Abstract
MicroRNA (MiR)-506 serves a vital role in several types of cancer. However, the role of miR-506 in bladder cancer (BCa) progression remains to be investigated. The present study demonstrated that miR-506 expression was downregulated in BCa tissues and cell lines. Meanwhile, overexpression of miR-506 inhibited cell proliferation, migration and invasion. Additionally, upregulated miR-506 increased E-cadherin, and reduced N-cadherin and Vimentin expression levels, as markers of epithelial-to-mesenchymal transition. RWD domain containing 4 (RWDD4) was revealed to be a miR506 target in BCa cells, and the downregulation of RWDD4 was found to suppress BCa cell proliferation, migration and invasion. In summary, miR-506 may suppress the aggressive properties of human BCa cells by targeting RWDD4, and thus may be a novel therapeutic target in human BCa.
Keywords: bladder cancer, miR-506, epithelial-to-mesenchymal transition, RWDD4
Introduction
Bladder cancer (BCa) is the most common malignancy of the urinary system and is associated with a high mortality rate (1). Approximately 90% of BCa cases manifest as transitional cell carcinoma (TCC) of the urinary tract. TCC can be clinically divided into two groups: Non-muscle-invasive TCC, which can be treated by transurethral resection and bladder irrigation chemotherapy with satisfactory clinical outcome in terms of prognosis; and muscle-invasive TCC, which even with radical cystectomy, adjuvant chemotherapy and other treatments, remains to have a high mortality rate due to its high incidence of metastasis (2). Therefore, there is an urgent need to investigate the biological characteristics of TCC to improve the therapeutic outcome of TCC.
The short (−22 nt) non-coding RNAs known as microRNAs (miRNA/miRs) regulate target gene expression post-transcriptionally (3–5). Recent studies show that miRNAs control genes involved in cell apoptosis, differentiation, stress responses, migration and the cell cycle, and that their dysregulation is related to the initiation and progression of many cancers. MiR-506, a miRNA cluster located in chromosome X, has been implicated in several types of cancer. Previous studies confirmed that miR-506 exerted anti-tumor effects in cervical, ovarian and breast cancers (6–9). By contrast, miR-506 played the role of an oncogene in lung cancer and melanoma (10,11). However, how miR-506 is involved in BCa remains largely unknown, and its specific role in BCa requires further study.
In the present study, we aimed to identify the biological roles of miR-506 in BCa. We reported that miR-506 expression was decreased in human BCa tissues and cell lines. Furthermore, overexpressed miR-506 had antitumor effects that manifested as inhibited BCa cell proliferation, migration, invasion and EMT, which were indicated to occur through targeting of RWD domain containing (4) (RWDD4).
Materials and methods
Cell lines
TCC cell lines (T24, J82, and UM-UC-3) and the bladder epithelial immortalized cell line SV-HUC-1 were provided by the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The T24 and SV-HUC-1 cells were maintained in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), and the others were cultured in MEM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 1.5 g/l NaHCO3 and 0.1 g/l sodium pyruvate. All the cells were cultured in a humidified incubator containing 5% CO2 at 37°C.
Tumor tissues
This study was approved by the ethical committee of the Fourth People's Hospital of Shenyang (ethics review approval no: 20140124-8). Informed consent was provided by all participants. A total of 40 patients who were pathologically diagnosed with muscle-invasive TCC were enrolled in this study. A total of 40 pairs of primary TCC tissues and paracarcinoma specimens were obtained from surgical resection specimens and stored in liquid nitrogen. Hematoxylin-eosin (HE) staining was performed and evaluated by at least two experienced pathologists.
Total RNA extraction and quantitative PCR
Total RNA from the TCC cell lines and tissues was extracted with 1 ml TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The relative expression of miR506 or RWDD4 was detected by a real-time PCR system using SYBR Green Master Mix (Takara Bio, Inc., Otsu, Japan) and quantified by the 2−ΔΔCq method (12), with target expression normalized to the level of U6 or GAPDH. The primers designed and used for the assay were as follows: MiR-506-forward: GACATGCATAAGGCACCCTTC MiR-506-reverse: GTGCAGGGTCCGAGGT U6-forward: CGCTTCGGCAGCACATATACU6-reverse: CAGGGGCCATCCTAATCTTRWDD4-forward: TGGTGATCCCAAAGCCTTCTRWDD4-reverse: CATCAACCCAGTTCCAGCCTGAPDH-forward: ACAACTTTGGTATCGTGGAAGGGAPDH-reverse: GCCATCACGCCACAGTTTC
Transfection
MiR-506 mimic and a negative control of the mimic (miR-Ctrl) were obtained from GenePharma (Shanghai, China). According to the manufacturer's instructions, T24 cells were transiently transfected with miR-506 mimic or miR-Ctrl by using 30 nM Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Cell viability assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used to measure cell viability following transfection. T24 cells transfected with miR-506 mimic or si-RWDD4 were seeded in 96-well dishes (1×104 cells per well) and cultured for 24, 48 and 72 h. Cells transfected with miR-Ctrl or si-Ctrl were used as negative controls. After incubation, 10 µl CCK-8 was added and the cells were incubated at 37°C for 3 h. Absorbance was then assessed at a wavelength of 450 nm.
Transwell migration assay
T24 cells transfected with miR-506 mimic or miR-Ctrl (1×105 cells/well) were resuspended in 200 µl medium without FBS and seeded in the upper chambers of a Transwell insert containing an 8-µm pore filter (BD Biosciences, Franklin Lakes, NJ, USA). The lower chamber was filled with medium supplemented with 10% FBS. After 72 h of incubation, cells on the lower side of the membrane were removed and fixed. The cells were then stained with 0.1% crystal violet and counted.
Invasion assay
T24 cells transfected with miR-506 mimic or miR-Ctrl were plated in the upper Transwell chambers containing Matrigel-treated 8-µm pore filters (BD Biosciences). The invasion assay was performed using the same procedures described for the Transwell migration assay.
Luciferase assay
T24 cells were co-transfected miR-506 and recombinant vectors. The transfection groups were established as follows: A: miR-506 mimics + pmirGLO-RWDD4 WT; B: miR-Ctrl + pmirGLO-RWDD4 WT; C: miR-506 mimics + pmirGLO-RWDD4 mutant; D: miR-Ctrl + pmirGLO-RWDD4 mutant. A total of 20 µl PLB lysate was added to each group and the groups were incubated at room temperature for 15 min. A Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) was used to compare the luciferase activities of the transfected cells. Relative fluorescence was determined as the ratio of firefly luciferase fluorescence to Renilla luciferase fluorescence.
Western blot analysis
T24 cells transfected with miR-506 mimic or miR-Ctrl were lysed and then processed for western blot analysis as previously described (13). The primary antibodies were used at 1:1,000 dilution with incubation at 4°C overnight, after which the blots were incubated with secondary antibodies (goat anti-mouse IgG-HRP, 1:10,000 and goat anti-rabbit IgG-HRP 1:15,000) for 2 h at room temperature. Immunodetection was performed using Super Signal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.) and detected with a Bio-Rad GelDoc XR+ system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Primary antibodies against E-cadherin, N-cadherin, Snail, and GAPDH were purchased from Cell Signaling Technology, Inc., (Danvers, MA, USA), and the primary RWDD4 antibody, goat anti-mouse IgG-HRP and goat anti-rabbit IgG-HRP were obtained from Abcam (Cambridge, MA, USA).
Statistical analysis
Data were statistically analyzed with IBM SPSS statistics software v21 (IBM Corp., Armonk, NY, USA) and expressed as the mean ± SD. To analyze miR-506 expression in human TCC cell lines, one-way analysis of variance (ANOVA) was used and multiple comparisons between the groups were performed using Dunnett's least significant difference (LSD) test. Differences the control group and the experimental group were compared by using the Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
MiR-506 expression is decreased in human TCC cell lines and tissues
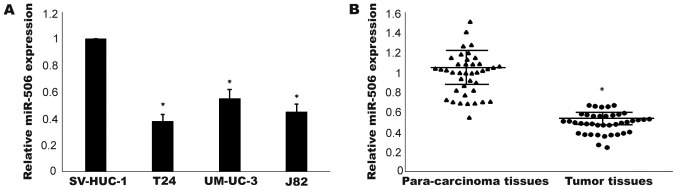
The expression levels of miR-506 in three TCC cell lines (T24, J82 and UM-UC-3) and a normal human bladder epithelial cell line (SV-HUC-1) were detected by RT-qPCR. As shown in Fig. 1A, compared with in the human normal bladder cells, miR-506 expression was downregulated in the TCC cell lines (P<0.05). T24 cells exhibiting the lowest expression level were used in the following experiments. Furthermore, as depicted in Fig. 1B, the expression level of miR-506 in TCC tissues was significantly lower than that in non-tumor tissues (P<0.05). Taken together, these results indicated that miR-506 was downregulated in TCC.
Figure 1.
Expression of miR-506 was downregulated in TCC. (A) The expression of miR-506 in three TCC cell lines (T24, J82, and UM-UC-3) and a normal human bladder epithelial cell line (SV-HUC-1) was detected by RT-qPCR. (B) MiR-506 expression in TCC specimens was compared with that in the corresponding paired non-tumor tissues. *P<0.05. TCC, transitional cell carcinoma.
MiR-506 suppresses TCC cell proliferation, invasion, migration and EMT
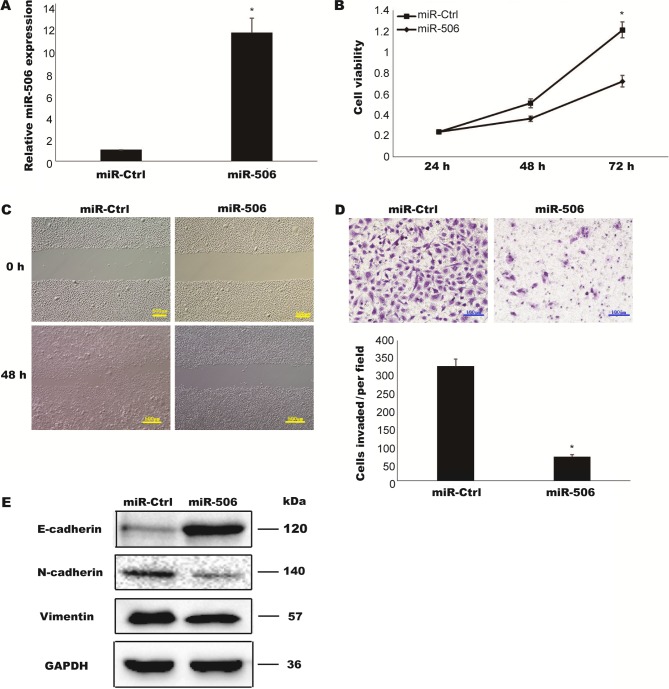
To assess the biological function of miR-506 in TCC, T24 cells were transiently transfected with miR-506 mimic, and the relative expression of miR-506 was determined to be successfully upregulated compared with in a negative control group (miR-Ctrl) (P<0.05; Fig. 2A). A CCK-8 assay was performed to determine the cell viabilities of the transfected cells. As presented in Fig. 2B, the proliferation rate of T24 cells overexpressing miR-506 was markedly decreased compared with that of the cells transfected with miR-Ctrl (P<0.05). In vitro Transwell assays were further utilized to examine whether miR-506 is involved in TCC cell invasion and migration. The overexpression of miR506 lead to decreases in the abilities of cells to invade and migrate (Fig. 2C and D). EMT is considered an early and key step in the metastatic cascade. The loss of epithelial protein E-cadherin and the increase of mesenchymal proteins N-cadherin and Vimentin are hallmarks of EMT (14). To analyze whether miR-506 suppressed cell migration through inhibition of EMT, the expressions of these markers were detected by western blotting. Compared with the cells transfected with miR-Ctrl, the overexpression of miR-506 lead to upregulation of E-cadherin and downregulation of N-cadherin and Vimentin (Fig. 2E). Taken together, these data indicated that miR-506 inhibited TCC cell proliferation, invasion, migration and EMT.
Figure 2.
MiR-506 inhibited TCC cell proliferation, invasion, migration and EMT. T24 cells were transfected with negative control miR (miR-Ctrl) or miR-506 mimic. (A) The relative expression increased in miR-506 was confirmed by RT-qPCR. (B) Cell proliferation was detected by CCK-8 assay. (C and D) Transwell migration and invasion assays were performed to investigate the effect of miR-506 on (C) cell migration (scale bar, 500 µm) and (D) invasion (scale bar, 100 µm). (E) The expressions of EMT-related proteins (E-cadherin, N-cadherin and Vimentin) were detected by western blotting using GAPDH as a loading control. Three independent experiments were performed. *P<0.05. TCC, transitional cell carcinoma.
RWDD4 is a target of miR506 in human TCC cell lines
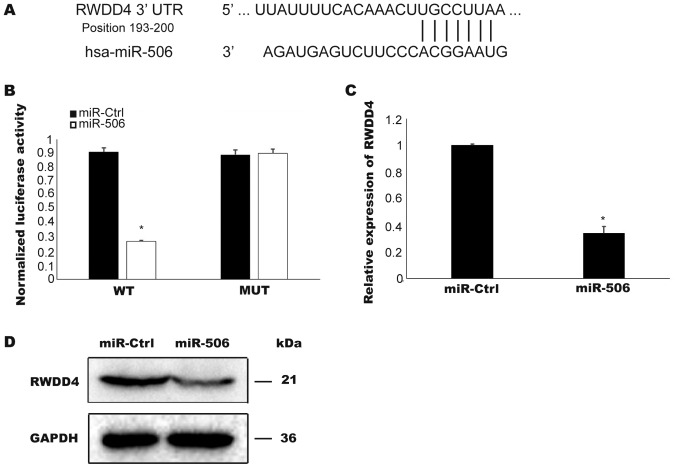
To determine the potential targets of miR506, TargetScan software was used to search for conserved sites. As depicted in Fig. 3A, the 3′UTR region of RWDD4 was identified to contain a highly conserved miR506 binding site. To confirm that RWDD4 is a target of miR506 in TCC cells, a luciferase activity assay was performed. Overexpressed miR506 suppressed the luciferase activities of pmirGLO-WT- RWDD4-3′-UTR plasmid in the T24 cells but did not affect the activities controlled by the mutant RWDD4-3′-UTR plasmid (P<0.05; Fig. 3B). Furthermore, overexpressed miR506 in the T24 cells inhibited the expression of RWDD4 mRNA and protein (P<0.05; Fig. 3C and D). Collectively these results confirmed RWDD4 as a target of miR506.
Figure 3.
RWDD4 is a biological target of miR-506 in human TCC. (A) The 3′UTR region of RWDD4 was determined to have a highly conserved miR506 binding site using TargetScan software. (B) Luciferase activities were compared between cells with overexpression of miR-506 controlled by pmirGLO-WT-RWDD4-3′-UTR and by the mutant RWDD4-3′-UTR in T24 cells. (C and D) T24 cells transfected with miR-Ctrl or miR-506. (C) Relative expression of RWDD4 detected by RT-qPCR. (D) RWDD4 protein expression level assessed by western blotting. Three independent experiments were performed. *P<0.05. RWDD4, RWD domain containing (4); TCC, transitional cell carcinoma.
Knockdown of RWDD4 inhibits TCC cell proliferation, migration and invasion
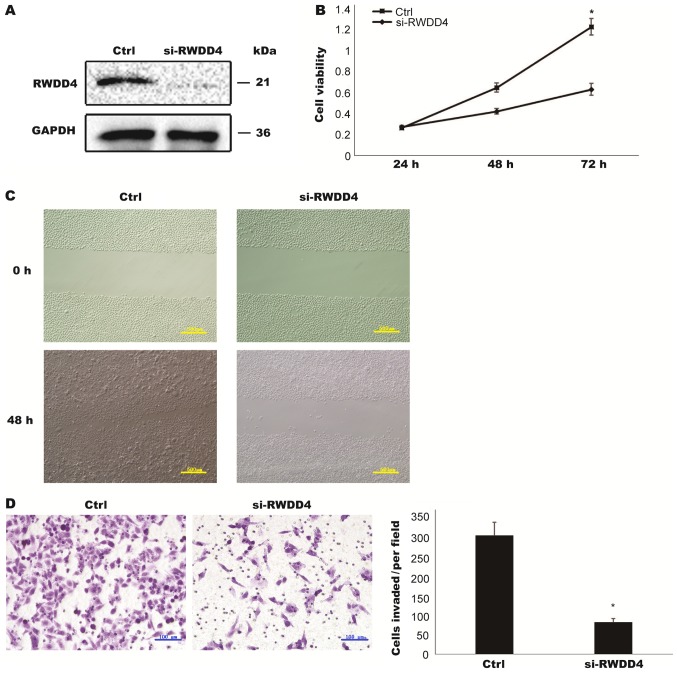
Many functions of the RWDD4 gene have not yet been fully elucidated. RWDD4 contains a RWD domain that is involved in protein-protein interactions (15). Compared with si-Ctrl, si-RWDD4 inhibited RWDD4 protein expression (Fig. 4A). The knockdown of RWDD4 inhibited TCC cell proliferation, migration and invasion, which was consistent with the results of miR-506 overexpression (Fig. 4B-D). Therefore, we concluded that miR-506 may inhibit TCC development through its relationship with RWDD4.
Figure 4.
Downregulated RWDD4 inhibited TCC cell proliferation, migration and invasion. T24 cells were transfected with negative control (si-Ctrl) or si-RWDD4. (A) Decreased RWDD4 protein expression level was confirmed by western blotting. (B) Cell proliferation was compared by CCK-8 assay. (C and D) Migration and invasion assays were performed to investigate the effect of RWDD4 downregulation on cell (C) cell migration (scale bar, 500 µm) and (D) invasion (scale bar, 100 µm). *P<0.05. RWDD4, RWD domain containing (4); TCC, transitional cell carcinoma.
Discussion
Short (−22 nt) non-coding miRNAs play essential roles in the regulation of gene expression post-transcriptionally. Recent studies showed that miRNAs play vital roles in the regulation of tumor growth and progression in various tumor types including TCC (16,17). The essential functions of miRNAs have prompted increasing investigations into the use of miRNA-targeted strategies for human cancer treatment (13,18). As invasive BCa is a disease that can affect the whole bladder (19), HE staining was performed before RT-qPCR detection to verify that the controls were non-tumor tissues pathologically. However, due to the characteristics of invasive BCa, there is still the possibility that genetic alterations were present in the non-tumor tissues. MiR-506 plays distinct roles in different cancers through the regulation of specific genes. Consistent with previous findings in breast, cervical and ovarian cancer, we revealed that miR-506 was notably decreased in human TCC cell lines and TCC tissues compared with in a normal human bladder cell line and paired non-tumor samples, respectively. In vitro studies also demonstrated that overexpressed miR-506 suppressed TCC cell proliferation, migration and invasion. Moreover, we identified RWDD4 to be a biological target of miR-506. Decreased RWDD4 expression via siRNA transfection showed comparable results to those obtained with miR-506 overexpression. Therefore, our findings suggested that miR-506 functioned as a tumor suppressor though targeting RWDD4 in human TCC.
The effects of therapies for advanced TCC are still not up to expectations due to the high incidence of metastasis in TCC (20). EMT, which was originally established as a process in normal cell differentiation, is divided into three types. Type 3 EMT is involved in formation of cancer stem cells and cancer progression (21). The loss of epithelial features and the acquisition of mesenchymal characteristics enable cancer cells to diffuse more rapidly and thus more invasive (22). Therefore, we also investigated the effects of miR-506 on the expression of proteins involved in the regulation of EMT. We found that the overexpression of miR-506 upregulated E-cadherin and downregulated N-cadherin and Vimentin. Several transcription factors including snail family zinc finger (Snail), twist basic helix-loop-helix transcription factor 1 (TWIST1), and zinc finger E-box-binding homeobox (ZEB1) have been reported to induce EMT by suppressing E-cadherin expression (23). To the best of our knowledge, only one previous study has revealed an association of RWDD4 with prostate cancer aggression (15). Consistent with this previous study, we found downregulated RWDD4 inhibited TCC cell proliferation, migration and invasion in vitro. Based on large-scale data analyses, cancer metastasis has been established as a complicated procedure driven by more than 100 genes (24,25). Whether and how RWDD4 is involved in the miR-506-mediated regulation of EMT remains to be clarified in further studies. However, the characterization of RWDD4 gene function in the present study has in part enriched our knowledge on cancer metastasis in TCC.
In conclusion, this study has identified that miR-506 plays critical roles in TCC cell proliferation, migration, invasion and EMT through targeting RWDD4. Our data warrant further investigation into the potential of miR-506 as a novel target in human TCC therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YH conceived and performed the all experiments.
Ethics approval and consent to participate
The present study was approved by the ethical committee of the Fourth People's Hospital of Shenyang (ethics review approval no: 20140124-8). Informed consent was provided to the all participants.
Patient consent for publication
All the patient, or parent, guardian or next of kin provided written informed consent for the publication of any associated data and accompanying images.
Competing interests
The author declares that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Leow JJ, Martin-Doyle W, Rajagopal PS, Patel CG, Anderson EM, Rothman AT, Cote RJ, Urun Y, Chang SL, Choueiri TK, Bellmunt J. Adjuvant chemotherapy for invasive bladder cancer: A 2013 apdated systematic review and meta-analysis of randomized trials. Eur Urol. 2014;66:42–54. doi: 10.1016/j.eururo.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 3.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-A brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2015;34:717–725. doi: 10.1038/onc.2014.9. [DOI] [PubMed] [Google Scholar]
- 7.Du J, Zheng X, Cai S, Zhu Z, Tan J, Hu B, Huang Z, Jiao H. MicroRNA506 participates in pancreatic cancer pathogenesis by targeting PIM3. Mol Med Rep. 2015;12:5121–5126. doi: 10.3892/mmr.2015.4109. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang D, Parker Kerrigan BC, Shmulevich I, Chen K, Sood AK, et al. MiR-506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol. 2014;233:308–318. doi: 10.1002/path.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora H, Qureshi R, Park WY. miR-506 regulates epithelial mesenchymal transition in breast cancer cell lines. PLoS One. 2013;8:e64273. doi: 10.1371/journal.pone.0064273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin M, Ren X, Zhang X, Luo Y, Wang G, Huang K, Feng S, Bao X, Huang K, He X, et al. Selective killing of lung cancer cells by miRNA-506 molecule through inhibiting NF-kB p65 to evoke reactive oxygen species generation and p53 activation. Oncogene. 2015;34:691–703. doi: 10.1038/onc.2013.597. [DOI] [PubMed] [Google Scholar]
- 11.Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z, Tice DA, Higgs BW, et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. 2012;31:1558–1570. doi: 10.1038/onc.2011.345. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Sun D, Tai J, Wang L. Ganoderic acid A inhibits proliferation and invasion, and promotes apoptosis in human hepatocellular carcinoma cells. Mol Med Rep. 2017;16:3894–3900. doi: 10.3892/mmr.2017.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 15.Winter JM, Gildea DE, Andreas JP, Gatti DM, Williams KA, Lee M, Hu Y, Zhang S, NISC Comparative Sequencing Program5. Mullikin JC, et al. Mapping complex traits in a diversity outbred f1 mouse population identifies germline modifiers of metastasis in human prostate cancer. Cell Syst. 2017;4:31–45 e6. doi: 10.1016/j.cels.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enokida H, Yoshino H, Matsushita R, Nakagawa M. The role of microRNAs in bladder cancer. Investig Clin Urol. 2016;57(Suppl 1):S60–S76. doi: 10.4111/icu.2016.57.S1.S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fendler A, Stephan C, Yousef GM, Kristiansen G, Jung K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol. 2016;13:734–752. doi: 10.1038/nrurol.2016.193. [DOI] [PubMed] [Google Scholar]
- 18.Bhardwaj A, Singh S, Singh AP. MicroRNA-based cancer therapeutics: Big hope from small RNAs. Mol Cell Pharmacol. 2010;2:213–219. doi: 10.4255/mcpharmacol.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanli O, Dobruch J, Knowles MA, Burger M, Alemozaffar M, Nielsen ME, Lotan Y. Bladder cancer. Nat Rev Dis Primers. 2017;3:17022. doi: 10.1038/nrdp.2017.22. [DOI] [PubMed] [Google Scholar]
- 20.Froehner M, Koch R, Heberling U, Novotny V, Oehlschlaeger S, Hubler M, Baretton GB, Hakenberg OW, Wirth MP. Decreased overall and bladder cancer-specific mortality with adjuvant chemotherapy after radical cystectomy: Multivariable competing risk analysis. Eur Urol. 2016;69:984–987. doi: 10.1016/j.eururo.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 21.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 22.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 24.Qian CN, Mei Y, Zhang J. Cancer metastasis: Issues and challenges. Chin J Cancer. 2017;36:38. doi: 10.1186/s40880-017-0206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei Y, Yang JP, Qian CN. For robust big data analyses: A collection of 150 important pro-metastatic genes. Chin J Cancer. 2017;36:16. doi: 10.1186/s40880-016-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.