Abstract
Resistance to apoptosis is a characteristic of cancer. Curcumin has become a potential anticancer drug for its pro-apoptotic effects, but the underlying mechanisms remain unclear. Furthermore, the Notch3-p53 signaling axis serves an important role in cell fate. The present study was designed to investigate the antitumor effect of curcumin by the Notch3-p53 axis in mouse myeloma P3X63Ag8 cells. The effects of curcumin on the viability of P3X63Ag8 cells were evaluated using an MTT assay. Quantitative expression of the Notch3-p53 signaling axis-associated genes was measured by reverse transcription-quantitative polymerase chain reaction, and western blot analysis was used to investigate the expression of proteins. Additionally, flow cytometry was used to measure the ratio of apoptosis. The results demonstrated that curcumin could significantly inhibit cell viability. No significant pro-apoptotic effect was observed when the concentration of curcumin was <30 µM. At 30 µM, curcumin-treated cells exhibited an apoptotic phenomenon, and the ratio of late apoptosis increased with the concentration of curcumin, and reached 28.4 and 51.8% in the medium- and high-dose groups, respectively. Curcumin inhibited the expression of Notch3, while the middle- and high-dose groups promoted p53. The expression of Notch3-responsive genes Hes family BHLH transcription factor 1 and Hes-related family transcription factor with YRPW motif 1 were notably promoted. Curcumin treatment significantly downregulated B-cell lymphoma-2 (Bcl-2) at the mRNA and protein levels, but upregulated Bcl-2-associated X. These data indicated that curcumin exhibited antitumor effects in mouse myeloma cells with induction of apoptosis by affecting the Notch3-p53 signaling axis.
Keywords: curcumin, apoptosis, Notch3, p53, P3X63Ag8 cell
Introduction
Curcumin is the primary active component of turmeric and is part of the ginger family (Zingiberaceae). Due to it having no deleterious effects and it having a broad-spectrum anticancer activity, curcumin has become one of the most promising anticancer phytochemicals that has been frequently studied for its molecular mechanism (1–4). Accumulating evidence has indicated that curcumin may significantly inhibit the proliferation of tumor cells and induce apoptosis (5–8). Additionally, a number of studies have determined that curcumin may trigger the intrinsic apoptotic pathway (9,10).
Multiple myeloma (MM) is a B-cell malignancy characterized by overabundance of monoclonal plasma cells in the bone marrow, which can cause osteolytic lesions (11). Previous studies demonstrated that direct contact between osteocytes and MM cells activates the Notch signaling pathway, increases Notch receptor expression in MM cells, particularly Notch3 and 4, and stimulates MM proliferation (12–14). The Notch signaling pathway is a highly conserved system that dictates cell fate and critically influences cell proliferation, differentiation and apoptosis (15). Notch3 is one of the four Notch receptors (Notch1-4) that are overexpressed in numerous cancer cells, including ovarian high-grade serous carcinoma (16), pancreatic cancer (17) and urothelial carcinoma (18). It has been reported that overexpression of Notch3 may be responsible for promoting tumor cell growth in human lung cancer types, including hepatocellular carcinoma and non-small cell lung cancers (19,20). A similar study determined that enforced expression of Notch3 could cause dysregulated hyperplasia of T cells (21). All these data indicated that Notch3 serves a crucial role in the anti-apoptosis of cancer cells. Components of the two families of basic helix-loop-helix transcription factors, Hes family BHLH transcription factor 1 (Hes1) and Hes-related family transcription factor with YRPW motif 1 (Hey1), are two of the main effectors of the Notch signaling pathway (22).
Tumor suppressor p53 is an important transcription factor that modulates cell death and survival (23). A previous study determined that Notch3 regulates p53 at the post-transcriptional level by controlling cyclin G1 expression and the cell signal transduction circuit in hepatocellular carcinoma (24). There are a number of potential mediators of p53-induced apoptosis, including murine double minute 2 (MDM2), first apoptosis signal receptor (Fas) and leucine repeat death domain containing protein (LRDD) (25). p53 has been demonstrated to function by direct interactions with, or activation of, B-cell lymphoma-2 (Bcl-2) family proteins in the mitochondria (26). The members of the Bcl-2 family regulate apoptosis by the permeabilization of the outer mitochondrial membrane (27).
Notch3 and p53 signaling pathways are important in cell fate (15,23). Curcumin is known to activate cell death signals and induce apoptosis in cancer cells by regulating multiple important cellular signaling pathways, including Notch (28); however, the effects of curcumin on Notch3 are less well studied. Therefore, the present study focused on the effect of curcumin on mouse myeloma cells and the underlying mechanisms mediated through the Notch3-p53 signaling pathway.
Materials and methods
Cell culture
Mouse myeloma P3X63Ag8 cells (Stem Cell Bank, Chinese Academy of Sciences, Shanghai, China) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and 1% penicillin-streptomycin (cat. no., 15140122, Gibico, Thermo Fisher Scientific, Inc., USA) and incubated in a humidified incubator containing 5% CO2 at 37°C. P3X63Ag8 cells were stimulated with curcumin (0, 30, 40 and 50 µM) for 24 h at 37°C with 5% CO2.
MTT assay
The cells were placed into 96-well plates (Eppendorf, Hamburg, Germany) at a density of 4×104/200 µl/well in DMEM (Hyclone; GE Healthcare Life Sciences) supplemented with 10% FBS and 1% penicillin-streptomycin, and cultured overnight at 37°C. Medium was replaced with fresh DMEM containing different concentrations (0, 10, 20, 30, 40, 50, 60, 70 and 80 µM) of curcumin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Following a further incubation for 24 h at 37°C, 20 µl MTT (2 mg/ml) was added to each well followed by a 4 h incubation at 37°C. Viability was determined with formazan crystal pellets dissolved in dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA), and the absorbance of the plate was determined with a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 570 nm. The results are expressed as percentage viability, calculated using the following formula:
Isolation of RNA and DNase treatment
Total RNA was extracted using TRIzol Reagent® (Sigma-Aldrich; Merck KGaA). All RNA extracts were treated with DNase using a RQ1 RNase-Free DNase kit (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocols.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay. Synthesis of complementary DNA (cDNA) was performed using the GoScript™ Reverse Transcription system (Promega Corporation), according to the manufacturer's protocols. Quantitative expression of the genes was conducted by qPCR using a CFX96 Touch Deep Well Real-Time PCR Detection system (Bio-Rad Laboratories, Inc.). Target genes were amplified using the primers in Table I. β-actin was employed as the endogenous control. The reaction mixture contained 6.25 µl 2X GoTaq®qPCR Master mix (Promega Corporation), 1 µl cDNA, 0.25 µl upstream and 0.25 µl downstream PCR primers, and nuclease-free water up to a final volume of 12.5 µl. Each reaction was conducted in triplicate. The RT-qPCR reaction conditions were subjected to an initial pre-degeneration step of 95°C for 3 min, followed by 39 cycles of 95°C for 20 sec, followed by 60°C for 30 sec. mRNA levels were quantified using the 2−ΔΔCq method (29) and normalized to those of β-actin. Fluorescence data were collected and analyzed with the Bio-Rad CFX Manager software 3.0 (Bio-Rad Laboratories, Inc.). The melting curves were produced following qPCR.
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| Gene symbol | NCBI Ref seq no. | Sequence (5′→3′) | Product length (bp) |
|---|---|---|---|
| β-actin | NM_007393.4 | 149 | |
| Forward | TGAGAGGGAAATCGTGCGTGAC | ||
| Reverse | GCTCGTTGCCAATAGTGATGACC | ||
| p53 | NM_001127233.1 | 179 | |
| Forward | ACAGGCAGACTTTTCGCCACAG | ||
| Reverse | CCGTCCCAGAAGGTTCCCACT | ||
| Bax | NM_007527.3 | 118 | |
| Forward | TGGAGATGAACTGGACAGCA | ||
| Reverse | GAAGTTGCCATCAGCAAACA | ||
| Bcl-2 | NM_009741.4 | 131 | |
| Forward | CGATTGTGGCAGTCCCTTA | ||
| Reverse | CCAGGATGAAGTGCTCAGGT | ||
| Hes1 | NM_008235.2 | 157 | |
| Forward | CAACACGACACCGGACAAAC | ||
| Reverse | GGAATGCCGGGAGCTATCTT | ||
| Hey1 | NM_010423.2 | 164 | |
| Forward | TGCAGTTAACTCCTCCTTGCC | ||
| Reverse | CGCCGAACTCAAGTTTCCATT |
Bcl-2, B-cell lymphoma 2; Bax, Bcl-2 associated X; Hes1, Hes family BHLH transcription factor 1; Hey1, Hes-related family transcription factor with YRPW motif 1.
Western blot analysis
P3X63Ag8 cells were stimulated with curcumin (0, 30, 40 or 50 µM) for 24 h at 37°C. Subsequently, the cells treated with indicated reagents were lysed in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with phenylmethylsulfonyl fluoride (Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 30 min on ice. Following centrifugation at 16,000 × g for 20 min at 4°C, the protein concentration was determined using a Bradford Protein Assay kit (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). Subsequently, 30 µg protein was subjected to 10% SDS-PAGE electrophoresis. The separated proteins were transferred to a nitrocellulose filter membrane using the Trans-Blot®SD Semi-Dry Transfer Cell (Bio-Rad Laboratories, Inc.), according to the manufacturer's protocols. Membranes were blocked with 5% skimmed milk powder at 4°C overnight and incubated with primary antibodies [anti-Notch3 rabbit polyclonal antibody (dilution, 1:1,000; cat. no., ab23426), anti-Bcl-2 associated X (Bax) rabbit monoclonal antibody (dilution, 1:8,000; cat. no., ab32503) and anti-Bcl-2 mouse monoclonal antibody (dilution, 1:500; cat. no., ab692) (all from Abcam, Cambridge, UK); anti-P53 mouse monoclonal antibody (dilution, 1:5,000; cat. no., GTX70214; GeneTex, Inc., Irvine, CA, USA); and anti-β-Actin mouse monoclonal antibody (dilution, 1:8,000; cat. no., HC201; Beijing Transgen Biotech Co., Ltd., Beijing, China)] in 5% skimmed milk in TBS with Tween 20 (TBST) for 1 h at room temperature. Following three washes with TBST for 5 min each, the membrane was incubated with the appropriate secondary antibody [goat anti-mouse IgG (H+L) horseradish peroxidase (HRP)-conjugated (dilution, 1:3,000; cat. no., BA1050; Wuhan Boster Biological Technology, Ltd., Wuhan, China) and goat anti-rabbit IgG (H+L) HRP-conjugated (dilution, 1:3,000; cat. no., BA1054; Wuhan Boster Biological Technology, Ltd.) secondary antibodies] at room temperature for 1 h, followed by washing three times with TBST for 10 min each time. The membrane was developed with a Western Bright enhanced chemiluminescent (ECL) kit (Advansta, Menlo Park, CA, USA).
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis assay
Cells were harvested by centrifugation at 1,000 × g for 5 min, and washed twice with 500 µl of PBS at room temperature. Annexin V-FITC/PI double staining kit (cat. no., KGA106; Jiangsu KeyGen Biotech Co., Ltd., Nanjing, China), according to the manufacturer's protocols, were employed to analyze apoptotic rate in cells. In brief, liquots of 1×106 cells were suspended in 500 µl binding buffer followed by the addition of 5 µl Annexin V-FITC and 5 µl PI at room temperature for 5 min. Following incubation in the dark at 37°C for 5 min, the cells were analyzed using a flow cytometer BD Accuri C6 Plus (BD Accuri™; BD Biosciences, Franklin Lakes, NJ, USA). A total of 10,000 events were measured per sample.
Statistical analysis
Differences between groups were analyzed using one-way analysis of variance and multiple comparison between the groups was performed using Tukey's post hoc test. Experimental data are expressed as the mean ± standard error of the mean of three independent experiments. P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed with SPSS version 10.0 (SPSS, Inc., Chicago, IL, USA).
Results
The effect of curcumin on the proliferation of P3X63Ag8 cells
To determine the effect of curcumin on cell viability, cells were treated with different concentrations of curcumin for 24 h, and viable cells were measured using an MTT assay. As depicted in Fig. 1, increasing concentrations of curcumin resulted in a significant decrease (P<0.01) in cell viability compared with the 0 µM group. Notably, curcumin treatment resulted in a reduced inhibition at 40 µM, compared with 30 µM. A total of 3 effective concentrations (30, 40 and 50 µM) were selected for all further mechanistic studies.
Figure 1.
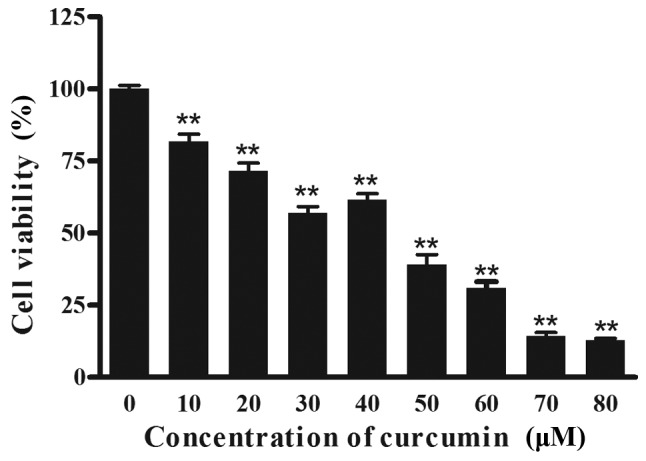
Effect of curcumin on the proliferation of P3X63Ag8 cells. The cells were treated with different concentrations of curcumin (0, 10, 20, 30, 40, 50, 60, 70 and 80 µM) for 24 h. The results are expressed as the mean ± standard error of the mean of eight independent experiments. **P<0.01 vs. the curcumin-untreated group.
The effect of curcumin on the mRNA and protein levels of P3X63Ag8 cells
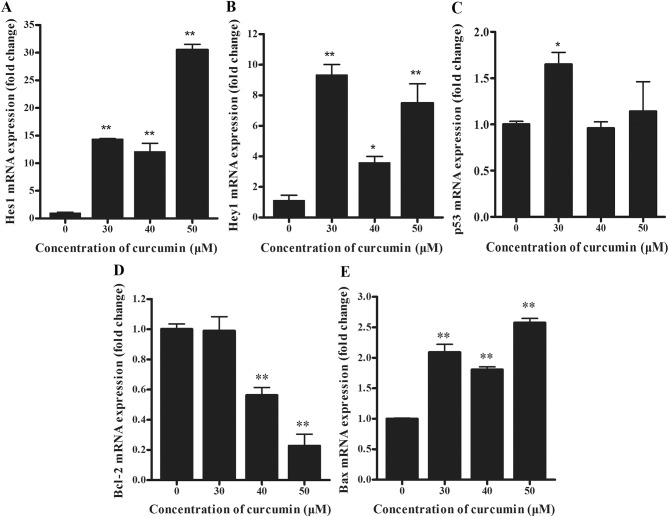
To determine the underlying molecular mechanisms of apoptosis of cells by curcumin, the mRNA levels of Notch3-p53 signaling axis genes were examined. Compared with the negative control group, the expression of Hes1 transcripts was significantly increased in curcumin-treated cells (Fig. 2A; P<0.01). Similarly, the expression of Hey1 mRNA copies was significantly increased in high-dose curcumin-treated cells compared with the negative control group (Fig. 2B; P<0.01). Furthermore, no significant differences were determined in the expression levels of p53 in 40 and 50 µM curcumin-treated cells, compared with the control, whereas levels increased significantly in 30 µM curcumin-treated cells (Fig. 2C; P<0.05). As depicted in Fig. 2D, Bcl-2 mRNA was expressed at significantly reduced levels in 40 and 50 µM curcumin-treated cells (P<0.01). By contrast, Bax transcripts were significantly overexpressed in curcumin-treated cells (Fig. 2E; P<0.01), compared with the control.
Figure 2.
(A) Hes1, (B) Hey1, (C) p53, (D) Bcl-2 and (E) Bax mRNA levels of curcumin-treated (0, 30, 40 and 50 µM) P3X63Ag8 cells for 24 h by reverse transcription-quantitative polymerase chain reaction. The results are expressed as the mean ± standard error of the mean of eight independent experiments. *P<0.05 and **P<0.01, compared with the curcumin-untreated group. Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X; Hes1, Hes family BHLH transcription factor 1; Hey1, Hes-related family transcription factor with YRPW motif 1.
The activation status of the Notch3-p53 signaling pathways in curcumin-stimulated P3X63Ag8 cells was investigated using western blotting. The 90 kDa band for Notch3 was detectable in samples (Fig. 3). As predicted, Notch3 was overexpressed in P3X63Ag8 cells, compared with β-actin, and significantly decreased in curcumin-treated cells. The results demonstrated that Bcl-2 (26 kDa) and Bax (21 kDa) protein expression levels (Fig. 3) were similar to their mRNA levels, while the p53 (53 kDa) expression level was not similar to its mRNA levels.
Figure 3.
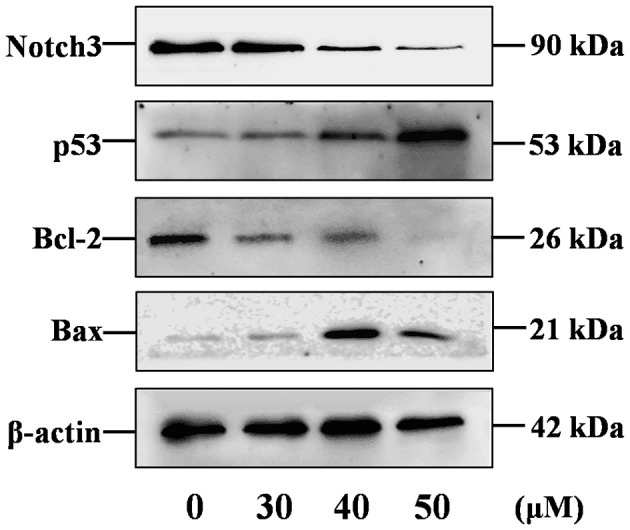
Western blotting of Notch3, p53, Bcl-2, Bax and β-actin in P3X63Ag8 cells stimulated with 0, 30, 40 and 50 µM curcumin for 24 h. Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X.
The effect of curcumin on the apoptosis of P3X63Ag8 cells
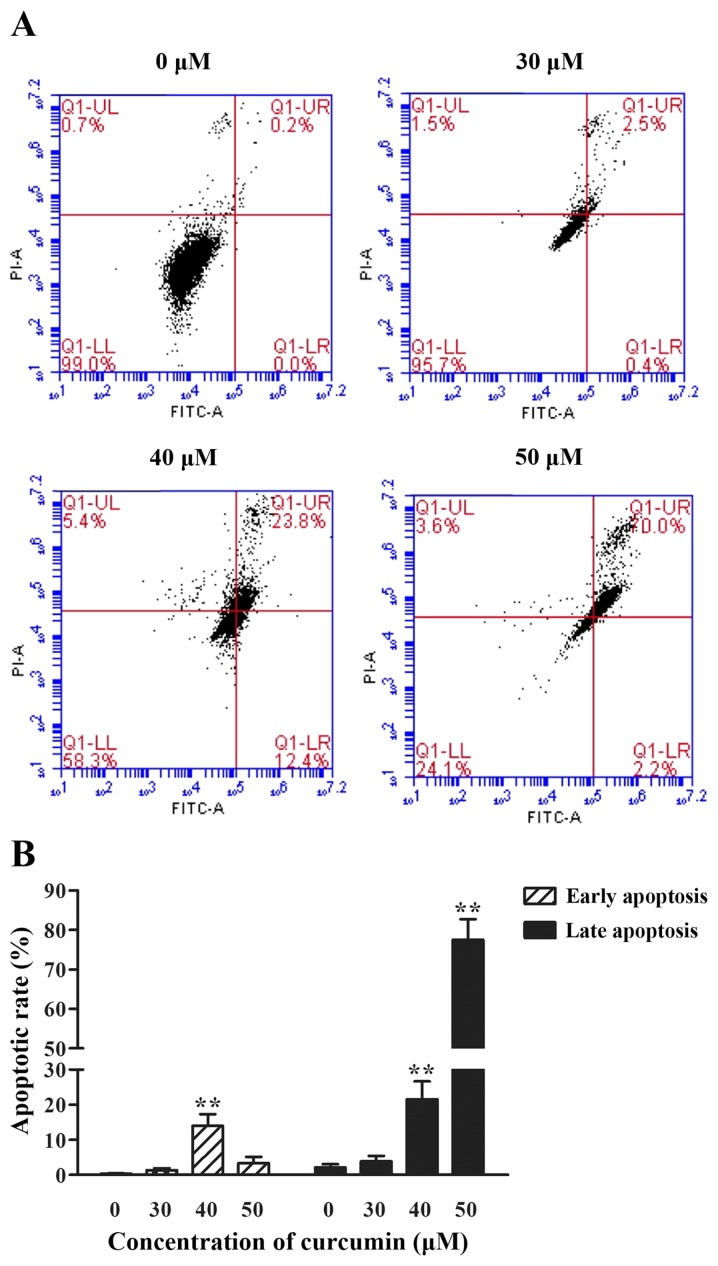
To examine whether curcumin could cause apoptosis, cells treated with different concentrations of curcumin (30, 40 and 50 µM) were stained with PI and FITC, and analyzed by fluorescence-activated cell sorting analysis. Curcumin induced apoptosis in a dose-dependent manner, with the exception in a decrease of early apoptotic rate at 50 µM. The percentage of late apoptosis cells was notably increased (P<0.01) compared with the 0 µM group. As depicted in Fig. 4, the percentage of early apoptosis cells reached the highest level when stimulated with 40 µM curcumin (P<0.01).
Figure 4.
The effect of curcumin on the apoptosis of P3X63Ag8 cells. (A) Flow cytometric dot plots of one representative experiment. Results are depicted as logarithmic fluorescence intensity on the x-axis (Annexin V-FITC) vs. y-axis (PI). A total of four quadrants represent necrotic cells (Q1-UL:AV-/PI+), late apoptotic cells (Q1-UR:AV+/PI+), early apoptotic cells (Q1-LR:AV+/PI-) and viable cells (Q1-LL:AV-/PI-). The numbers indicate the percentage of cells in each quadrant and a minimum of 10,000 events were read. (B) The histogram depicts the percentage of early and late apoptosis cells ± standard error of three independent experiments. **P<0.01 vs. the curcumin-untreated group. Cells were treated with 0, 30, 40 and 50 µM curcumin for 24 h. FITC, fluorescein isothiocyanate PI, propidium iodide.
Discussion
Apoptosis is a multi-step, multi-pathway cell-death program that is inherent in every cell of the body. A characteristic of cancer cells is anti-apoptosis; therefore, cancer treatment is primarily dependent on inducing apoptosis (30). Curcumin has been frequently used as a spice, food additive and herbal medicine in Asia, and has not been demonstrated to cause any toxicity (31). A number of studies indicated anticancer effects for curcumin, and the mechanisms underlying these effects may be the modulation of multiple oncogenic signaling transduction elements (32–34). In the present study, it was determined that curcumin-induced mouse myeloma P3X63Ag8 cell apoptosis is mediated by modulation of the molecular targets p53, Bcl-2 and Bax. It was also demonstrated, with RT-qPCR and western blotting, that curcumin regulates the Notch3 signaling pathway. Additionally, it was observed that curcumin dose-dependently induces apoptosis in P3X63Ag8 cells, as indicated by Annexin V-FITC/PI staining.
Notch3 overexpression has been observed in a number of tumor types, including T-cell acute lymphoblastic leukemia (35), triple-negative breast cancer (36) and prostate cancer (37). Notch3 activation favors cell proliferation, resistance to apoptotic stimuli, metastatic capability and maintenance of stem cell-like features (38). The results of the present study demonstrated that the Notch3 protein was overexpressed in mouse myeloma P3X63Ag8 cells, as predicted. Furthermore, curcumin treatment significantly decreased Notch3 protein expression, which is associated with the induction of apoptosis. p53 is a tumor suppressor and transcription factor (39). When activated, p53 induces cell cycle arrest in G1 or G2, p53 also induces cell death by apoptosis (39). The association between Notch3 and p53 has been studied in hepatocellular carcinoma (24,40). The results of the present study demonstrated that curcumin significantly decreased Notch3 expression, promoting cell apoptosis, which is consistent with the anticancer effects of sorafenib and valproic acid (41). In the present study, the mRNA and protein expression level of p53 demonstrated a comparative trend, which indicated that p53 may depend on the post-transcriptional regulation of curcumin. The explanation of the post-transcriptional regulation of curcumin on p53 has been reported. Curcumin could upregulate scaffold/matrix attachment region 1, decreasing histone acetylation at H3K9, a lysine residue 9 on histone H3, and H3K18, a lysine residue 18 on histone H3, resulting in the inhibition of E6 promoter, which interrupts degradation of E6-mediated p53 in HPV18-infected cervical carcinoma (42). In the present study, the protein expression level of p53 is significantly increased in the middle- and high-dose groups, while the low-dose group demonstrated no significant difference. We hypothesized that the apoptosis caused by the low-dose group may not depend on p53 activation, which is different to the middle- and high-dose groups. This may be the reason for reduced inhibition at 40 µM, compared with 30 µM. From the flow cytometry results (Fig. 4), it was also speculated that the middle-dose group causes an early appearance of apoptotic cells, which is comparative to the other groups. The mechanism underlying this remains to be further investigated.
The present study indicated that Hes1 lies downstream of Notch3 and mediates Notch3 signaling-induced proliferation (43). Notably, Notch-mediated cell death has been associated with upregulation of Hes1 expression (44). Hey1, a well-known transcriptional target of Notch signaling, may be a pathway of Notch3 in sustaining mature vessel structure and vascular integrity (45). Furthermore, the expression of Hes1 and Hey1 could induce cell death through the Notch signaling pathway (15). The Bcl-2 family is able to promote or inhibit apoptosis by direct action on permeabilization of the mitochondrial outer membrane (27). Additionally, it is well known that p53 can directly regulate the expression of Bcl-2 and Bax (26,46,47). To define the role of curcumin on the impact of the Notch3-p53 signaling pathway on apoptosis, the p53-responsive genes Bcl-2 and Bax were examined at the mRNA and protein expression levels (Figs. 2 and 3). Additionally, the Notch3-responsive gene Hes1 and Hey1 mRNA expression levels were measured. Curcumin inhibited the expression of Bcl-2 while promoting the expression of Bax, Hes1 and Hey1, indicating that the apoptosis effects of curcumin on P3X63Ag8 cells depend on the Notch3-p53 signaling pathway.
In conclusion, the results of the present study indicated that curcumin exhibited antitumor effects in mouse myeloma cells by inducing apoptosis through effects on the Notch3-p53 signaling pathway. Further studies are warranted to investigate the involvement of the Notch3-p53 signaling pathway in the mechanism underlying curcumin suppressing MM. Furthermore, the effect of inducing cell apoptosis by curcumin with inhibition of the Notch3-p53 signaling pathway will be the focus of our future investigations.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant No. 31572484; Beijing, China).
Availability of data and materials
All data generated or analyzed during the present study are included in this article.
Authors' contributions
XH conceived and designed the experiments. Data collection and experiments were performed by YZ and XL. JZ, ZX, HD and YH analyzed the data. YZ, XL and YH contributed to the writing of the manuscript. All authors revised and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent to participate
Not applicable.
Competing interests
The authors declare no conflict of interest.
References
- 1.Zhang X, Wang R, Chen G, Dejean L, Chen QH. The Effects of curcumin-based compounds on proliferation and cell death in cervical cancer cells. Anticancer Res. 2015;35:5293–5298. [PubMed] [Google Scholar]
- 2.Hu A, Huang JJ, Li RL, Lu ZY, Duan JL, Xu WH, Chen XP, Fan JP. Curcumin as therapeutics for the treatment of head and neck squamous cell carcinoma by activating SIRT1. Sci Rep. 2015;5:13429. doi: 10.1038/srep13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakrishna A, Kumar MH. Evaluation of synergetic anticancer activity of berberine and curcumin on different models of A549, Hep-G2, MCF-7, jurkat, and K562 cell lines. Biomed Res Int. 2015;2015:354614. doi: 10.1155/2015/354614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ting CY, Wang HE, Yu CC, Liu HC, Liu YC, Chiang IT. Curcumin triggers DNA damage and inhibits expression of DNA repair proteins in human lung cancer cells. Anticancer Res. 2015;35:3867–3873. [PubMed] [Google Scholar]
- 5.Mukhopadhyay A, Banerjee S, Stafford LJ, Xia C, Liu M, Aggarwal BB. Curcumin-induced suppression of cell proliferation correlates with down-regulation of cyclin D1 expression and CDK4-mediated retinoblastoma protein phosphorylation. Oncogene. 2002;21:8852–8861. doi: 10.1038/sj.onc.1206048. [DOI] [PubMed] [Google Scholar]
- 6.Li ZC, Zhang LM, Wang HB, Ma JX, Sun JZ. Curcumin inhibits lung cancer progression and metastasis through induction of FOXO1. Tumour Biol. 2014;35:111–116. doi: 10.1007/s13277-013-1013-7. [DOI] [PubMed] [Google Scholar]
- 7.Choudhuri T, Pal S, Agwarwal ML, Das T, Sa G. Curcumin induces apoptosis in human breast cancer cells through p53-dependent Bax induction. FEBS Lett. 2002;512:334–340. doi: 10.1016/S0014-5793(02)02292-5. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Wang L, Song R, Shen Y, Sun Y, Gu Y, Shu Y, Xu Q. Targeting sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2 by curcumin induces ER stress-associated apoptosis for treating human liposarcoma. Mol Cancer Ther. 2011;10:461–471. doi: 10.1158/1535-7163.MCT-10-0812. [DOI] [PubMed] [Google Scholar]
- 9.Chang PY, Peng SF, Lee CY, Lu CC, Tsai SC, Shieh TM, Wu TS, Tu MG, Chen MY, Yang JS. Curcumin-loaded nanoparticles induce apoptotic cell death through regulation of the function of MDR1 and reactive oxygen species in cisplatin-resistant CAR human oral cancer cells. Int J Oncol. 2013;43:1141–1150. doi: 10.3892/ijo.2013.2050. [DOI] [PubMed] [Google Scholar]
- 10.Rashmi R, Santhosh Kumar TR, Karunagaran D. Human colon cancer cells differ in their sensitivity to curcumin-induced apoptosis and heat shock protects them by inhibiting the release of apoptosis-inducing factor and caspases. FEBS Lett. 2003;538:19–24. doi: 10.1016/S0014-5793(03)00099-1. [DOI] [PubMed] [Google Scholar]
- 11.Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC. Multiple myeloma. Lancet. 2009;374:324–339. doi: 10.1016/S0140-6736(09)60221-X. [DOI] [PubMed] [Google Scholar]
- 12.Delgado-Calle J, Anderson J, Cregor MD, Hiasa M, Chirgwin JM, Carlesso N, Yoneda T, Mohammad KS, Plotkin LI, Roodman GD, Bellido T. Bidirectional notch signaling and osteocyte-derived factors in the bone marrow microenvironment promote tumor cell proliferation and bone destruction in multiple myeloma. Cancer Res. 2016;76:1089–1100. doi: 10.1158/0008-5472.CAN-15-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA. Pathogenesis of bone disease in multiple myeloma: From bench to bedside. Blood Cancer J. 2018;8:7. doi: 10.1038/s41408-017-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jundt F, Pröbsting KS, Anagnostopoulos I, Muehlinghaus G, Chatterjee M, Mathas S, Bargou RC, Manz R, Stein H, Dörken B. Jagged1-induced notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- 15.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 16.Park JT, Chen X, Tropè CG, Davidson B, Shih IeM, Wang TL. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to carboplatin. Am J Pathol. 2010;177:1087–1094. doi: 10.2353/ajpath.2010.100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang T, Vo K, Washington K, Berlin J. The role of Notch3 signaling pathway in pancreatic cancer. J Clin Oncol. 2007;25:21049–21049. [Google Scholar]
- 18.Zhang H, Liu L, Liu C, Pan J, Lu G, Zhou Z, Chen Z, Qian C. Notch3 overexpression enhances progression and chemoresistance of urothelial carcinoma. Oncotarget. 2017;8:34362–34373. doi: 10.18632/oncotarget.16156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gramantieri L, Giovannini C, Lanzi A, Chieco P, Ravaioli M, Venturi A, Grazi GL, Bolondi L. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int. 2007;27:997–1007. doi: 10.1111/j.1478-3231.2007.01544.x. [DOI] [PubMed] [Google Scholar]
- 20.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, Dang TP. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 21.Bellavia D, Campese AF, Checquolo S, Balestri A, Biondi A, Cazzaniga G, Lendahl U, Fehling HJ, Hayday AC, Frati L, et al. Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc Natl Acad Sci USA. 2002;99:3788–3793. doi: 10.1073/pnas.062050599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 23.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 24.Giovannini C, Minguzzi M, Baglioni M, Fornari F, Giannone F, Ravaioli M, Cescon M, Chieco P, Bolondi L, Gramantieri L. Suppression of p53 by Notch3 is mediated by Cyclin G1 and sustained by MDM2 and miR-221 axis in hepatocellular carcinoma. Oncotarget. 2014;5:10607–10620. doi: 10.18632/oncotarget.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasty P, Campisi J, Sharp ZD. Do p53 stress responses impact organismal aging? Transl Cancer Res. 2016;5:685–691. doi: 10.21037/tcr.2016.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 27.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 28.Subramaniam D, Ponnurangam S, Ramamoorthy P, Standing D, Battafarano RJ, Anant S, Sharma P. Curcumin induces cell death in esophageal cancer cells through modulating notch signaling. PLoS One. 2012;7:e30590. doi: 10.1371/journal.pone.0030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Igney FH, Krammer PH. Death and anti-death: Tumour resistance to apoptosis. Nat Rev Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 31.Ammon HP, Wahl MA. Pharmacology of curcuma longa. Planta Med. 1991;57:1–7. doi: 10.1055/s-2006-960004. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Cheng X, Gao Y, Bao J, Guan H, Lu R, Yu H, Xu Q, Sun Y. Induction of ROS-independent DNA damage by curcumin leads to G2/M cell cycle arrest and apoptosis in human papillary thyroid carcinoma BCPAP cells. Food Funct. 2016;7:315–325. doi: 10.1039/C5FO00681C. [DOI] [PubMed] [Google Scholar]
- 33.Bhullar KS, Jha A, Rupasinghe HP. Novel carbocyclic curcumin analog CUR3d modulates genes involved in multiple apoptosis pathways in human hepatocellular carcinoma cells. Chem Biol Interact. 2015;242:107–122. doi: 10.1016/j.cbi.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Gali-Muhtasib H, Hmadi R, Kareh M, Tohme R, Darwiche N. Cell death mechanisms of plant-derived anticancer drugs: Beyond apoptosis. Apoptosis. 2015;20:1531–1562. doi: 10.1007/s10495-015-1169-2. [DOI] [PubMed] [Google Scholar]
- 35.Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, Stoppacciaro A, Tiveron C, Tatangelo L, Giovarelli M, et al. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedrosa AR, Graça JL, Carvalho S, Peleteiro MC, Duarte A, Trindade A. Notch signaling dynamics in the adult healthy prostate and in prostatic tumor development. Prostate. 2016;76:80–96. doi: 10.1002/pros.23102. [DOI] [PubMed] [Google Scholar]
- 38.Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, Santini D, Ceccarelli C, Chieco P, Bonafé M. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- 39.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giovannini C, Gramantieri L, Chieco P, Minguzzi M, Lago F, Pianetti S, Ramazzotti E, Marcu KB, Bolondi L. Selective ablation of Notch3 in HCC enhances doxorubicin's death promoting effect by a p53 dependent mechanism. J Hepatol. 2009;50:969–979. doi: 10.1016/j.jhep.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 41.Zhu W, Liang Q, Yang X, Yu Y, Shen X, Sun G. Combination of sorafenib and Valproic acid synergistically induces cell apoptosis and inhibits hepatocellular carcinoma growth via down-regulating Notch3 and pAkt. Am J Cancer Res. 2017;7:2503–2514. [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty S, Das K, Saha S, Mazumdar M, Manna A, Chakraborty S, Mukherjee S, Khan P, Adhikary A, Mohanty S, et al. Nuclear matrix protein SMAR1 represses c-Fos-mediated HPV18 E6 transcription through alteration of chromatin histone deacetylation. J Biol Chem. 2014;289:29074–29085. doi: 10.1074/jbc.M114.564872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Y, Zhang Y, Jiang H, Zhu Y, Liu L, Feng W, Yang L, Wang Y, Li M. Activation of Notch3 promotes pulmonary arterial smooth muscle cells proliferation via Hes1/p27Kip1 signaling pathway. FEBS Open Biol. 2015;5:656–660. doi: 10.1016/j.fob.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zweidler-McKay PA, He Y, Xu L, Rodriguez CG, Karnell FG, Carpenter AC, Aster JC, Allman D, Pear WS. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106:3898–3906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaucker A, Mercurio S, Sternheim N, Talbot WS, Marlow FL. Notch3 is essential for oligodendrocyte development and vascular integrity in zebrafish. Dis Model Mech. 2013;6:1246–1259. doi: 10.1242/dmm.012005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haldar S, Negrini M, Monne M, Sabbioni S, Croce CM. Down-regulation of bcl-2 by p53 in breast cancer cells. Cancer Res. 1994;54:2095–2097. [PubMed] [Google Scholar]
- 47.Harn HJ, Ho LI, Liu CA, Liu GC, Lin FG, Lin JJ, Chang JY, Lee WH. Down regulation of bcl-2 by p53 in nasopharyngeal carcinoma and lack of detection of its specific t(14;18) chromosomal translocation in fixed tissues. Histopathology. 1996;28:317–323. doi: 10.1046/j.1365-2559.1996.d01-431.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this article.