced using the Illumina MiSeq™ system (Illumina, Inc., San Diego, CA, USA).
Sequence analysis
All original sequence reads containing ‘N’ were discarded and sequences with <400 base pairs were omitted. All sequences were extracted using each sample's barcode and the barcode and primers were removed from the original sequences using BioEdit software (version 7.0.9.0; http://www.mbio.ncsu.edu/bioedit/bioedit.html). All sequences were aligned using the ‘chimera.pueseus’ command of mothur software (version 1.35.1; http://www.mothur.org/wiki/Download_mothur) (22). Sequences were assigned to an operational taxonomic unit (OTU) with 97% sequence homology. Small subunit ribosomal RNA sequence data and taxonomic information from the SILVA SSURef database (version 123) (23) were downloaded directly from the mothur website (https://www.mothur.org/).
An α-diversity analysis was performed using mothur (version 1.35.1) and included the following indexes: OTU number, Chao (24), ACE (25), Shannon (26) and Simpson (27). OTU numbers were divided on the basis of a similarity distance cut-off value of 0.03. The representative sequences of each OTU were aligned with the sequences of established taxonomic information obtained from the National Center for Biotechnology Information (NCBI) GenBank database using Clustal X software (version 1.83) (28). Phylogenetic trees were created using the neighbor-joining method (29) with MEGA software (version 5.0) (30) and the bootstrap number was 1,000. Finally, principle component analysis (PCA) was performed to evaluate variation between the bacterial composition from different patients and different samples.
Nucleotide sequences
The PCR product sequencing data obtained in the current study were deposited in the Sequence Read Archive of NCBI under the accession number SRP120060.
Results
Sequence information and diversity indices
Following the removal of replicates, duplicates and sequences with low quality or shortened read lengths (<400 bp), a total of 112,117 high-quality sequences with an average length of 424 bp were obtained. Diversity indices of OTU, Chao, ACE, Shannon and Simpson were determined and no significant differences were identified in the mean index for all samples (Table II). The coverage of all samples was >98%, indicating that the numbers of sequences were sufficient to demonstrate the diversity of the samples.
Table II.
Sequence numbers, lengths and α-diversity indexes for all samples (cut-off value, 0.03).
| Sample | Sequence, n | Mean length, bp | OTU number | ACE index | Chao index | Shannon index | Simpson index | Coverage |
|---|---|---|---|---|---|---|---|---|
| Patient 1_B | 2428 | 429 | 14 | 22 | 29 | 0.31 | 0.89 | 1 |
| Patient 1_C | 4308 | 429 | 37 | 165 | 75 | 1.07 | 0.51 | 1 |
| Patient 1_N | 1602 | 429 | 9 | 12 | 11 | 0.61 | 0.7 | 1 |
| Patient 1_A | 3641 | 427 | 48 | 144 | 84 | 1.52 | 0.3 | 0.99 |
| Patient 2_B | 2581 | 429 | 14 | 25 | 17 | 0.19 | 0.94 | 1 |
| Patient 2_C | 2607 | 430 | 14 | 14 | 14 | 0.93 | 0.56 | 1 |
| Patient 2_N | 2302 | 429 | 49 | 113 | 77 | 1.46 | 0.35 | 0.99 |
| Patient 2_A | 3322 | 427 | 67 | 146 | 96 | 1.88 | 0.27 | 0.99 |
| Patient 3_B | 3679 | 416 | 99 | 208 | 147 | 2.87 | 0.11 | 0.99 |
| Patient 3_C | 2393 | 418 | 97 | 162 | 132 | 2.67 | 0.13 | 0.99 |
| Patient 3_N | 3285 | 423 | 107 | 244 | 154 | 1.24 | 0.64 | 0.99 |
| Patient 3_A | 2261 | 419 | 35 | 133 | 69 | 1.65 | 0.27 | 0.99 |
| Patient 4_B | 3324 | 429 | 34 | 141 | 61 | 1.39 | 0.32 | 0.99 |
| Patient 4_C | 3269 | 417 | 49 | 163 | 112 | 2.34 | 0.15 | 0.99 |
| Patient 4_N | 3131 | 428 | 67 | 109 | 122 | 0.63 | 0.82 | 0.99 |
| Patient 4_A | 2809 | 428 | 33 | 82 | 46 | 1.45 | 0.34 | 0.99 |
| Patient 5_B | 3863 | 422 | 78 | 140 | 105 | 2.42 | 0.18 | 0.99 |
| Patient 5_C | 6224 | 424 | 98 | 192 | 162 | 2.61 | 0.12 | 0.99 |
| Patient 5_N | 6990 | 428 | 112 | 204 | 172 | 1.96 | 0.29 | 0.99 |
| Patient 5_A | 4434 | 418 | 58 | 137 | 87 | 2.16 | 0.23 | 1 |
| Patient 6_B | 4390 | 424 | 120 | 164 | 168 | 2.4 | 0.24 | 0.99 |
| Patient 6_C | 6399 | 427 | 92 | 220 | 170 | 1.72 | 0.41 | 0.99 |
| Patient 6_N | 6384 | 424 | 85 | 180 | 135 | 2.77 | 0.09 | 0.99 |
| Patient 6_A | 2996 | 422 | 95 | 206 | 161 | 2.3 | 0.21 | 0.99 |
| Patient 7_B | 3471 | 415 | 134 | 256 | 187 | 2.54 | 0.17 | 0.99 |
| Patient 7_C | 3004 | 418 | 156 | 221 | 243 | 3.4 | 0.09 | 0.98 |
| Patient 7_N | 3607 | 413 | 137 | 311 | 252 | 2.56 | 0.16 | 0.98 |
| Patient 7_A | 3076 | 426 | 114 | 149 | 136 | 1.58 | 0.53 | 0.99 |
| Patient 8_B | 2785 | 429 | 50 | 233 | 113 | 1.77 | 0.28 | 0.99 |
| Patient 8_C | 2756 | 429 | 23 | 27 | 26 | 0.73 | 0.68 | 1 |
| Patient 8_N | 2820 | 429 | 33 | 71 | 56 | 1.06 | 0.55 | 1 |
| Patient 8_A | 1706 | 421 | 34 | 120 | 69 | 1.57 | 0.37 | 0.99 |
N, normal tissue; C, cancerous tissue; B, before surgery; A, after surgery; bp, base pair; OTU, operational taxonomic unit.
Bacterial composition of stool samples prior to and following surgery
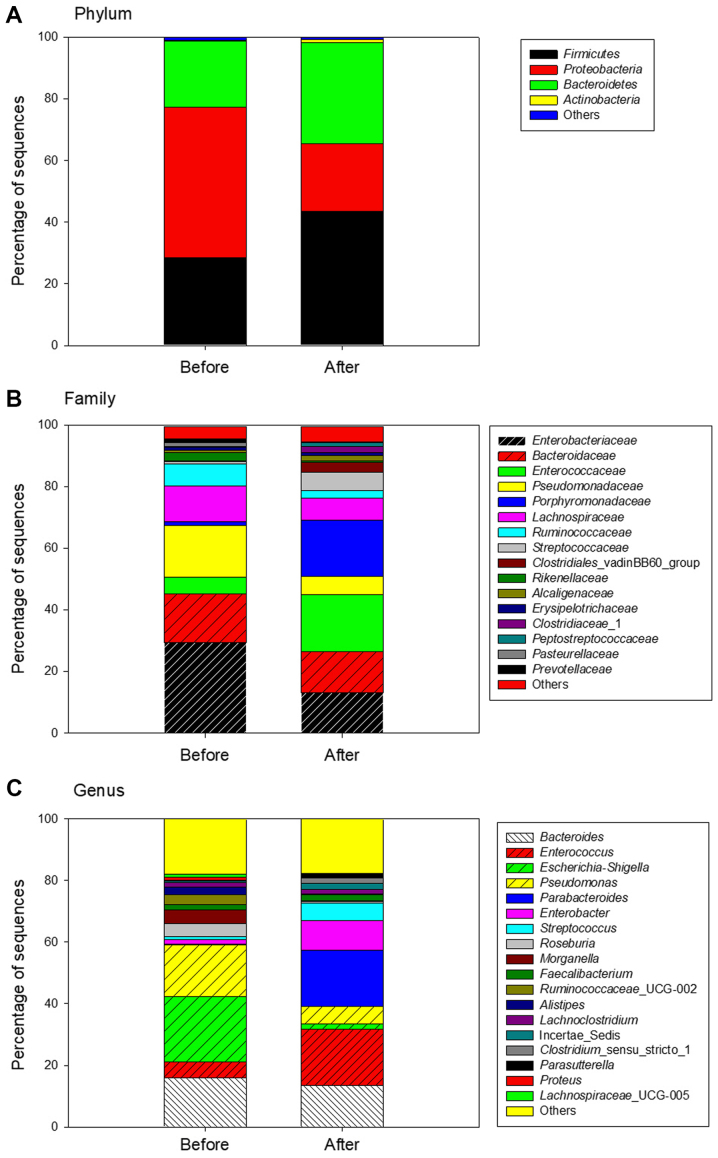
Differences were identified between the bacterial composition of the stool samples obtained prior to and following surgery. At the phylum level, a total of 10 different bacterial phyla were identified in preoperative stool and 15 different bacterial phyla were revealed in postoperative stool. In all stool samples >98% of sequences belonged to the Proteobacteria, Firmicutes and Bacteroidetes, however, differences were identified in the proportions of the species present. Following surgery, the proportions of the phyla Firmicutes and Bacteroidetes increased, from 28 to 44% and from 21 to 33%, respectively. However, the proportion of Proteobacteria markedly decreased, from 49 to 22% (Fig. 1A).
Figure 1.
Bacterial composition of all stool samples obtained from patients prior to and following surgery. The bacterial community structures of stool samples were analyzed at the (A) phylum level, (B) family level and (C) genus level.
At the family level, the most predominant sequences (>10%) belonged to Enterobacteriaceae (29%) and Bacteroidaceae (16%) prior to surgery. By contrast, following surgery the most predominant sequences belonged to Enterococcaceae (18%), Porphyromonadaceae (18%), Bacteroidaceae (13%) and Enterobacteriaceae (13%) (Fig. 1B). The percentage of the family Enterobacteriaceae decreased, but the family Enterococcaceae increased and became the most abundant family following surgery.
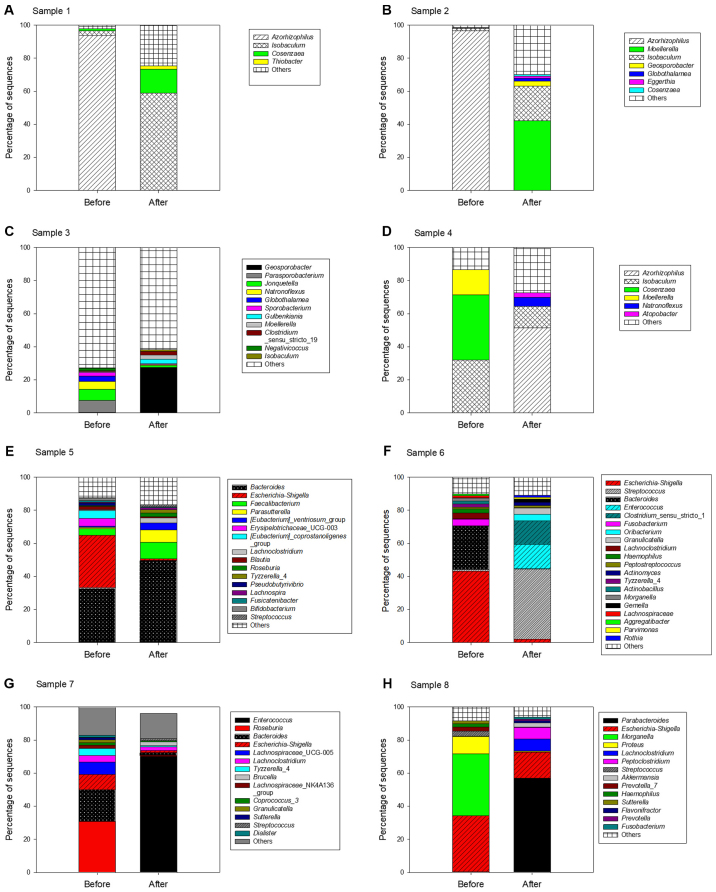
Finally, at the genus level, the most predominant bacterial genera were Escherichia-Shigella (21%), Pseudomonas (17%) and Bacteroides (16%) prior to surgery. However, following surgery, the most predominant bacterial genera were Enterococcus (18%), Parabacteroides (18%) and Bacteroides (13%) (Fig. 1C). Whereas, the abundance of Escherichia-Shigella significantly decreased from 21 to 1.7%, the abundance of Enterococcus and Parabacteroides markedly increased following surgery. The bacterial composition of each patient was evidently different; therefore, the sequences of the stool samples from each patient were analyzed at the genus level (Fig. 2).
Figure 2.
Bacterial compositions of stool samples obtained from each patient prior to and following surgery. (A-H) The bacterial composition of stool samples from every patient was analyzed at the genus level.
Bacterial differences between cancer tissue and adjacent normal tissue
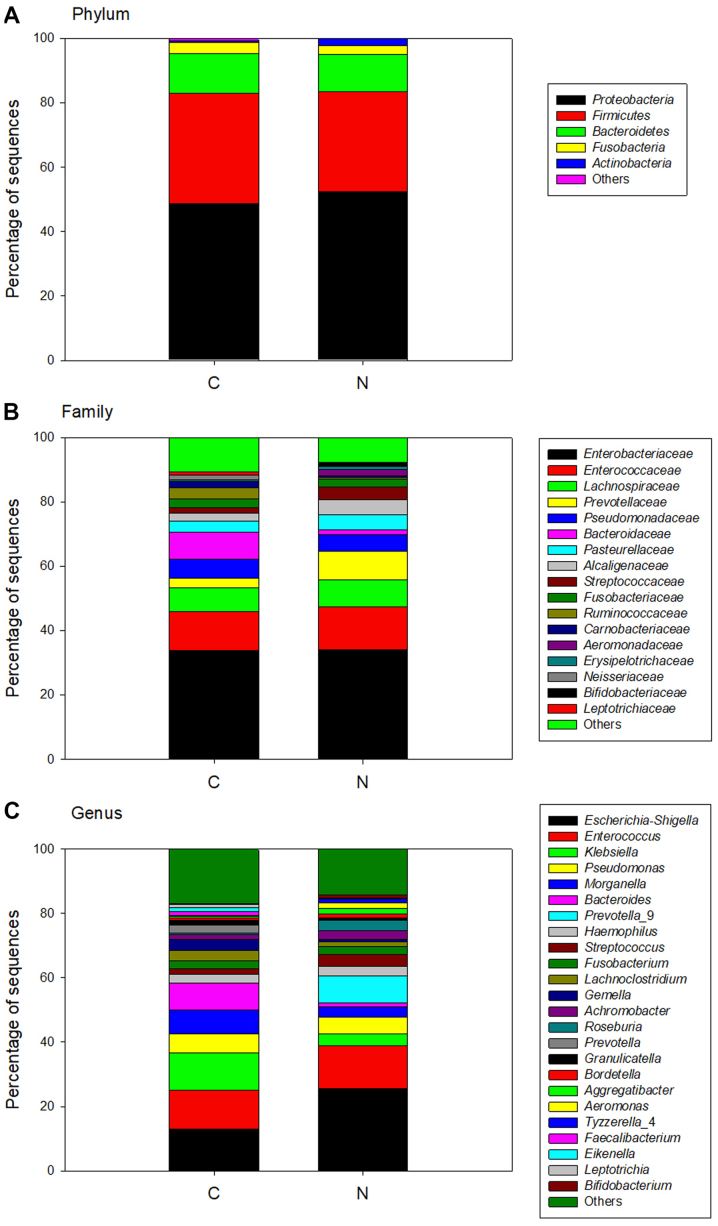
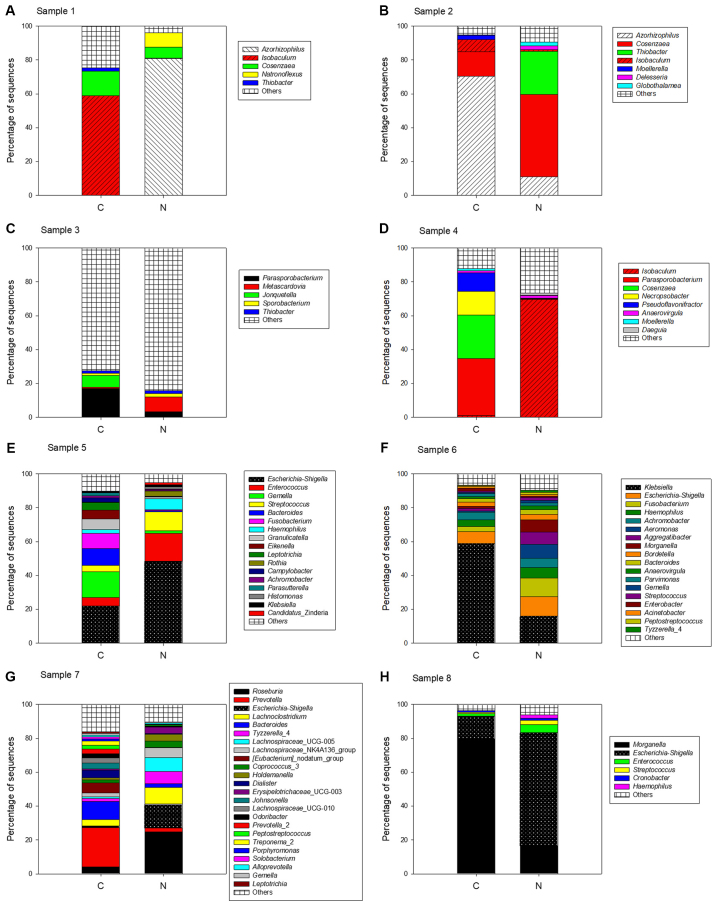
Comparison of the sequences identified in cancer tissue and adjacent normal tissue revealed that both bacterial community structures were similar. At the phylum level, a total of 18 different bacterial phyla were identified in cancer tissues and 15 different bacterial phyla were identified in adjacent normal tissue. The majority of sequences were from Proteobacteria (49 and 52% in cancer and adjacent normal tissue, respectively), Firmicutes (34 and 31% in cancer and adjacent normal tissue, respectively) and Bacteroidetes (12% in cancer tissue and adjacent normal tissue; Fig. 3A). At the family level, the majority of the sequences were restricted to Enterobacteriaceae (34% in cancer tissue and in adjacent normal tissue) and Enterococcaceae (12% in cancer tissue and in adjacent normal tissue); the percentages were similar in both tissue types (Fig. 3B). At the genus level, the predominant genera in cancer tissues were Escherichia-Shigella (13%), Enterococcus (12%) and Klebsiella (12%), whereas in the adjacent normal tissues, the main genera were Escherichia-Shigella (26%) and Enterococcus (13%), but the abundance of Klebsiella was low (Fig. 3C). The bacterial community structures of cancer tissue and adjacent normal tissue were analyzed and compared in every patient to further investigate the differences (Fig. 4). The results demonstrated that the bacterial composition of each patient was markedly different.
Figure 3.
Bacterial composition of all tissue samples obtained from the patients. All sequences were analyzed at the (A) phylum level, (B) family level and (C) genus level. C, cancerous tissue; N, adjacent normal tissue.
Figure 4.
Bacterial compositions of all tissue samples obtained from each patient. (A-H) All sequences of every patient were analyzed at the genus level. C, cancerous tissue; N, adjacent normal tissue.
Phylogenetic analysis
A phylogenetic tree was constructed using the OTU sequences based on a 3% cut-off value and the associated sequences were obtained from the NCBI GenBank database. A total of 44 OTUs with a sequence percentage >1% were selected for constructing the phylogenetic tree (Fig. 5). The OTUs were named based on the number of sequences; therefore, the lower the OTU number, the higher its abundance. The most abundant OTU0001 demonstrated 100% identity to E. coli. OTU0002, 0012 and 0024 were similar to Enterococcus faecalis and Enterococcus faecium. A high similarity was identified between OTU0003 and Pseudomonas azotoformans. OTU0004 and Klebsiella pneumoniae demonstrated 99% identity. OTU0005 was similar to Morganella morganii. OTU0010, OTU0013, OTU0014 and OTU0023 were similar to Streptococcus tigurinus, B. fragilis, Bacteroides dorei and Parabacteroides distasonis, respectively.
Figure 5.
Phylogenetic tree of 16S ribosomal RNA gene sequences based on a cut-off value of 0.03. A total of 44 OTUs were used to construct a phylogenetic tree. The OTU names were based on the number of sequences, therefore, the lower the number, the higher its abundance. OTU, operational taxonomic unit.
PCA
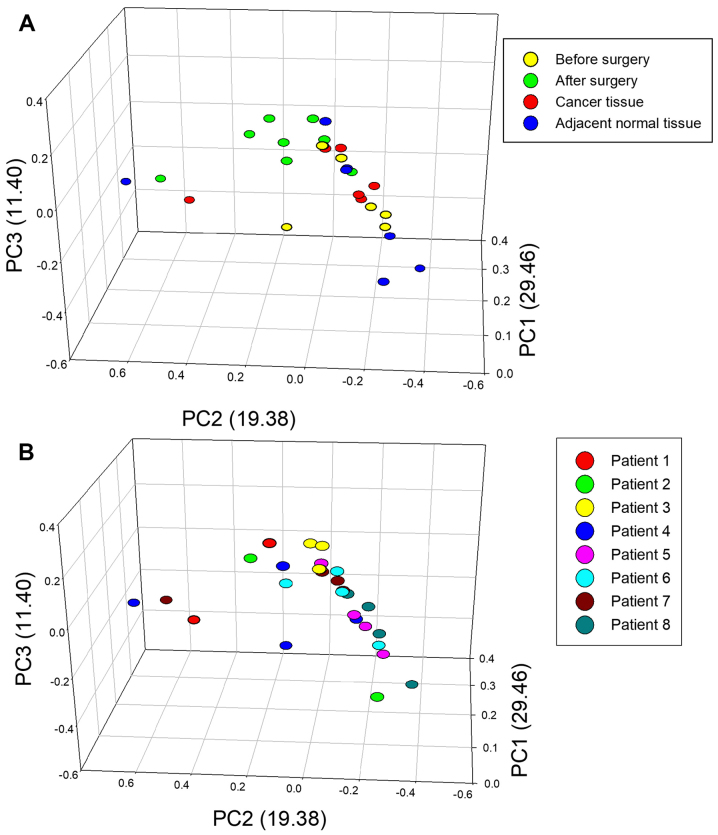
PCA on weighted UniFrac dissimilarities was also performed to evaluate variation between the bacterial composition from different patients and different samples (Fig. 6). The total percentage of principle components PC1, PC2 and PC3 was 60.24%, which could better explain the distribution of the bacterial composition. The dots represent different samples, and the dots from same plane showed the different samples had similar bacterial composition. The results demonstrated that the postoperative stool samples (green circles) were clustered together and are more similar compared with preoperative stool samples (yellow circles) and cancer tissue (red circles). However, the bacteria of adjacent normal tissue (blue circles) were distinct from other samples (Fig. 6A). For the majority of patients, including patients 3, 5, 6, 7 and 8, the four samples from the same patient were clustered together (Fig. 6B).
Figure 6.
PCA based on different sample sources and different patients. PCA was performed based on (A) different sample sources, including cancer tissue, adjacent normal tissue and stool samples obtained prior to and following surgery, and (B) different patients. PCA, principle component analysis; PC, principle component.
Discussion
The current study provided information regarding the intestinal bacteria community of both cancer tissue and adjacent normal tissue, as well as the bacterial intestinal status of patients with CRC prior to and following surgery. It was demonstrated that the bacterial composition of stool samples prior to and following surgery were markedly different and that the bacterial composition greatly improved following surgery. However, the bacterial community structures of cancer tissues and adjacent normal tissues prior to surgery were similar, which suggested that the bacterial composition of adjacent normal tissue may not be as healthy as previously considered.
The dominant bacteria of all samples at the phylum level were identified as Proteobacteria, Firmicutes and Bacteroidetes. Proteobacteria has previously been demonstrated to have a higher abundance in intestinal inflammation and in patients with tumors (4,31,32). In the current study, the abundance of bacteria from the phylum Proteobacteria decreased in stool samples following surgery, while those in the phylum Firmicutes increased. Previous studies have indicated that the incidence and mortality rates of patients of African-American background with CRC are higher compared with other racial and ethnic groups in the USA and these patients demonstrate a lower five-year survival rate compared with Caucasian patients (33–36). Compared with the intestinal microbes and metabolites of Caucasian patients, African-American patients exhibit significantly lower levels of short chain fatty acids and significantly higher levels of Firmicutes species (37). Another study that compared the intestinal microbes of patients with CRC and healthy individuals suggested that the phylum Firmicutes is more abundant in patients with CRC (3).
Enterobacteriaceae (phylum Proteobacteria) are normal commensal bacteria in the gut of humans, however, the family consists of numerous genera of bacteria that are potentially pathogenic, including Salmonella, Shigella, Escherichia, Proteus and Klebsiella (38). It has been reported that Enterobacteriaceae is more abundant in patients with inflammatory bowel disease (IBD) or CRC compared with healthy individuals (39–41). The Enterobacteriaceae bacteria, including Proteus and Klebsiella are associated with colitis in T-bet−/− Rag2−/− ulcerative colitis mice, due to intestinal inflammation that contributes to the high abundance of Enterobacteriaceae (42,43). In the current study, Enterobacteriaceae abundance decreased following surgery, in contrast to Enterococcaceae and Porphyromonadaceae, which increased. Mira-Pascual et al (41) have identified that the proportion of the family Enterococcaceae is higher in patients with polyps compared with healthy controls, suggesting that this could be used as a biomarker to detect polyps, which are a risk factor for CRC. Enterococcaceae includes four genera; Enterococcus, Melissococcus, Tetragenococcus and Vagococcus, and the genus Enterococcus has been most extensively studied (44). E. faecalis has been identified at a higher proportion in patients with CRC compared with healthy individuals (45,46). In addition, E. faecalis has been revealed to induce IBD in interleukin-10 knockout mice (47). Sun et al (48) revealed that the family Porphyromonadaceae is significantly enriched in an inflammatory group of 1,2-dimethylhydrazine (DMH)-induced mice compared with hyperproliferation and adenoma groups. The authors suggested that the family Porphyromonadaceae serves a protective role by mediating the microbiota balance in the murine gut. The current study demonstrated that following surgery, samples from patients contained a higher percentage of potential pathogens, including members of Enterobacteriaceae and Enterococcaceae. This may be due to the tumor resection time being too short to detect a change of bacteria or due to the use of antibiotics following surgery. Therefore, future studies should collect more samples at different time points following surgery.
The current study identified that the predominant genera were Escherichia-Shigella and Enterococcus in all samples analyzed. The 16S rRNA genes of Escherichia and Shigella have a high similarity, therefore more detailed classified information requires identification of other functional genes. The Escherichia-Shigella genera are considered to be potential pathogens of CRC; Escherichia-Shigella species are typically abundant in the colon and may promote tumor formation (3,40). The genera Escherichia-Shigella may produce bacteriocins in the intestine (49). Bacteriocins are antibacterial and are induced during apoptosis, however, to the best of our knowledge, their association with eukaryotic hosts remains unclear. An in vitro study identified several types of bacteriocins that are considered to promote tumor cell proliferation (50). E. coli was revealed to be more prevalent in stage III/IV CRC compared with stage I CRC and an association was identified between poor prognosis of CRC and colonization of mucosa by E. coli (51). The current study identified a large number of Escherichia-Shigella species in the stool samples of patients 5, 6, 7 and 8 prior to surgery, and the proportions decreased following surgery, which may suggest that the factors causing inflammation and tumor formation were reduced within the intestinal microflora following surgery.
Enterococcus species are also commensal bacteria within the human intestinal tract, serving as facultative anaerobes (52). Enterococcus are considered to benefit from an inflammatory niche in the intestine (53,54). A number of studies have demonstrated that the abundance of Enterococcus is higher in stool samples from patients with CRC compared with healthy controls (45,46,55). In addition, Enterococcus often develop antibiotic resistance due to abuse of antibiotics and may cause fatal nosocomial infections in humans (40,56,57). In the current study, the genera Escherichia-Shigella was abundant in stool samples prior to surgery, cancer tissues and adjacent normal tissues, while the genus Enterococcus was abundant in stool samples following surgery, cancer tissue and adjacent normal tissue. These two genera contain potential pathogens of CRC, which may indicate a poor prognosis or longer recovery time for patients with a high abundance.
In the present study, it was found that the composition of the bacterial community in every patient was different, and certain novel bacteria have been revealed in the current study that have rarely been identified in previous intestinal bacterial studies, including Azorhizophilus, Isobaculum, Moellerella, Streptococcus and Klebsiella. Previously, Azorhizophilus has not been detected in the gut; however it was identified at a higher level in the current study in patients 1, 2 and 4. Azorhizophilus, a member of the Pseudomonadaceae family, is a nitrogen-fixing bacteria associated with the legume species (58). Recently, the intestinal microbiota was also detected by the presence of genes associated with nitrogen-fixing bacteria, in populations that eat a large number of legumes (59). Isobaculum was also detected at a high level in patients 1, 2 and 4. Isobaculum is phylogenetically similar to lactic acid bacteria and the type of glucose metabolism is also similar, where acetate and lactate are produced (60). This suggests that Isobaculum may be a potential probiotic bacterium. Moellerella is a commensal bacterium in the gut but is often isolated from clinical specimens of patients with diarrhea (61,62). This bacterial genus was increased in the stool sample from patient 2 following surgery, which may suggest an unhealthy intestinal state. Another potential pathogenic bacteria identified was Streptococcus, which has a close association with the occurrence of CRC and intestinal inflammation (63,64). Finally, Klebsiella was detected in cancer tissues at 12% but was not detected in stool samples. Klebsiella is a classical respiratory pathogen (65), therefore the reason for its high abundance in cancer tissue is unclear.
According to phylogenetic analysis, the sequences of OTU0001 were similar to E. coli. It has previously been indicated that colorectal carcinoma mucosa, but not normal colonic mucosa, is colonized by intracellular E. coli (66). The DNA repair gene MUTYH in intestinal epithelial cells is a homologue of the E. coli gene mutY and is associated with CRC (67,68). The sequences of OTU0002, OTU0012 and OTU0024 were similar to E. faecalis or E. faecium. These two bacteria are common bacteria in the gut and resistance of these two strains frequently occurs due to antibiotics abuse (56,57). In addition, F. nucleatum promotes chemoresistance to CRC by modulating autophagy (69). F. nucleatum was identified in OTU0017 in the current study but the sequence ratio was very low. Furthermore, the sequences of OTU0010 and S. tigurinus were identified to be similar. S. tigurinus is associated with numerous clinical infections, including endocarditis, meningitis and oral infection, and in the USA, the spread of infectious disease is commonly associated with this strain (70–72). The sequences of OTU0005 demonstrated 100% identity to M. morganii, which can infect insects (73). Murphy et al (74) performed a study on neonatal fecal microbiota and identified that M. morganii is often associated with sulfhemoglobinemia. M. morganii is an opportunistic pathogen that may cause serious infection, particularly in immunocompromised hosts, including neonates (75). The immune systems of patients with CRC are impaired following surgery and during chemotherapy, therefore, infection with M. morganii must be strictly avoided. The sequences of OTU0023 were similar to P. distasonis, which is a normal intestinal bacterium (76). A previous study demonstrated that P. distasonis may be a colitis-promoting species (77). There were nine major OTUs that were prevalent in the intestinal bacteria and similar to Bacteroides species, including B. dorei and B. fragilis. B. dorei is dominant in the intestinal tracts of children with type I diabetes, with the highest level observed in 7.6-month-old infants, which suggests that it may be associated with a solid diet (78). B. fragilis is considered to be associated with intestinal inflammation and intestinal tumors due to the production of enterotoxin (79).
PCA revealed that the bacterial community structures were similar within the stool samples of all patients following surgery. However, the bacterial community structures between the stool samples prior to surgery and tissue samples from each patient were even more similar. There are two possible explanations for this result. Firstly, the impact of CRC is very complex. An obvious association was not identified from intestinal microbiota analysis alone, but several factors, including diet, genetics and lifestyle are likely associated with CRC. Secondly, the stool samples from all patients following surgery may be more consistent in their bacterial community structures since patients consumed a lighter diet and used antibiotics following surgery.
In summary, certain bacterial species have previously been identified to serve a role in the occurrence and development of CRC, including Streptococcus species, Helicobacter pylori, E. faecalis, B. fragilis, Clostridium septicum and E. coli (63). Almost all these bacteria were detected in the current study, which emphasizes the importance of studying the association between these bacteria and CRC to prevent infection of patients with CRC in the future. A number of conclusions have been made by the current study. Firstly, in terms of the intestinal microbiota present, cancer-adjacent tissues are not as healthy as previously considered. Furthermore, following surgery, there is an increase in patients' bacteria diversity index and the species of the bacterial community may be more abundant. The current study suggested that the bacterial community of the intestinal tract of patients with CRC may not be healthy. Finally, a number of drug-resistant bacteria were identified in the intestinal tract following surgery, which may possibly be caused by routine treatment with antibiotics. In the future, it may be necessary to study safe and effective clinical anti-infection methods, including the use of probiotics.
Acknowledgements
The authors would like to thank Miss Xiangxiang Liu and Miss Xue Yang, Faculty of Life Science and Technology, Kunming University of Science and Technology (Kunming, China), for performing part of the experimental work.
Glossary
Abbreviations
- CRC
colorectal cancer
- PCA
principle component analysis
- OTU
operational taxonomic unit
Funding
The current study was funded by the Fundamental Application Research Foundation of Yunnan Province [grant no. 2017FE468(−122)].
Availability of data and materials
The datasets generated during the current study are available in the Sequence Read Archive of NCBI under the accession number SRP120060 and all data analyzed during this study are included in the published article.
Authors' contributions
CJL and XRL contributed to the conception and design of the study. YS and BW assisted with the collection of samples. YLZ acquired the data. XRL, EY and YYL analyzed the data. YLZ, CJL, XRL, EY and YYL contributed to the writing, reviewing and/or revision of the submitted manuscript.
Ethics approval and consent to participate
This study was reviewed and approved by the Ethics Committee of The First People's Hospital of Yunnan Province and informed consent was obtained from all participants prior to engaging in any study activities.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Van Nhieu Tran J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tannock GW. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol. 2008;1(Suppl 1):S15–S18. doi: 10.1038/mi.2008.54. [DOI] [PubMed] [Google Scholar]
- 7.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 8.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 9.Burt RW. Colon cancer screening. Gastroenterology. 2000;119:837–853. doi: 10.1053/gast.2000.16508. [DOI] [PubMed] [Google Scholar]
- 10.Platz EA, Willett WC, Colditz GA, Rimm EB, Spiegelman D, Giovannucci E. Proportion of colon cancer risk that might be preventable in a cohort of middle-aged US men. Cancer Causes Control. 2000;11:579–588. doi: 10.1023/A:1008999232442. [DOI] [PubMed] [Google Scholar]
- 11.Watson AJ, Collins PD. Colon cancer: A civilization disorder. Dig Dis. 2011;29:222–228. doi: 10.1159/000323926. [DOI] [PubMed] [Google Scholar]
- 12.Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33:954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arthur JC, Perezchanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobhani I, Amiot A, Le Baleur Y, Levy M, Auriault ML, Van Nhieu JT, Delchier JC. Microbial dysbiosis and colon carcinogenesis: Could colon cancer be considered a bacteria-related disease? Therap Adv Gastroenterol. 2013;6:215–229. doi: 10.1177/1756283X12473674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaac A, Kochubiei O, Vizir M. Colorectal cancer treatment. KhNMU. 2014 [Google Scholar]
- 18.Xinli L, Dachang W, Cuili Z, Yi X. Side effects of antibiotics on the intestinal microflora by PCR-DGGE. Pak J Pharm Sci. 2013;26:339–343. [PubMed] [Google Scholar]
- 19.Schmidt TM, Delong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wuyts J, Peer YVD. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: Formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. doi: 10.2307/2531532. [DOI] [PubMed] [Google Scholar]
- 25.Chao A, Hwang WH, Chen YC, Kuo CY. Estimating the number of shared species in two communities. Statistica Sinica. 2000;10:227–246. [Google Scholar]
- 26.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 27.Simpson EH. Measurement of diversity. Nature. 1949 Apr 30;163 doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 28.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 31.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen-Vercoe E, Jobin C. Fusobacterium and enterobacteriaceae: Important players for CRC? Immunol Lett. 2014;162:54–61. doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Artinyan A, Mailey B, Christopher S, Lee W, McKenzie S, Chen SL, Bhatia S, Pigazzi A, Garcia-Aguilar J. An interaction of race and ethnicity with socioeconomic status in rectal cancer outcomes. Ann Surg. 2011;253:647–654. doi: 10.1097/SLA.0b013e3182111102. [DOI] [PubMed] [Google Scholar]
- 34.Day GE, Provost E, Lanier AP. Alaska native mortality rates and trends. Public Health Rep. 2009;124:54–64. doi: 10.1177/003335490912400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanier AP, Day GE, Kelly JJ, Provost E. Disparities in cancer mortality among Alaska Native people, 1994–2003. Alaska Med. 2008;49:120–125. [PubMed] [Google Scholar]
- 36.Yothers G, Sargent DJ, Wolmark N, Goldberg RM, O'Connell MJ, Benedetti JK, Saltz LB, Dignam JJ, Blackstock AW, ACCENT Collaborative Group Outcomes among black patients with stage II and III colon cancer receiving chemotherapy: An analysis of ACCENT adjuvant trials. J Natl Cancer Inst. 2011;103:1498–1506. doi: 10.1093/jnci/djr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hester CM, Jala VR, Langille MG, Umar S, Greiner KA, Haribabu B. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J Gastroenterol. 2015;21:2759–2769. doi: 10.3748/wjg.v21.i9.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruno ME, Rogier EW, Frantz AL, Stefka AT, Thompson SN, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor in intestinal epithelial cells by enterobacteriaceae: Implications for mucosal homeostasis. Immunol Invest. 2010;39:356–382. doi: 10.3109/08820131003622809. [DOI] [PubMed] [Google Scholar]
- 39.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stecher B. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MBP-0008-2014. [DOI] [PubMed] [Google Scholar]
- 41.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, Moris F, Rodrigo L, Mira A, Collado MC. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 42.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Holzapfel WH, Wood BJB. The family enterococcaceae. John Wiley Sons Ltd; 2014. [Google Scholar]
- 45.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 47.Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002;160:2253–2257. doi: 10.1016/S0002-9440(10)61172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun T, Liu S, Zhou Y, Yao Z, Zhang D, Cao S, Wei Z, Tan B, Li Y, Lian Z, Wang S. Evolutionary biologic changes of gut microbiota in an ‘adenoma-carcinoma sequence’ mouse colorectal cancer model induced by 1,2-Dimethylhydrazine. Oncotarget. 2017;8:444–457. doi: 10.18632/oncotarget.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohoutova D, Smajs D, Moravkova P, Cyrany J, Moravkova M, Forstlova M, Cihak M, Rejchrt S, Bures J. Escherichia coli strains of phylogenetic group B2 and D and bacteriocin production are associated with advanced colorectal neoplasia. BMC Infect Dis. 2014;14:733. doi: 10.1186/s12879-014-0733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farkas-Himsley H, Yu H. Purified colicin as cytotoxic agent of neoplasia: Comparative study with crude colicin. Cytobios. 1985;42:193–207. [PubMed] [Google Scholar]
- 51.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Déchelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 52.Peterson CT, Vaughn AR, Sharma V, Chopra D, Mills PJ, Peterson SN, Sivamani RK. Effects of turmeric and curcumin dietary supplementation on human gut microbiota: A double-blind, randomized, placebo-controlled pilot study. J Evid Based Integr Med. 2018;23:2515690X18790725. doi: 10.1177/2515690X18790725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLos Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 56.Hendrickx AP, Top J, Bayjanov JR, Kemperman H, Rogers MR, Paganelli FL, Bonten MJ, Willems RJ. Antibiotic-driven dysbiosis mediates intraluminal agglutination and alternative segregation of enterococcus faecium from the intestinal epithelium. MBio. 2015;6:e01346–e01315. doi: 10.1128/mBio.01346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez-Diaz AM, Cuartero C, Rodriguez JD, Lozano S, Alonso JM, Rodríguez-Domínguez M, Tedim AP, Del Campo R, López J, Cantón R, Ruiz-Garbajosa P. The rise of ampicillin-resistant Enterococcus faecium high-risk clones as a frequent intestinal colonizer in oncohaematological neutropenic patients on levofloxacin prophylaxis: A risk for bacteraemia? Clin Microbiol Infect. 2016;22(59):e51–e58. doi: 10.1016/j.cmi.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 58.Becking JH. The family azotobacteraceae. Springer; New York: 2006. [Google Scholar]
- 59.Chen L, Zhang YH, Huang T, Cai YD. Gene expression profiling gut microbiota in different races of humans. Sci Rep. 2016;6:23075. doi: 10.1038/srep23075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins MD, Hutson RA, Foster G, Falsen E, Weiss N. Isobaculum melis gen. nov., sp. nov., a Carnobacterium-like organism isolated from the intestine of a badger. Int J Syst Evol Microbiol. 2002;52:207–210. doi: 10.1099/00207713-52-1-207. [DOI] [PubMed] [Google Scholar]
- 61.Aller AI, Castro C, Medina MJ, González MT, Sevilla P, Morilla MD, Corzo JE, Martín-Mazuelos E. Isolation of Moellerella wisconsensis from blood culture from a patient with acute cholecystitis. Clin Microbiol Infect. 2009;15:1193–1194. doi: 10.1111/j.1469-0691.2009.03046.x. [DOI] [PubMed] [Google Scholar]
- 62.Stock I, Falsen E, Wiedemann B. Moellerella wisconsensis: Identification, natural antibiotic susceptibility and its dependency on the medium applied. Diagn Microbiol Infect Dis. 2003;45:1–11. doi: 10.1016/S0732-8893(02)00483-2. [DOI] [PubMed] [Google Scholar]
- 63.Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dubrow R, Edberg S, Wikfors E, Callan D, Troncale F, Vender R, Brand M, Yapp R. Fecal carriage of Streptococcus bovis and colorectal adenomas. Gastroenterology. 1991;101:721–725. doi: 10.1016/0016-5085(91)90531-O. [DOI] [PubMed] [Google Scholar]
- 65.Farhadi T, Fakharian A, Ovchinnikov RS. Virtual screening for potential inhibitors of CTX-M-15 protein of Klebsiella pneumoniae. Interdiscip Sci. 2018;10:694–703. doi: 10.1007/s12539-017-0222-y. [DOI] [PubMed] [Google Scholar]
- 66.Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, Lochs H. Association between intraepithelial escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. doi: 10.1016/S0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 67.Poulsen ML, Bisgaard ML. MUTYH associated polyposis (MAP) Curr Genomics. 2008;9:420–435. doi: 10.2174/138920208785699562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khan AA, Cash P. E. coli and colon cancer: Is mutY a culprit? Cancer Lett. 2013;341:127–131. doi: 10.1016/j.canlet.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(548–563):e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bourassa L, Clarridge JE., III Clinical significance and characterization of streptococcus tigurinus isolates in an adult population. J Clin Microbiol. 2015;53:3574–3579. doi: 10.1128/JCM.01551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim B, Huh HJ, Chung DR, Kim WS, Ki CS, Lee NY. The first case of concurrent infective endocarditis and spondylitis caused by streptococcus tigurinus. Ann Lab Med. 2015;35:654–656. doi: 10.3343/alm.2015.35.6.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michelena A, Bonavila C, Zubeltzu B, Goenaga MA. Endocarditis due to Streptococcus tigurinus: Presentation of a case and a review of the literature. Enferm Infecc Microbiol Clín. 2015;33:575–576. doi: 10.1016/j.eimc.2015.01.006. (In Spanish) [DOI] [PubMed] [Google Scholar]
- 73.Skowron MA, Zebrowska J, Wegrzyn G, Skowron PM. MmoSTI restriction endonuclease, isolated from Morganella morganii infecting a tropical moth, Actias selene, cleaving 5′-|CCNGG-3′ sequences. J Appl Genetics. 2016;57:143–149. doi: 10.1007/s13353-015-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murphy K, Ryan C, Dempsey EM, O'Toole PW, Ross RP, Stanton C, Ryan CA. Neonatal sulfhemoglobinemia and hemolytic anemia associated with intestinal morganella morganii. Pediatrics. 2015;136:1641–1645. doi: 10.1542/peds.2015-0996. [DOI] [PubMed] [Google Scholar]
- 75.Shahbazi E, Mollasalehi H, Minai-Tehrani D. Development and evaluation of an improved quantitative loop-mediated isothermal amplification method for rapid detection of morganella morganii. Talanta. 2019;191:54–58. doi: 10.1016/j.talanta.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 76.Koh GY, Kane A, Lee K, Xu Q, Wu X, Roper J, Mason JB, Crott JW. Parabacteroides distasonis attenuates toll-like receptor 4 signaling and Akt activation and blocks colon tumor formation in high-fat diet-fed azoxymethane-treated mice. Int J Cancer. 2018 Apr 26; doi: 10.1002/ijc.31559. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 77.Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D. Pglyrp-regulated gut microflora prevotella falsenii, parabacteroides distasonis and bacteroides eggerthii enhance and alistipes finegoldii attenuates colitis in mice. PLos One. 2016;11:e0146162. doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis-Richardson AG, Ardissone AN, Dias R, Simell V, Leonard MT, Kemppainen KM, Drew JC, Schatz D, Atkinson MA, Kolaczkowski B, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ignacio A, Fernandes MR, Avila-Campos MJ, Nakano V. Enterotoxigenic and non-enterotoxigenic bacteroides fragilis from fecal microbiota of children. Braz J Microbiol. 2015;46:1141–1145. doi: 10.1590/S1517-838246420140728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available in the Sequence Read Archive of NCBI under the accession number SRP120060 and all data analyzed during this study are included in the published article.